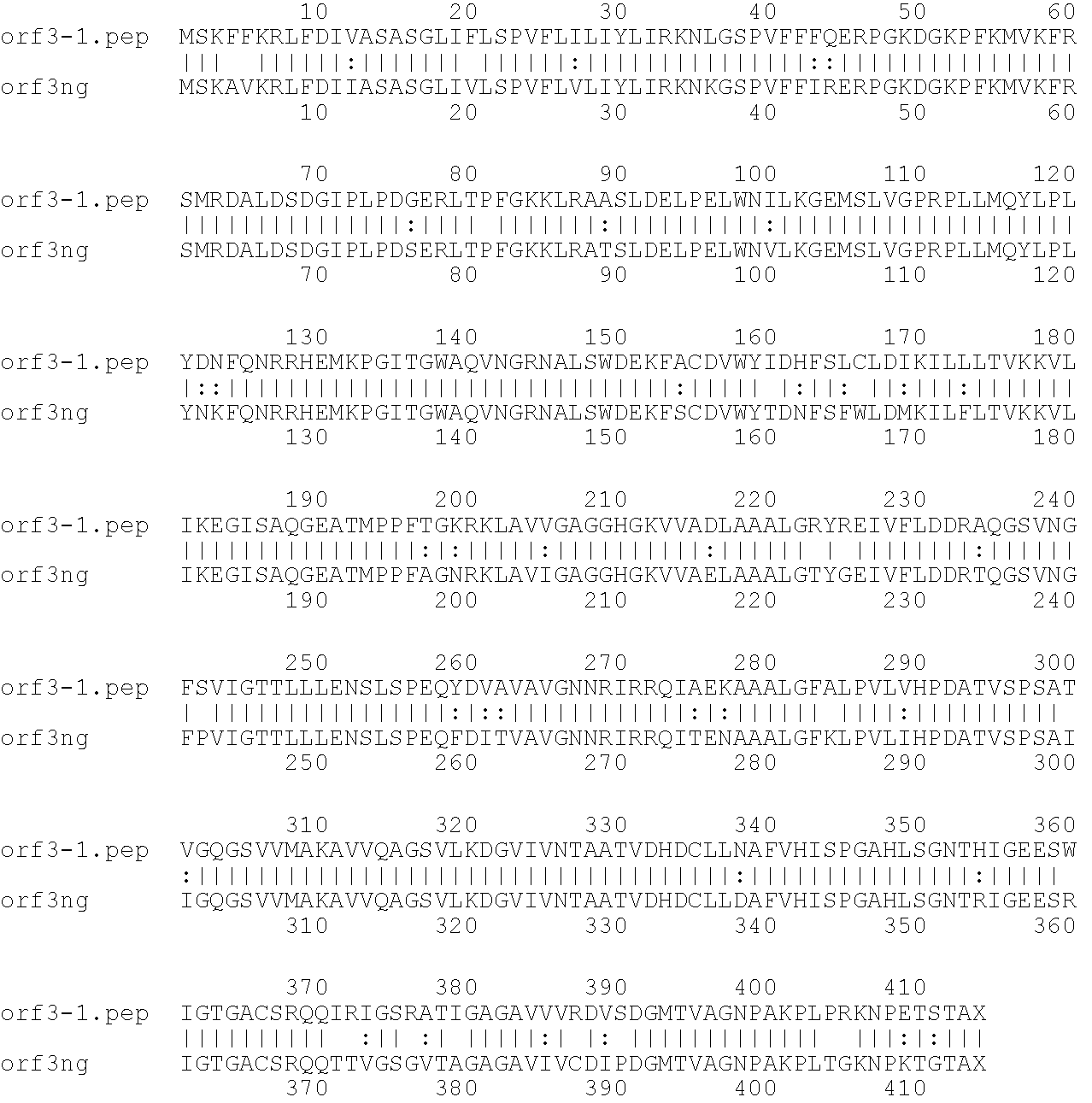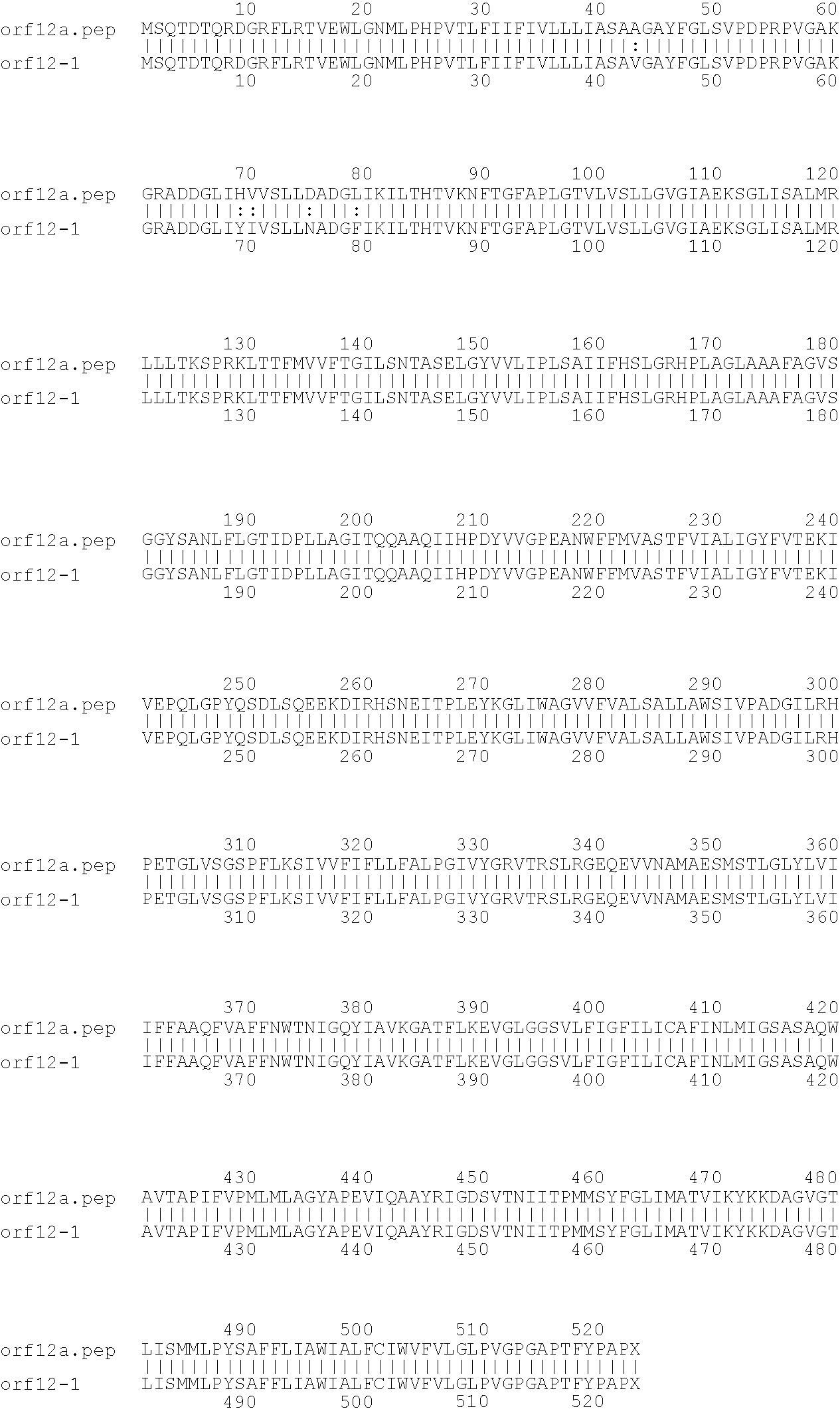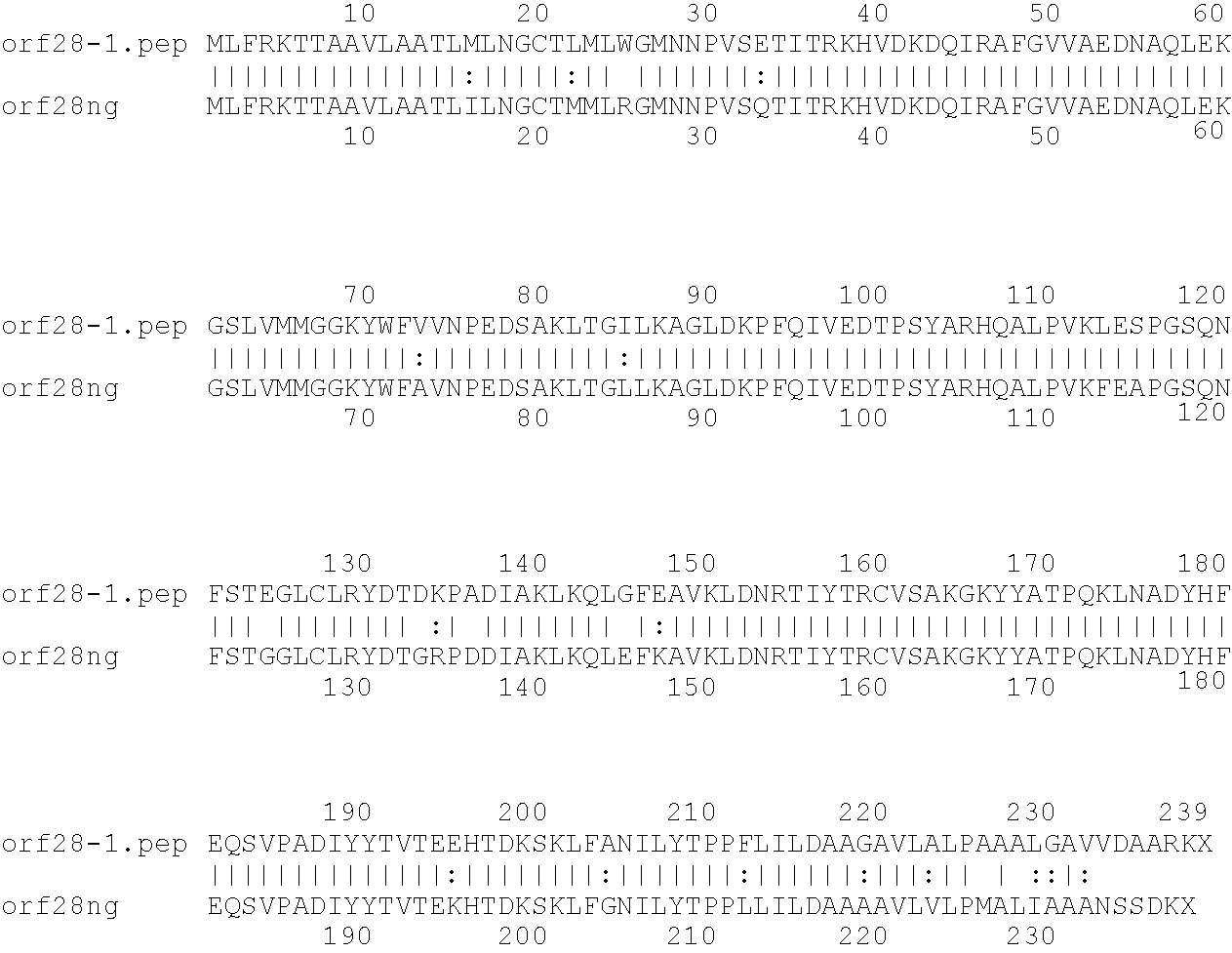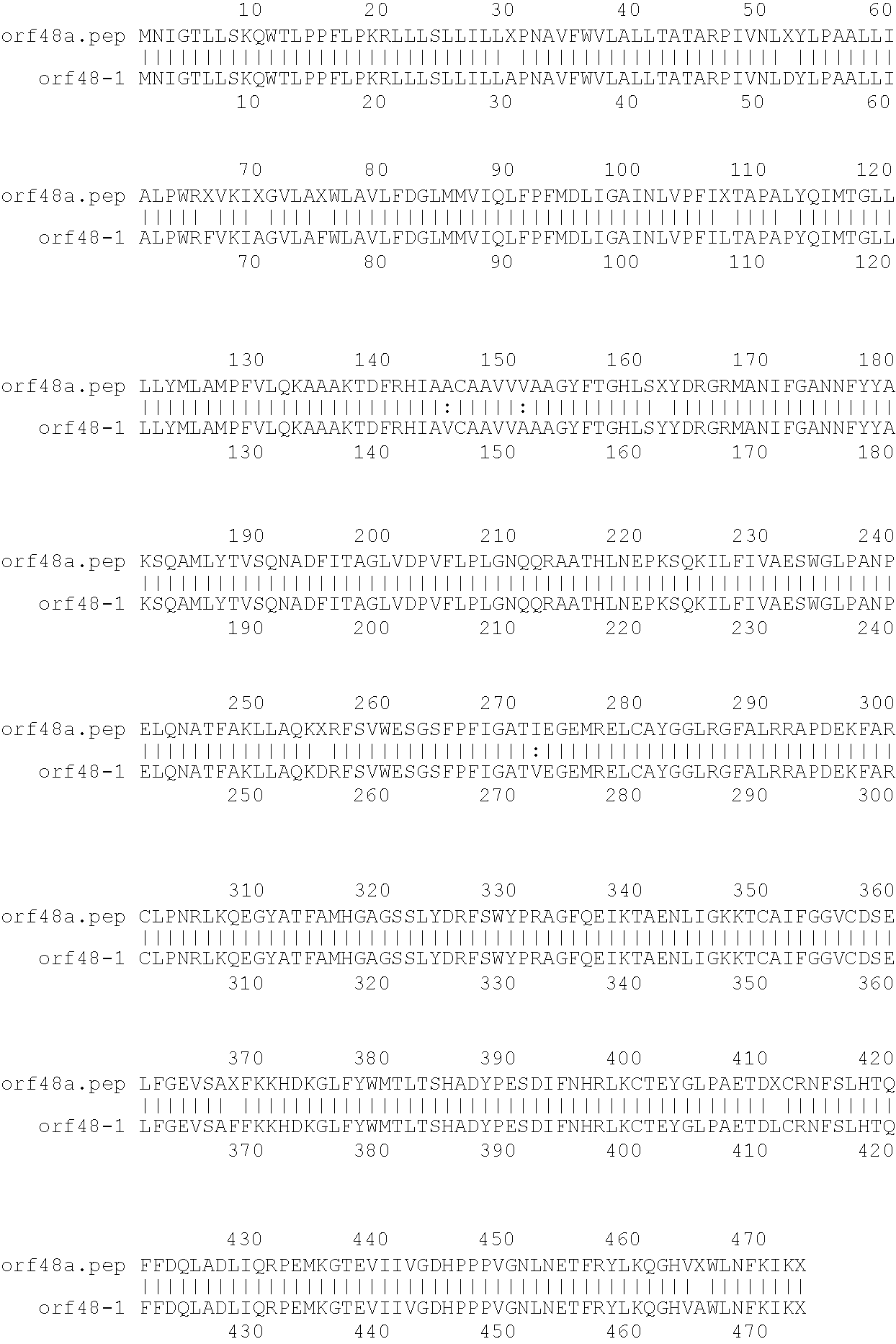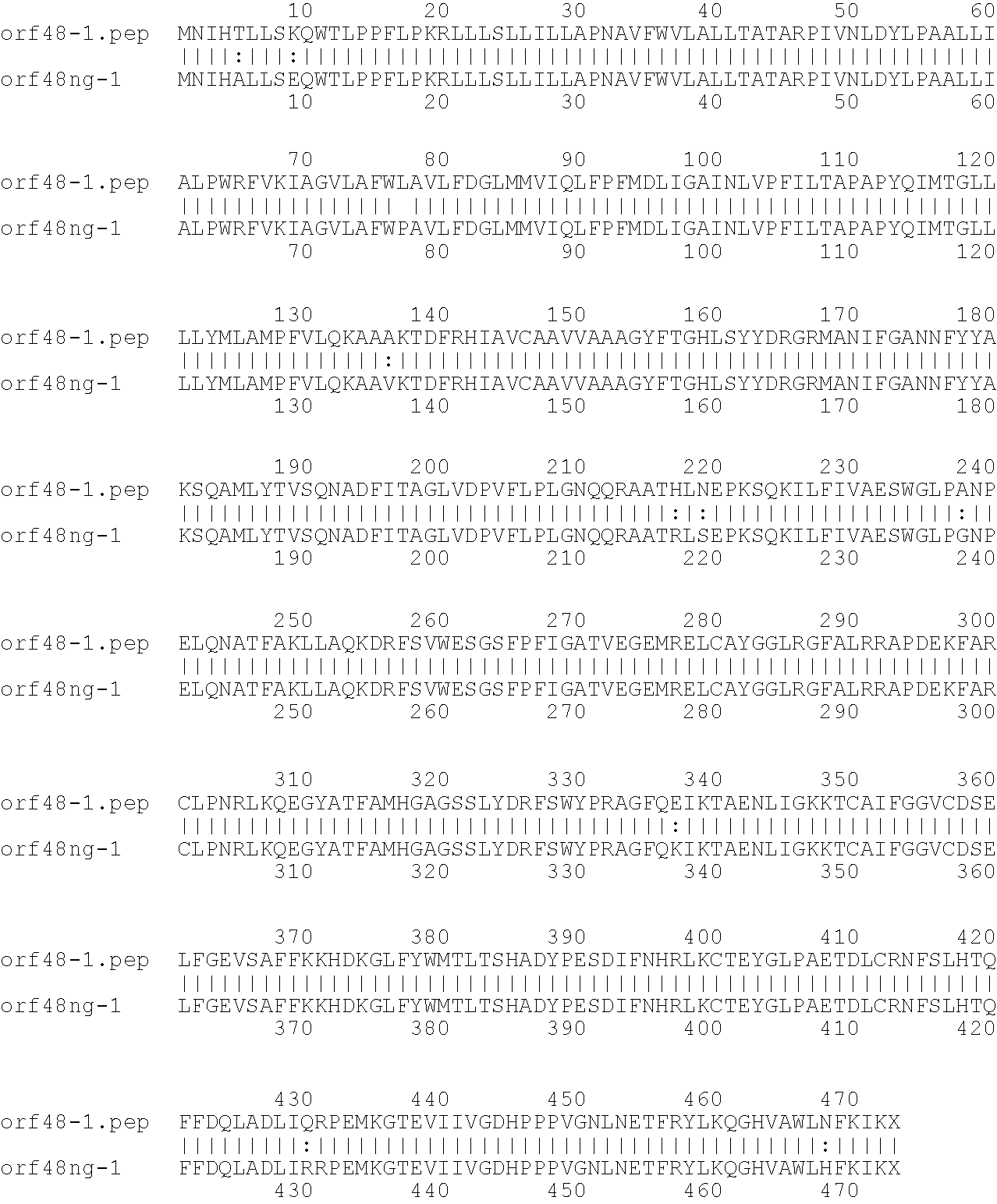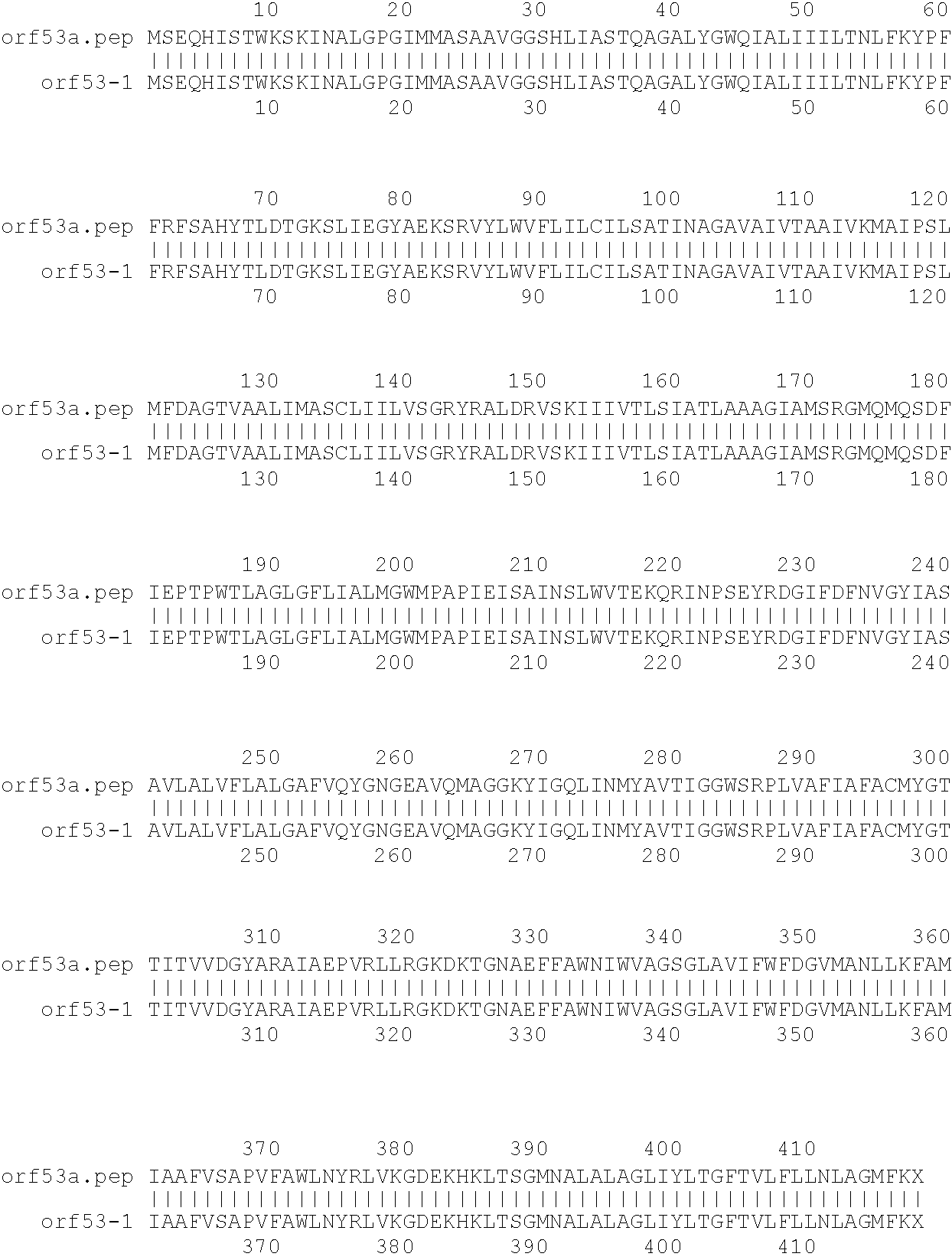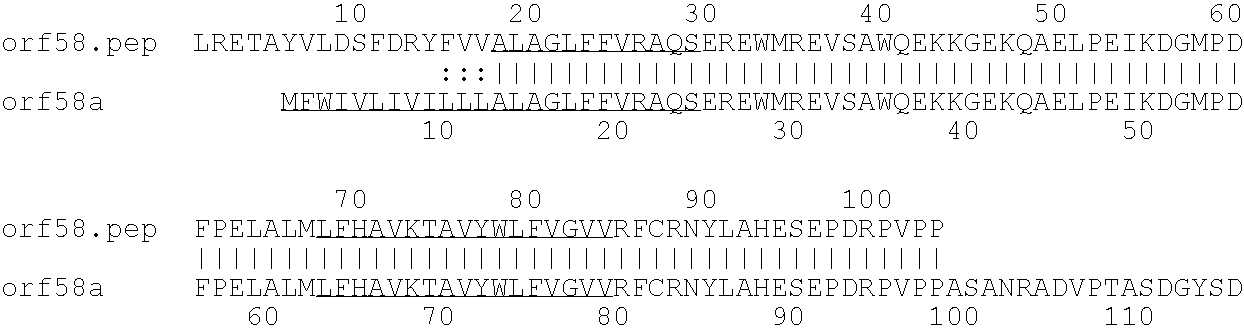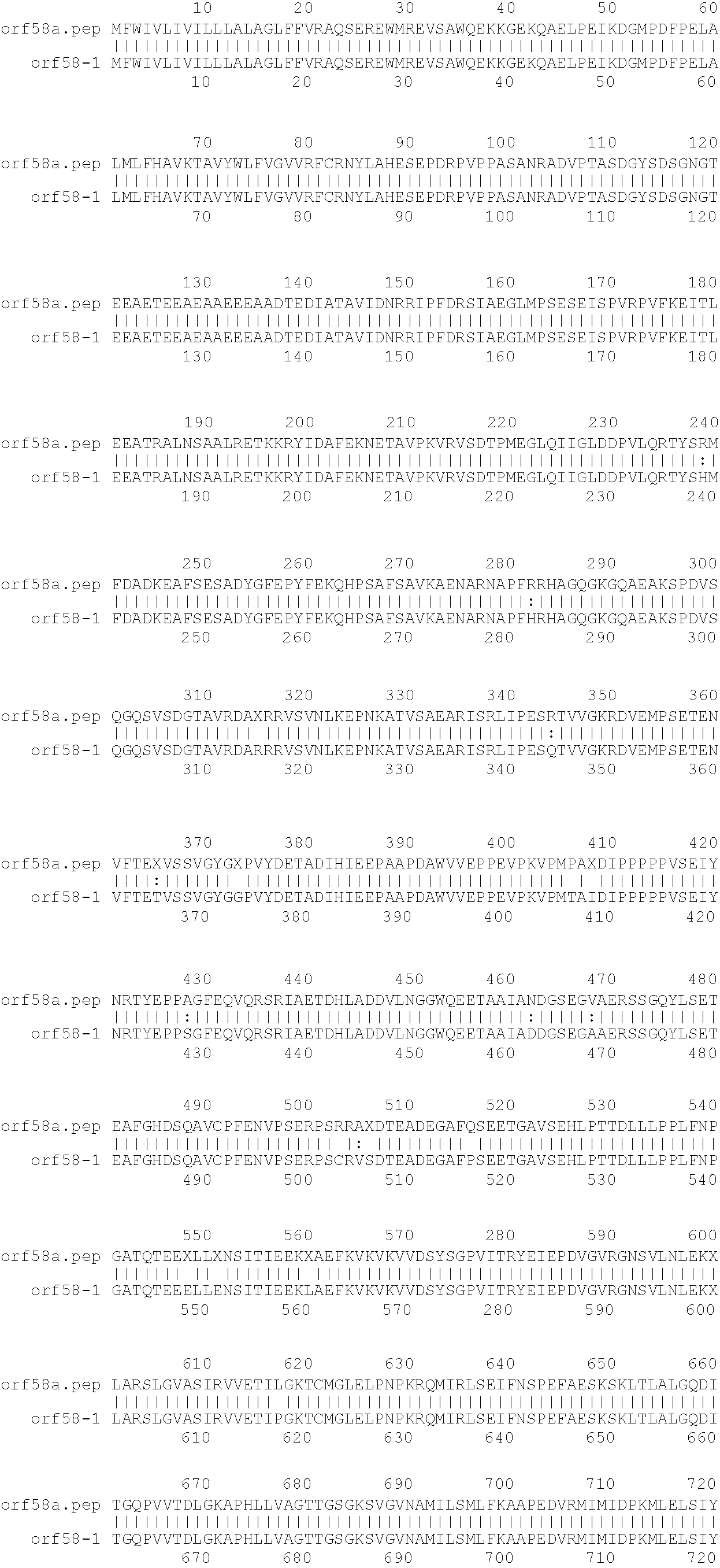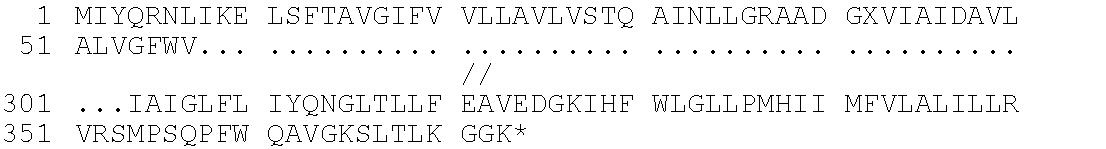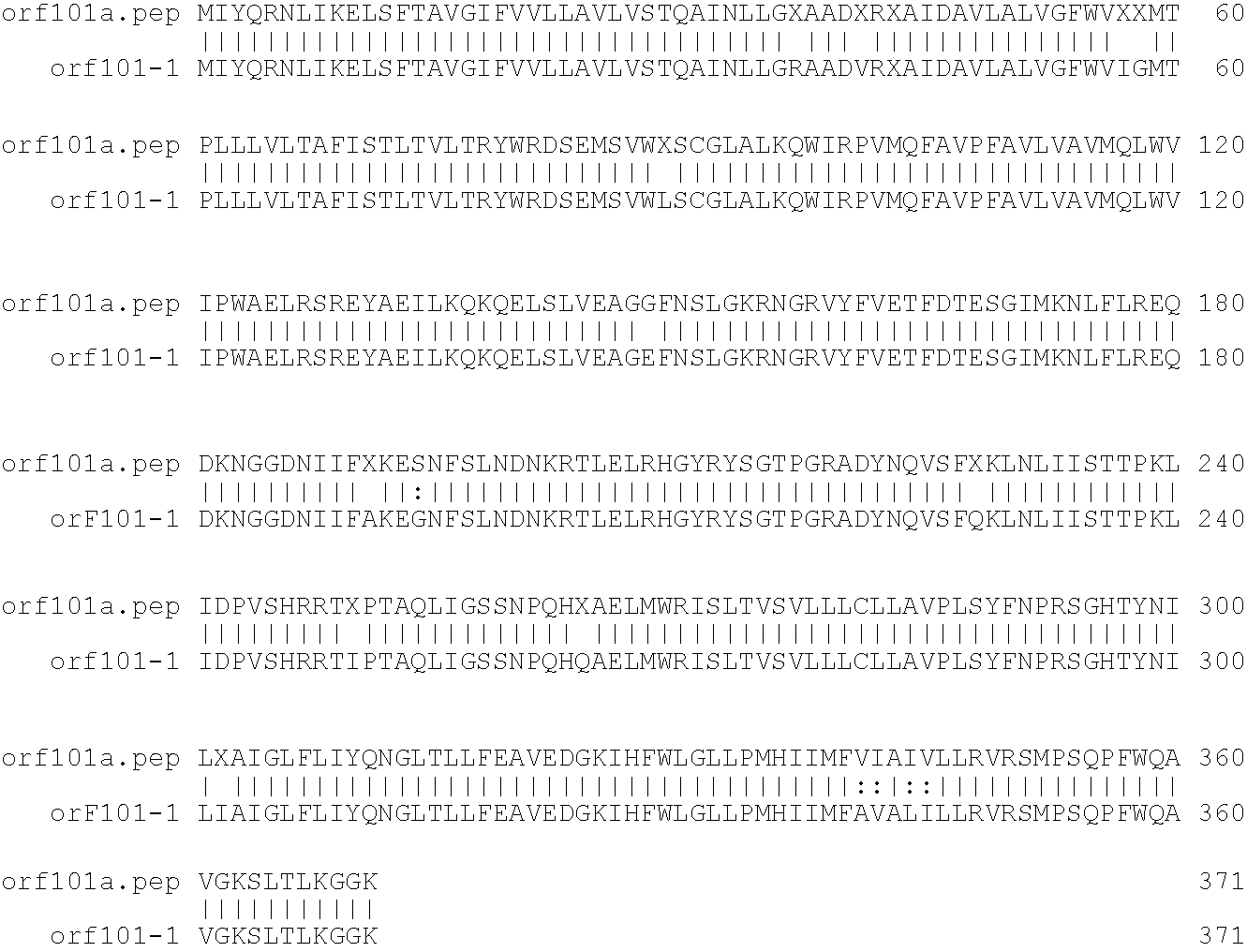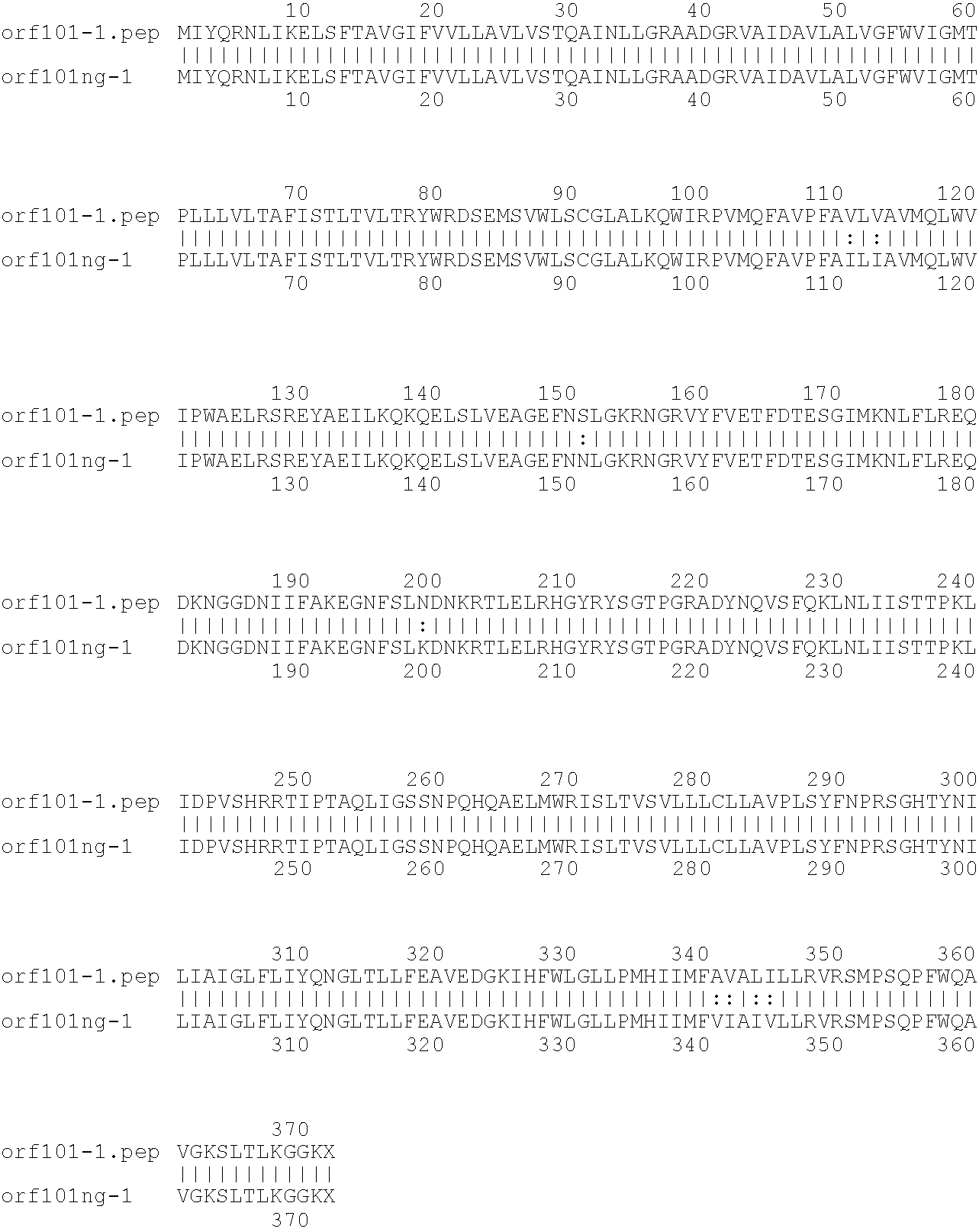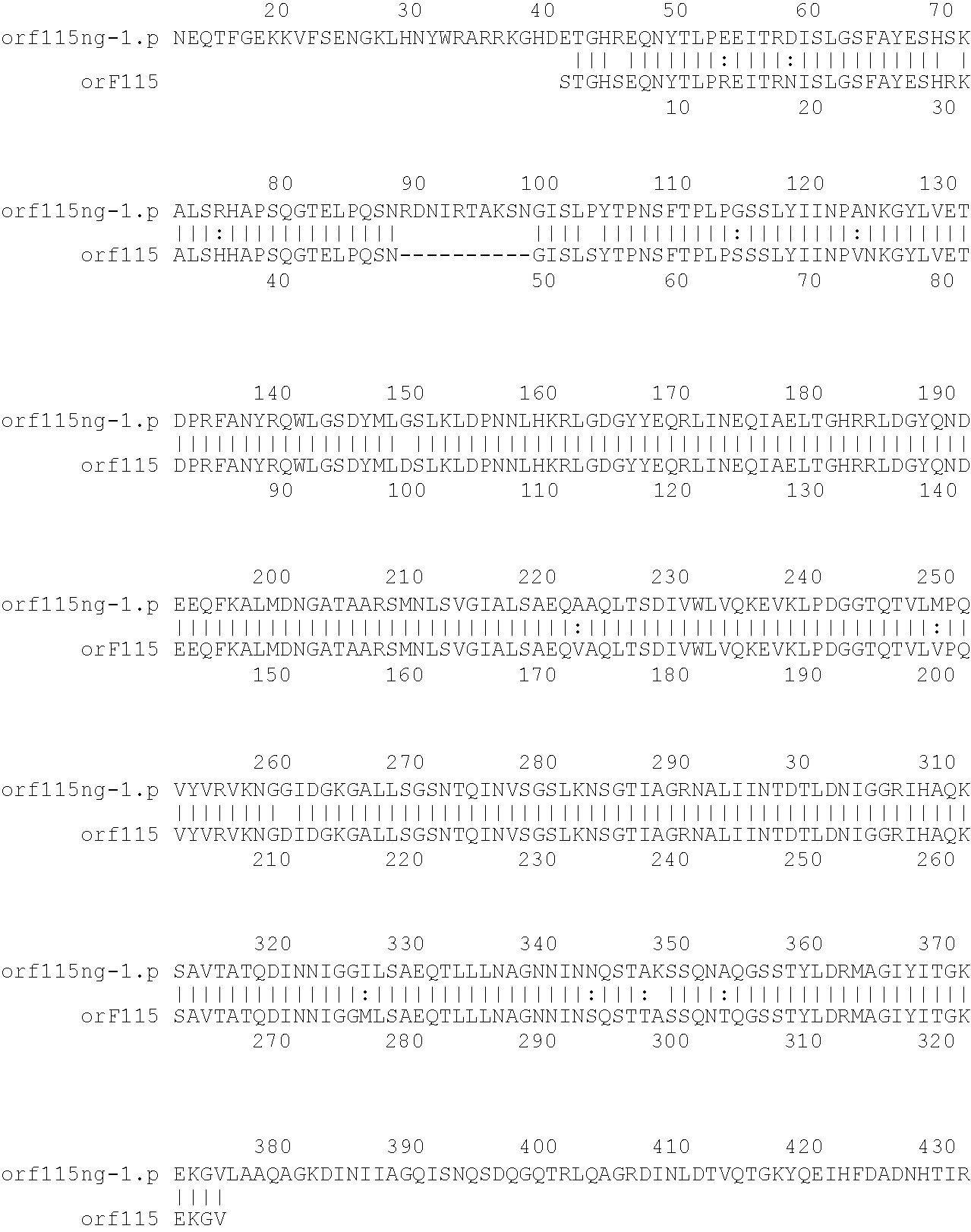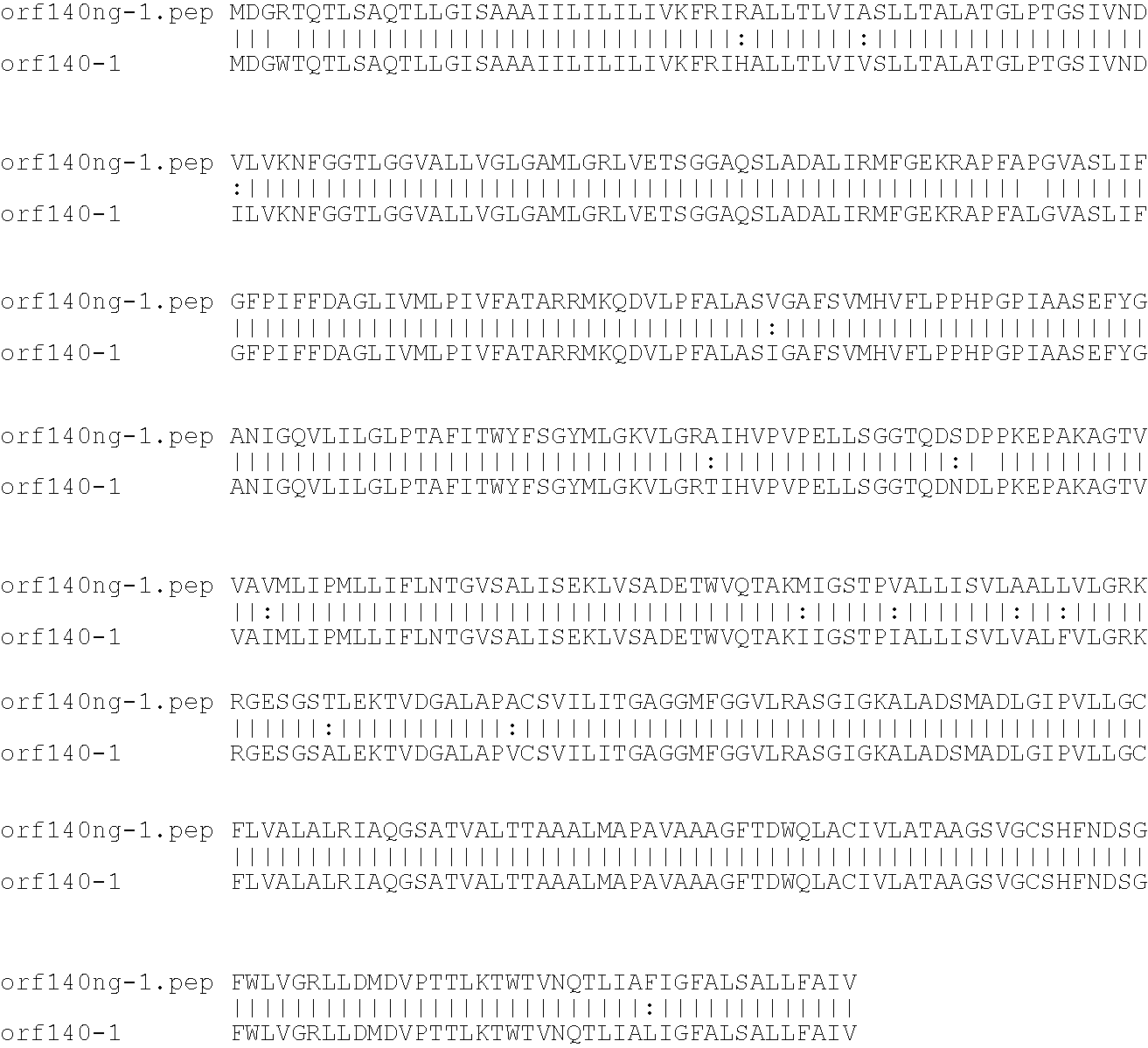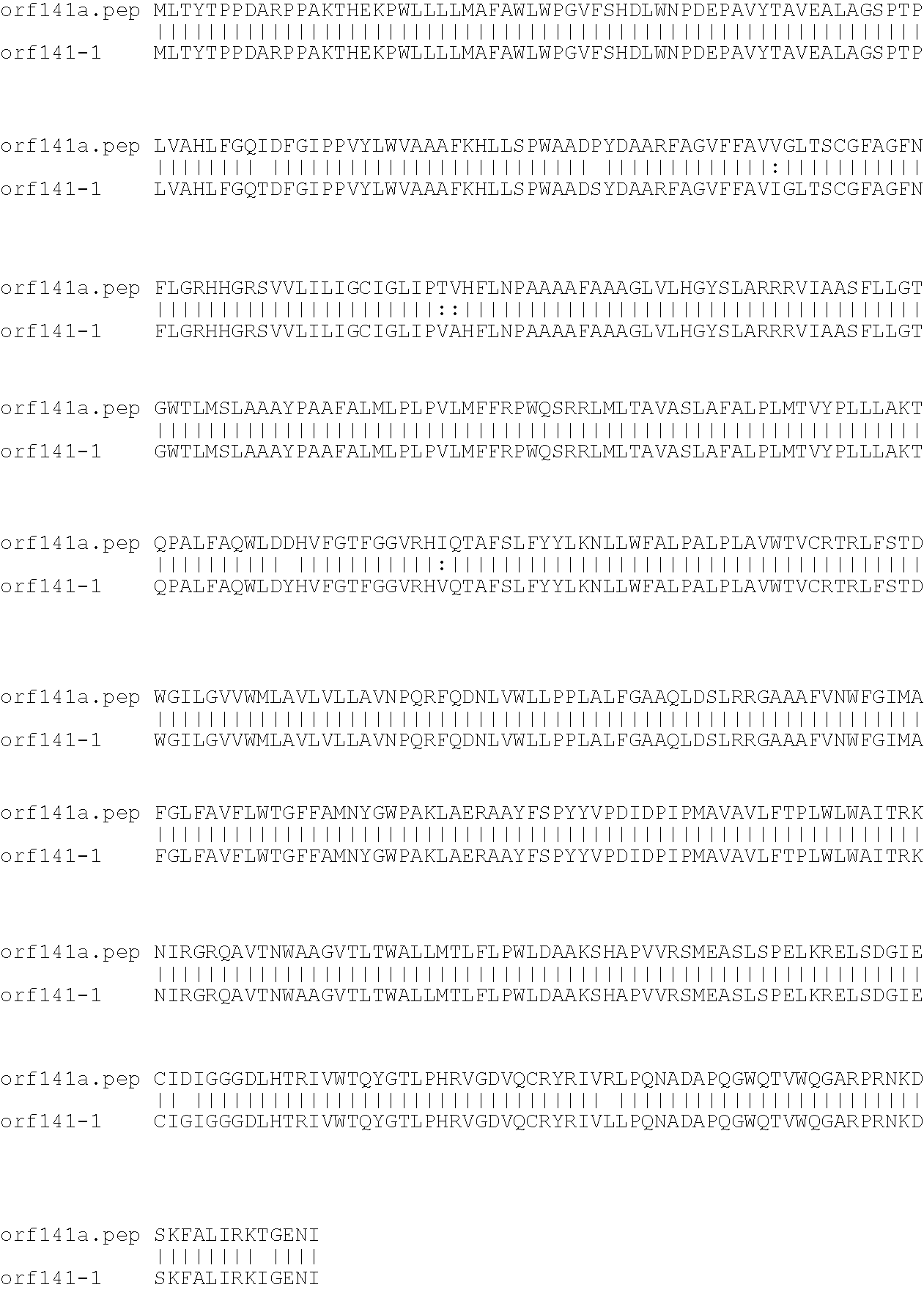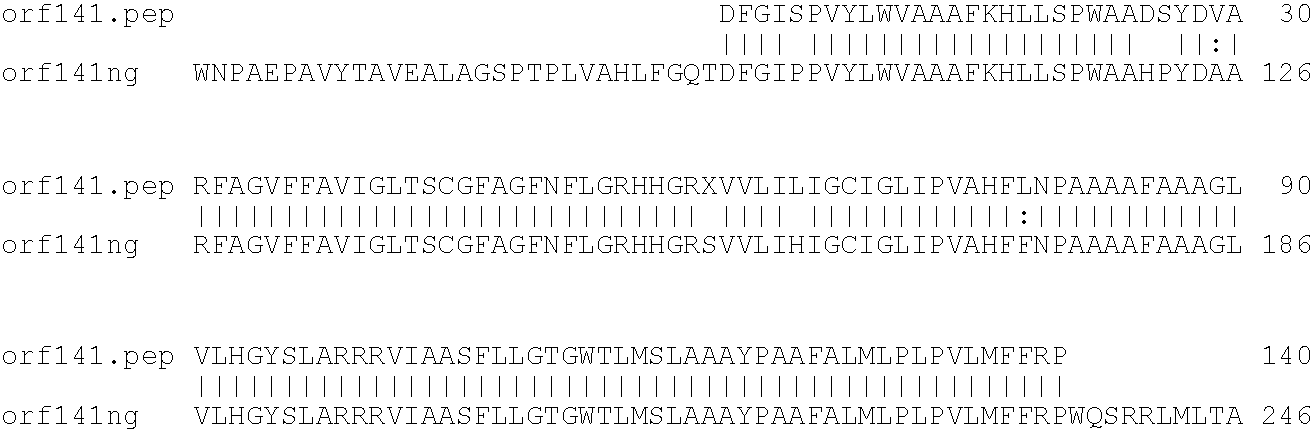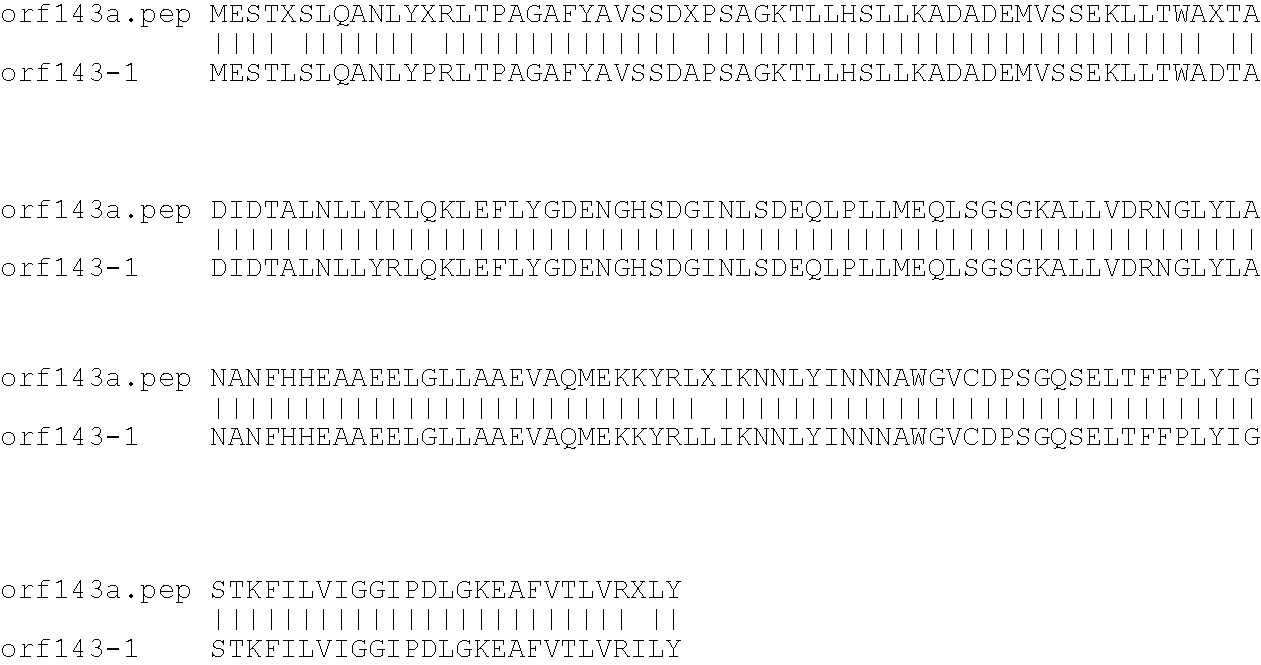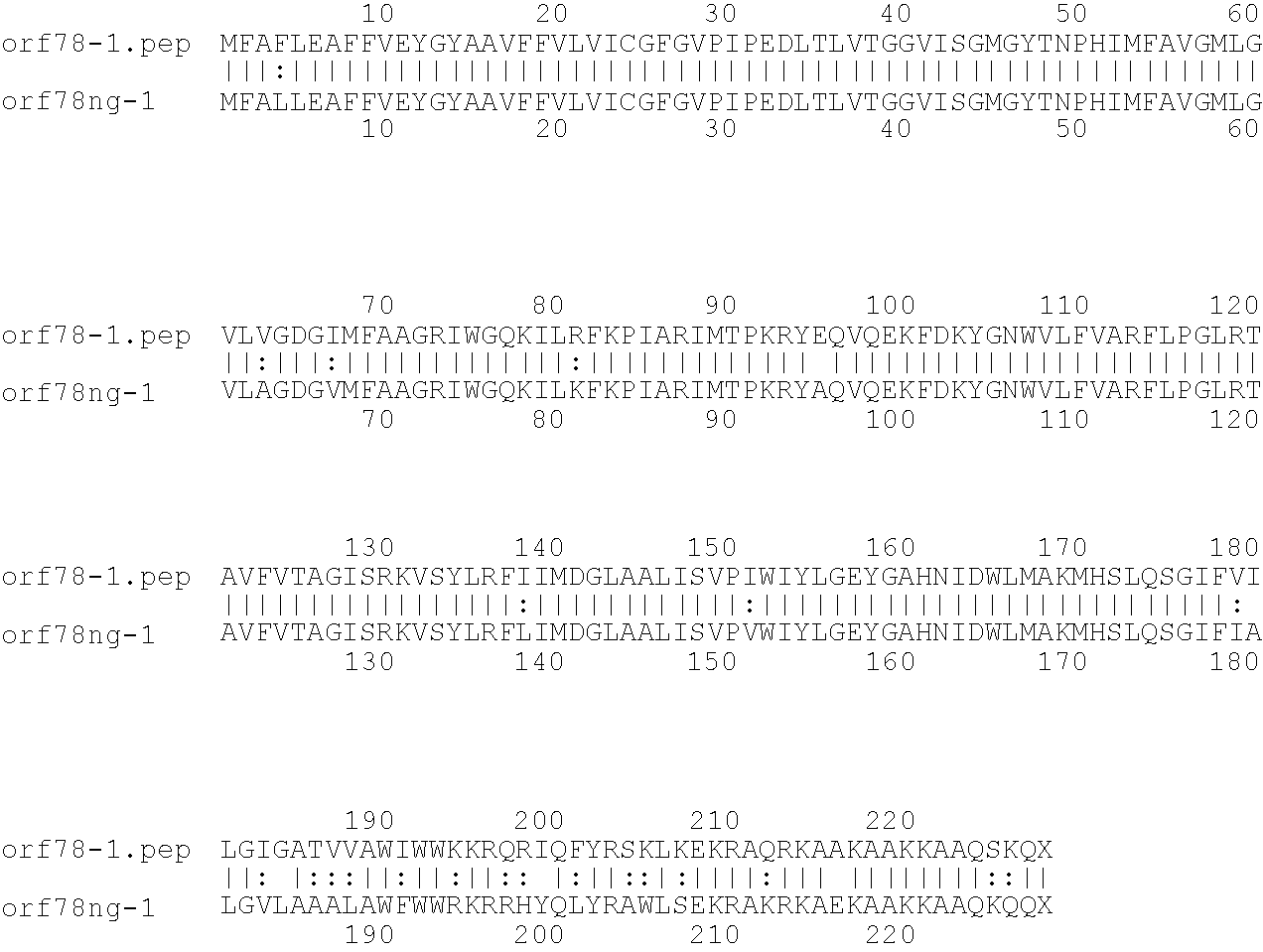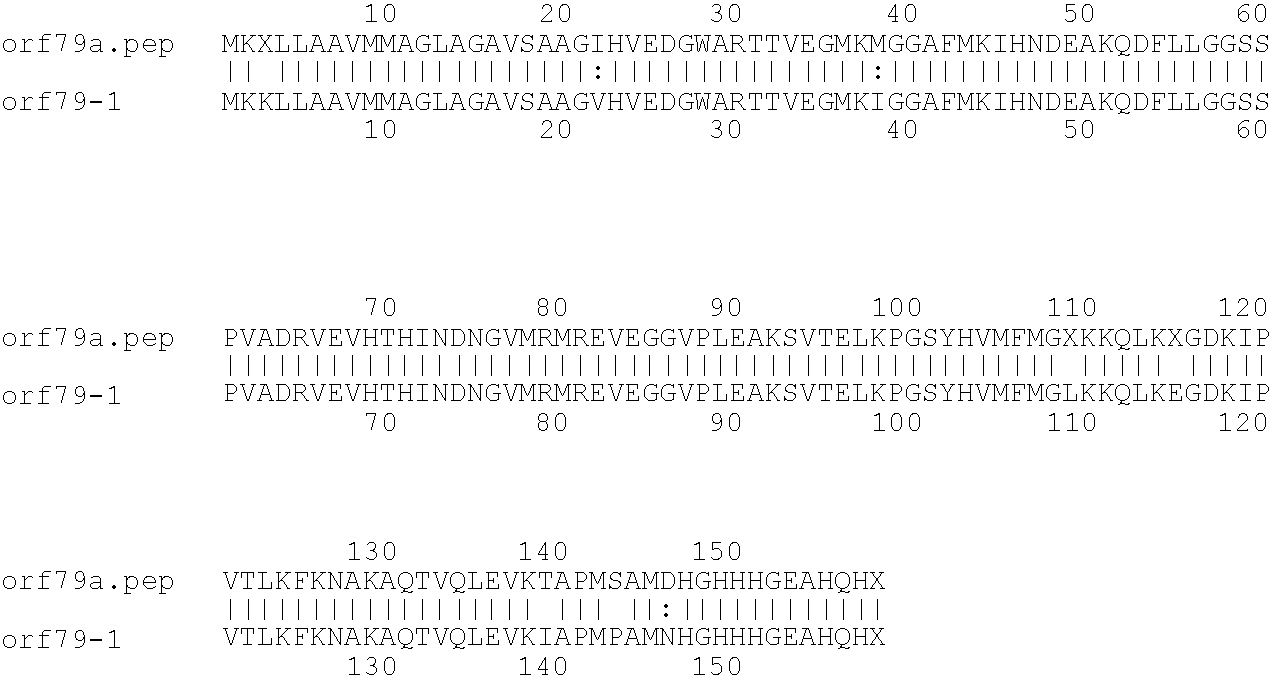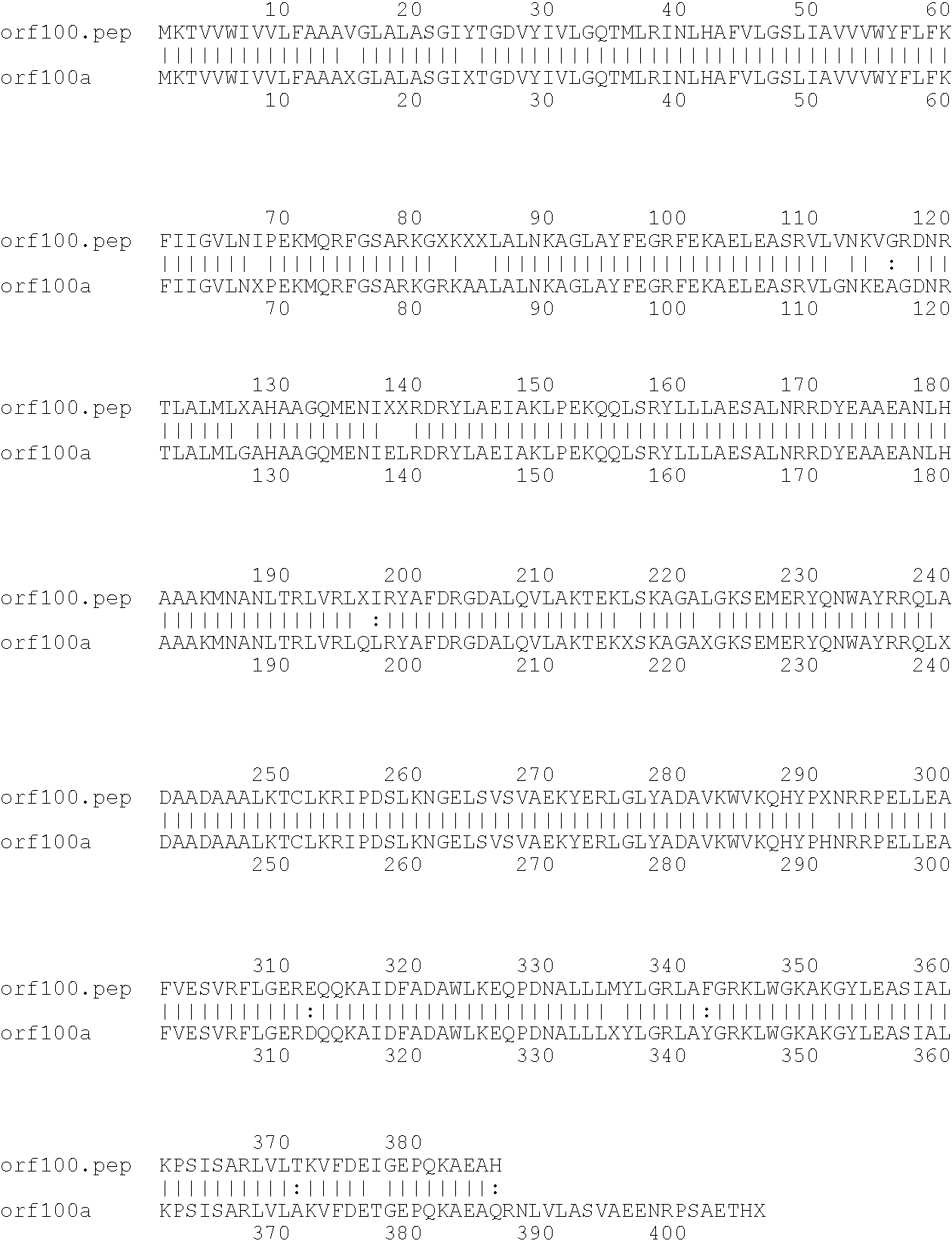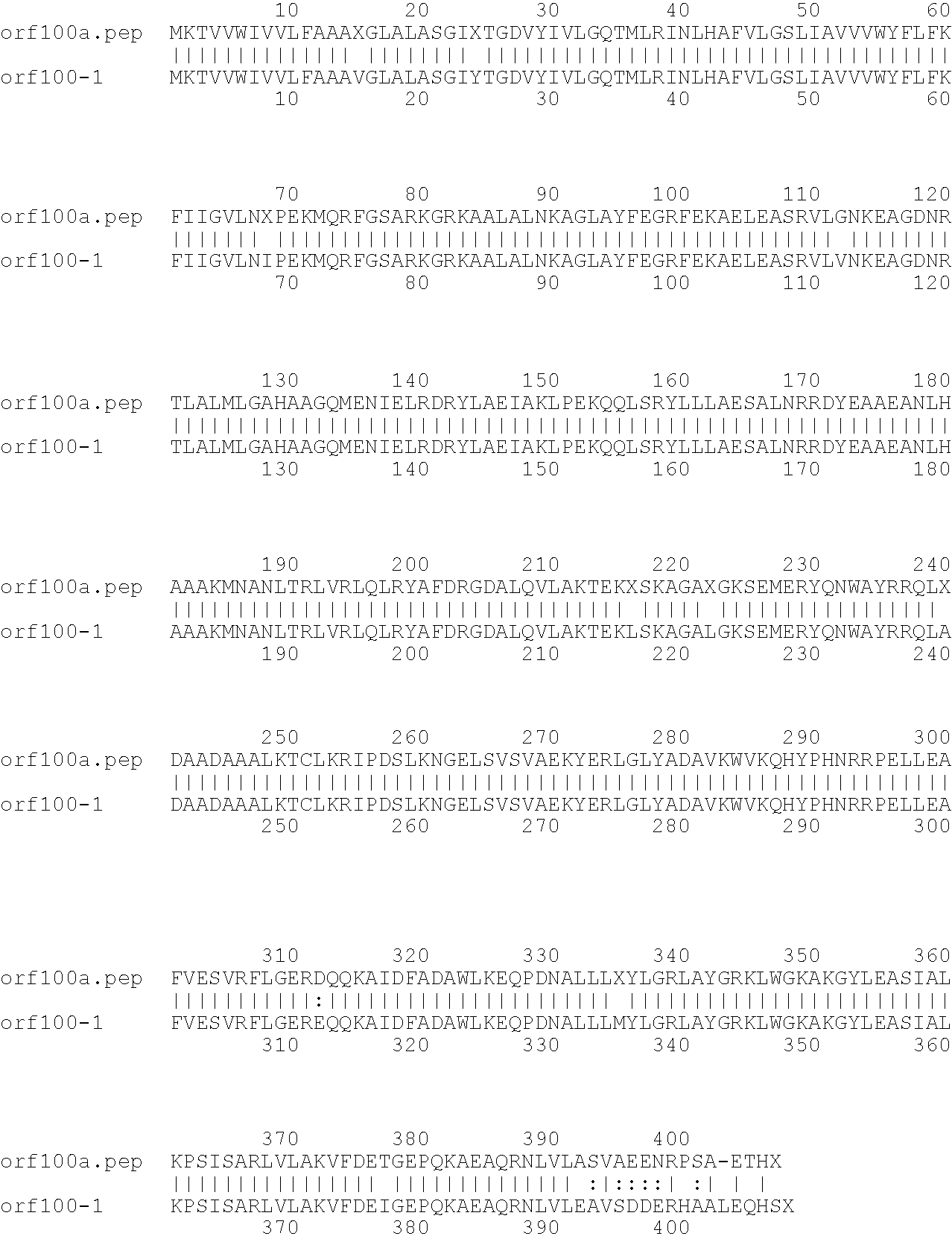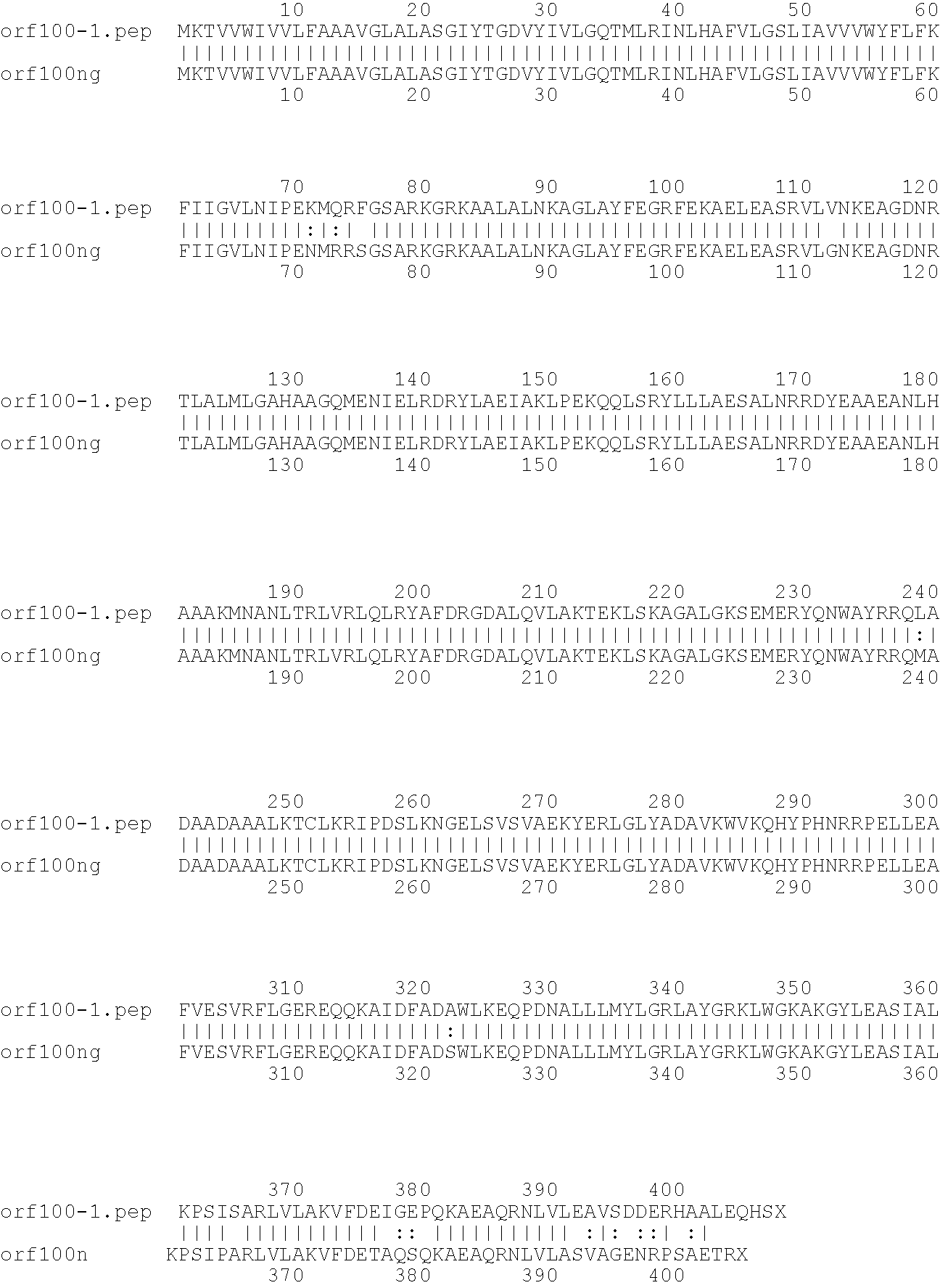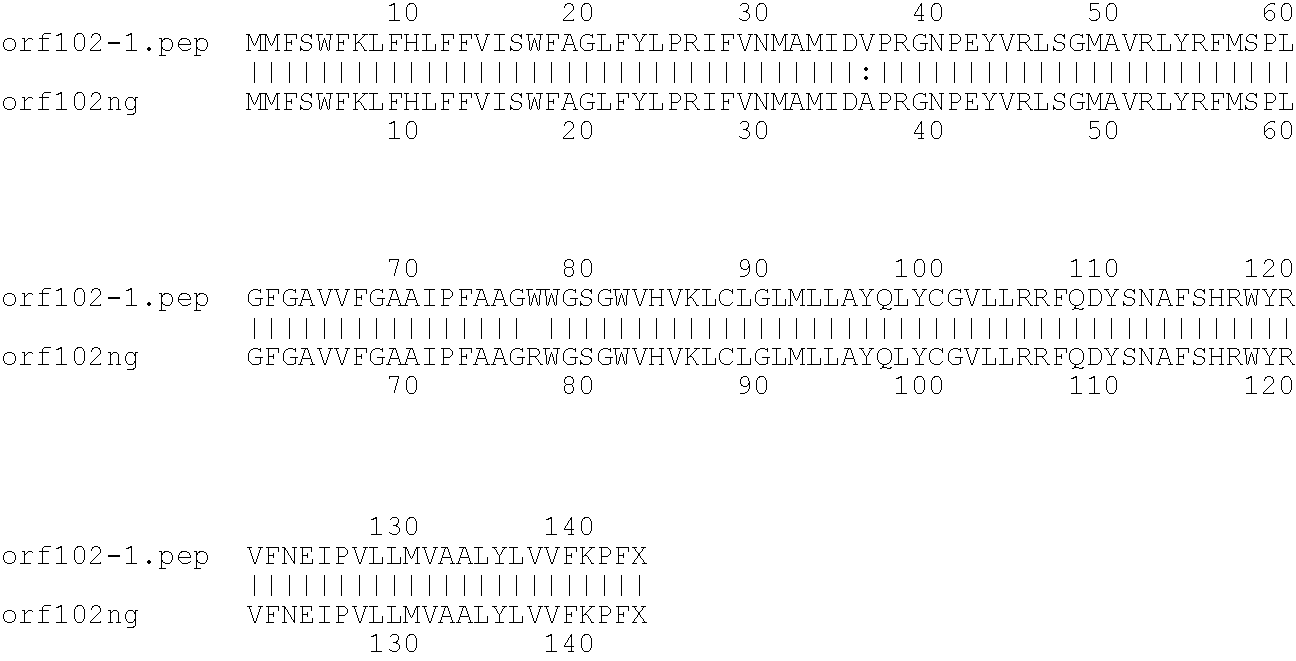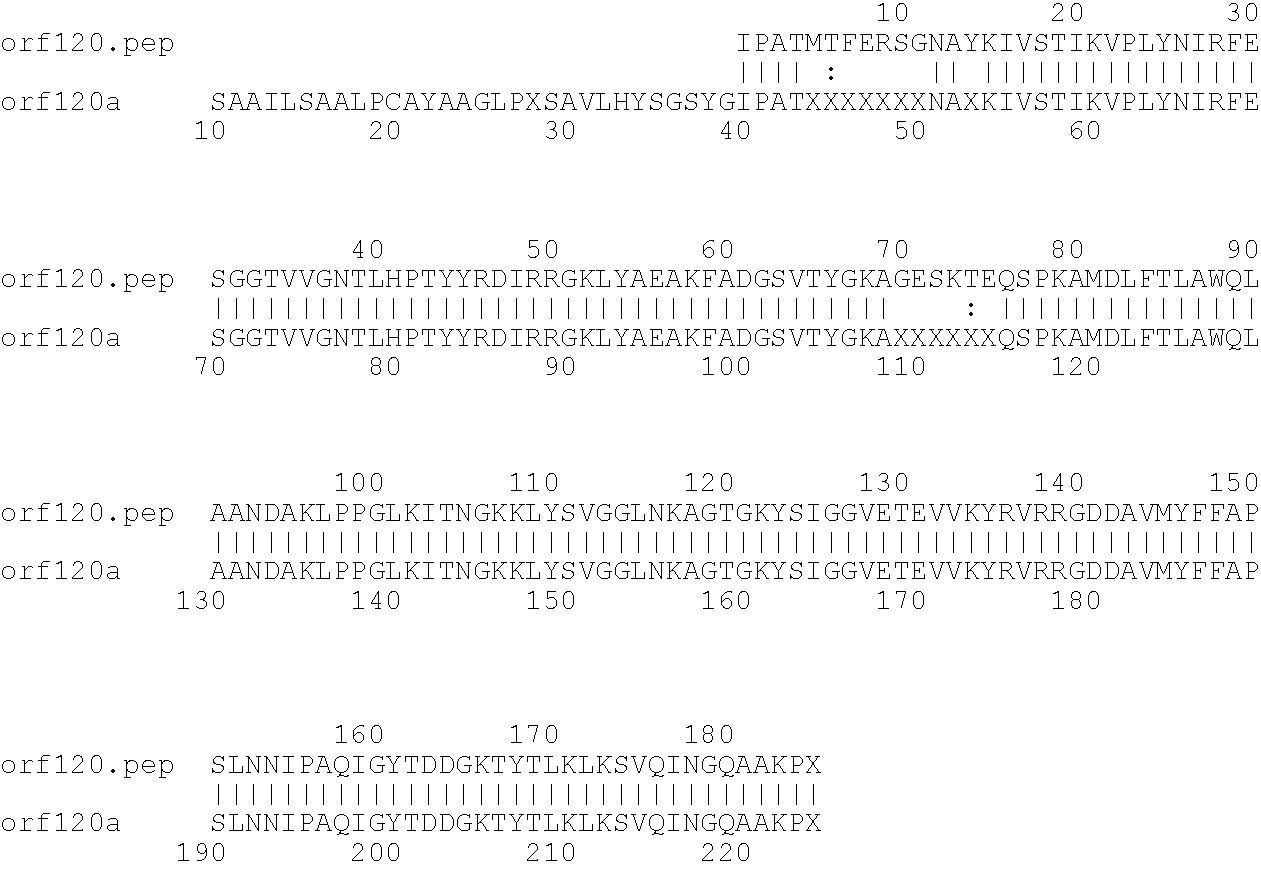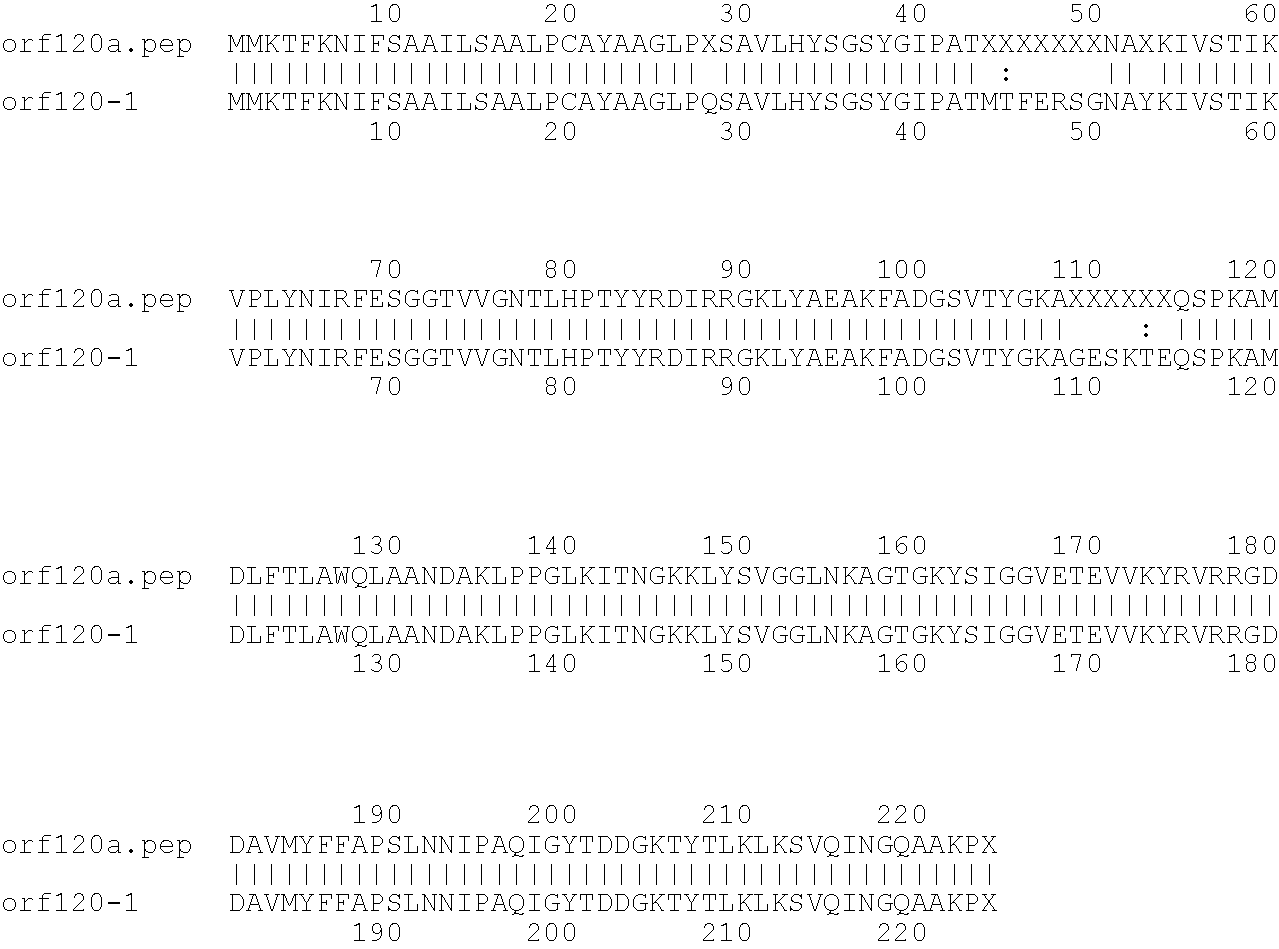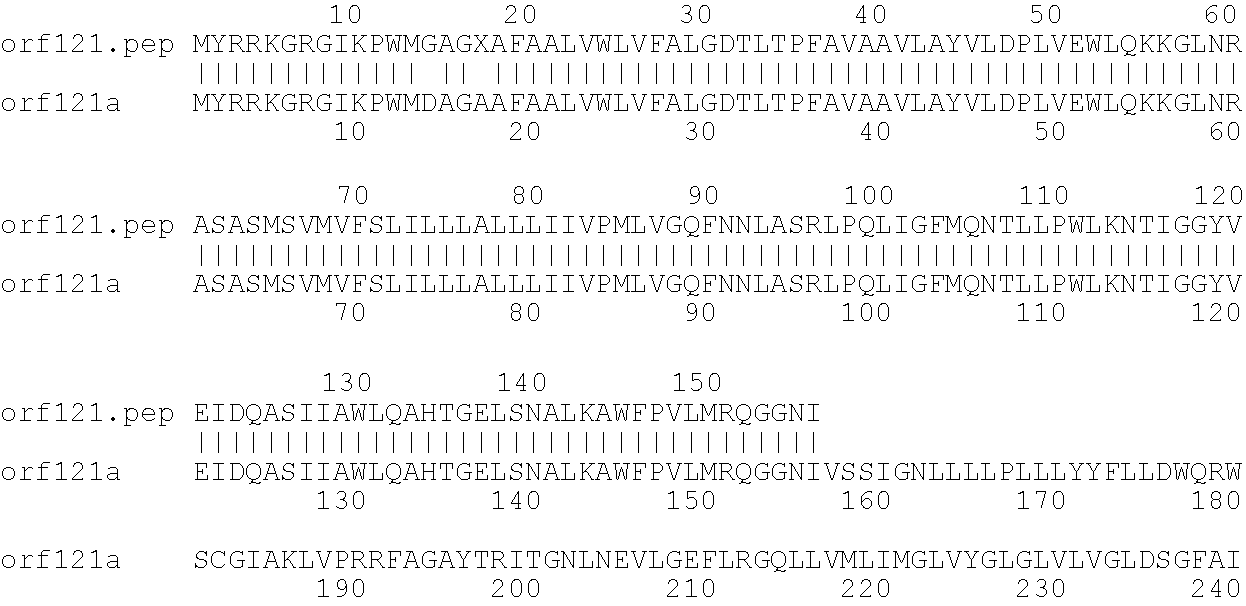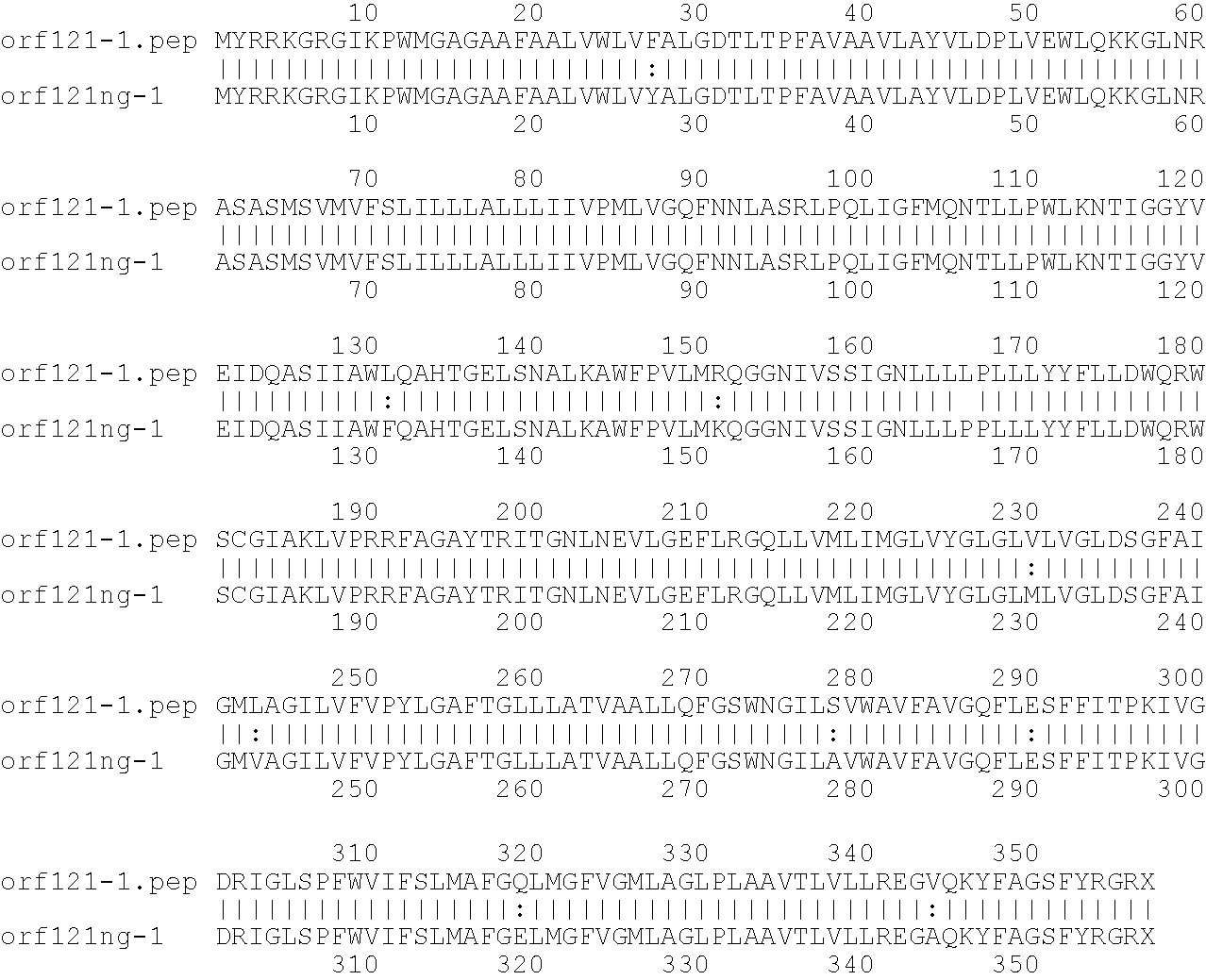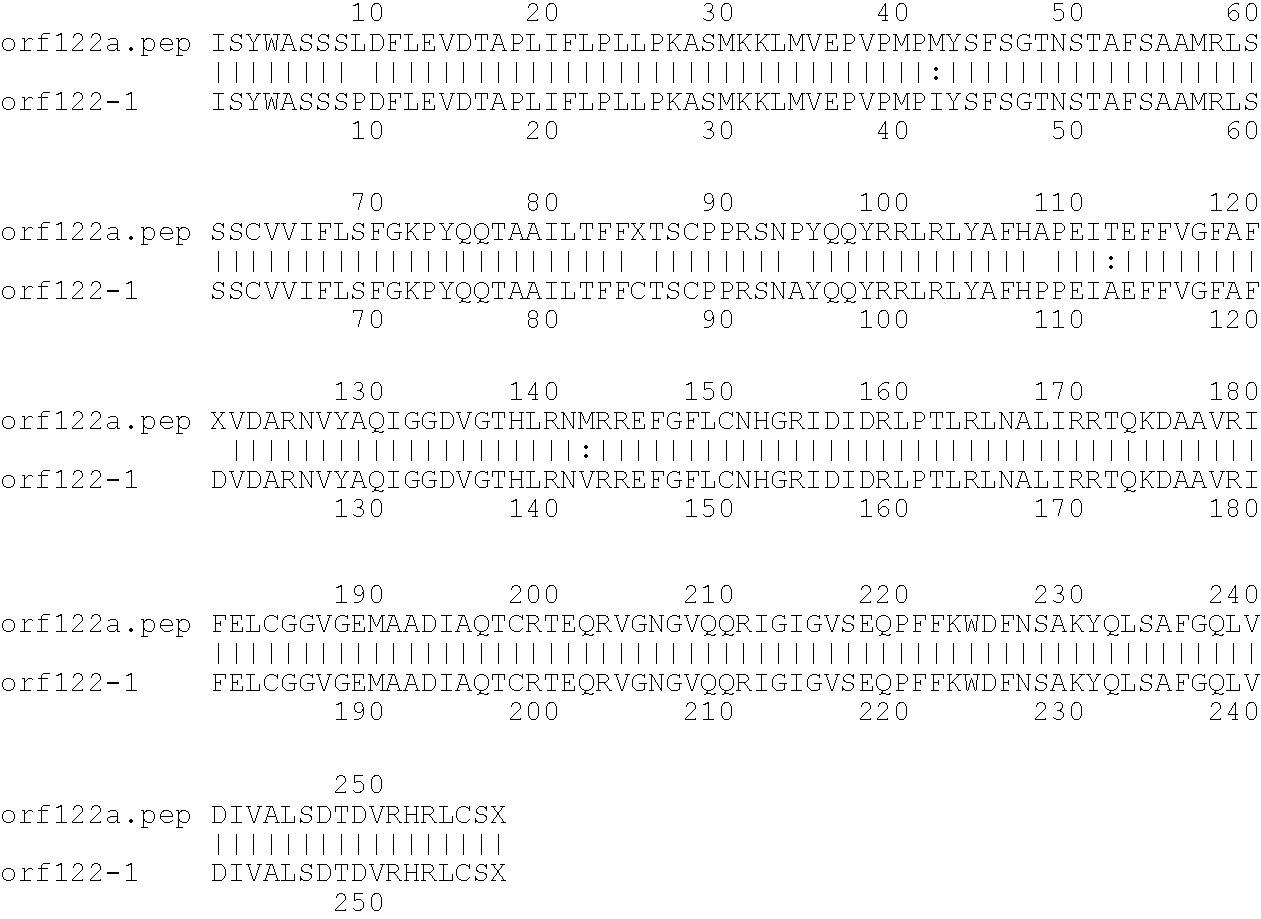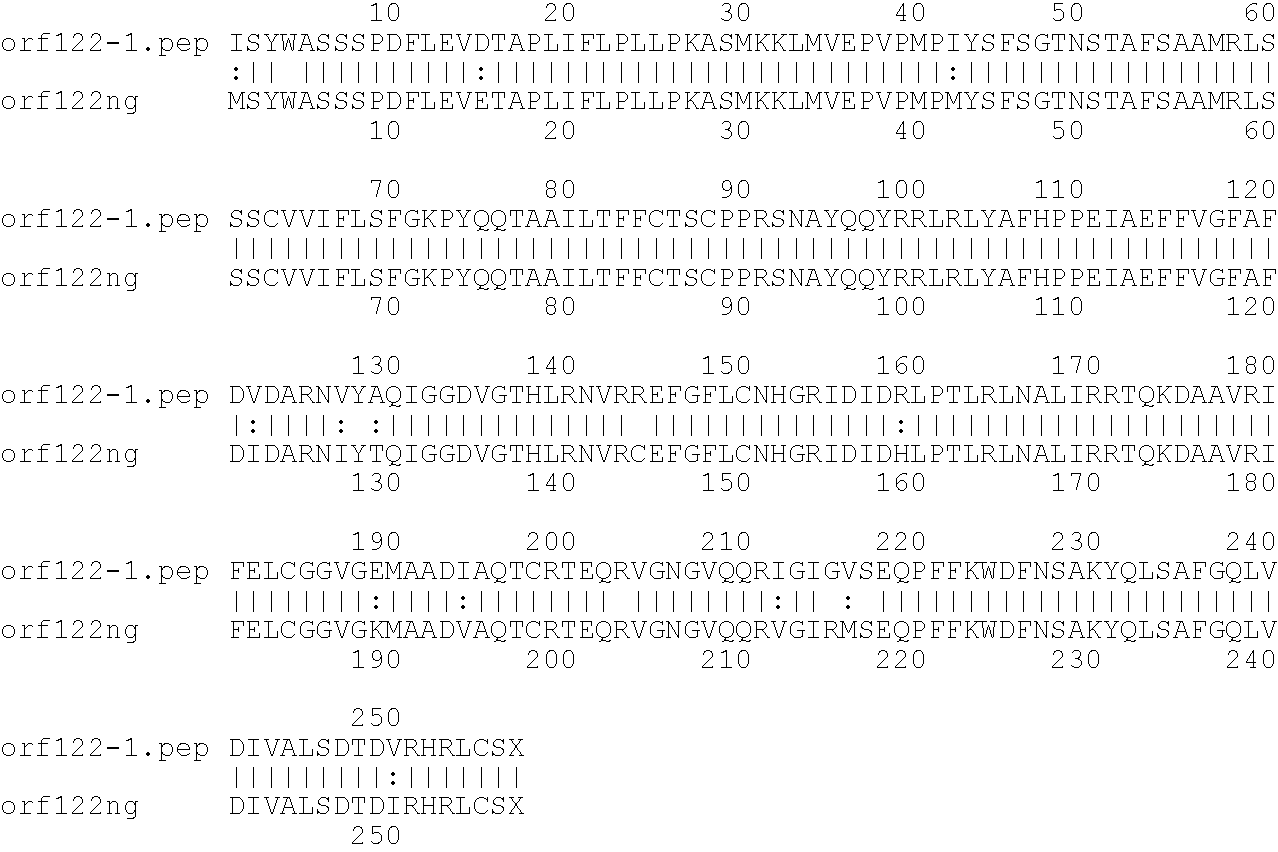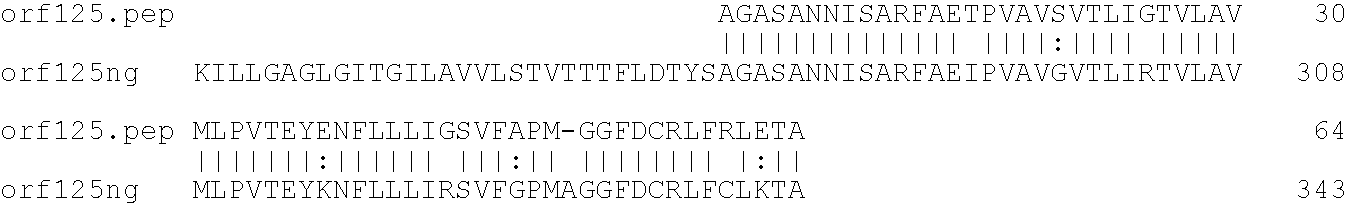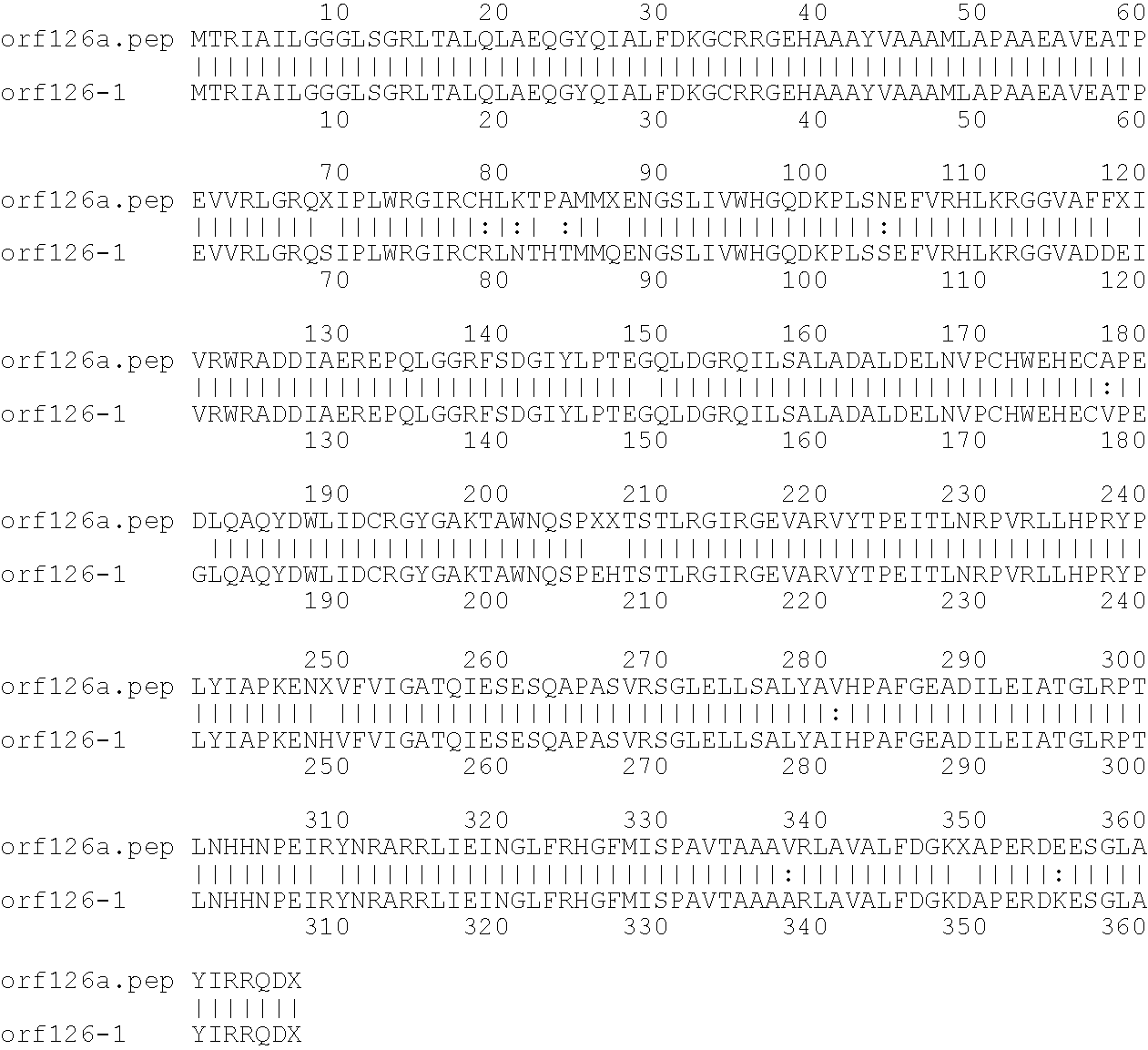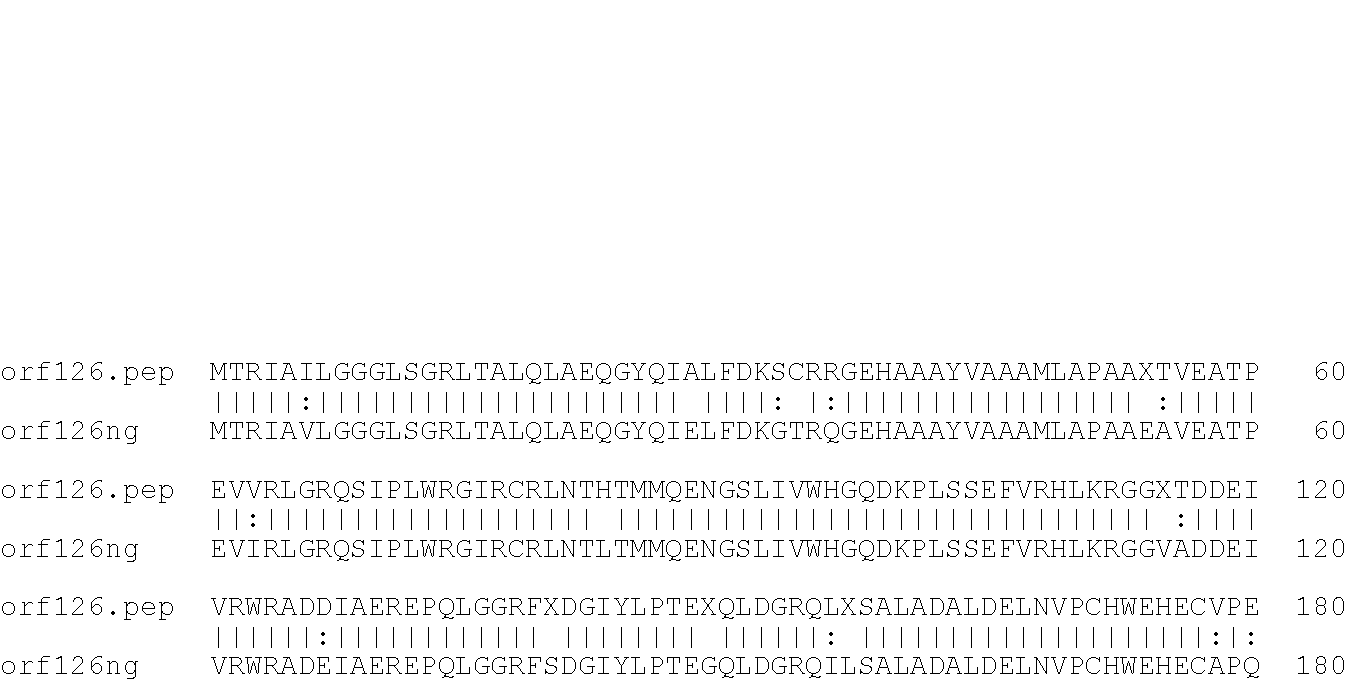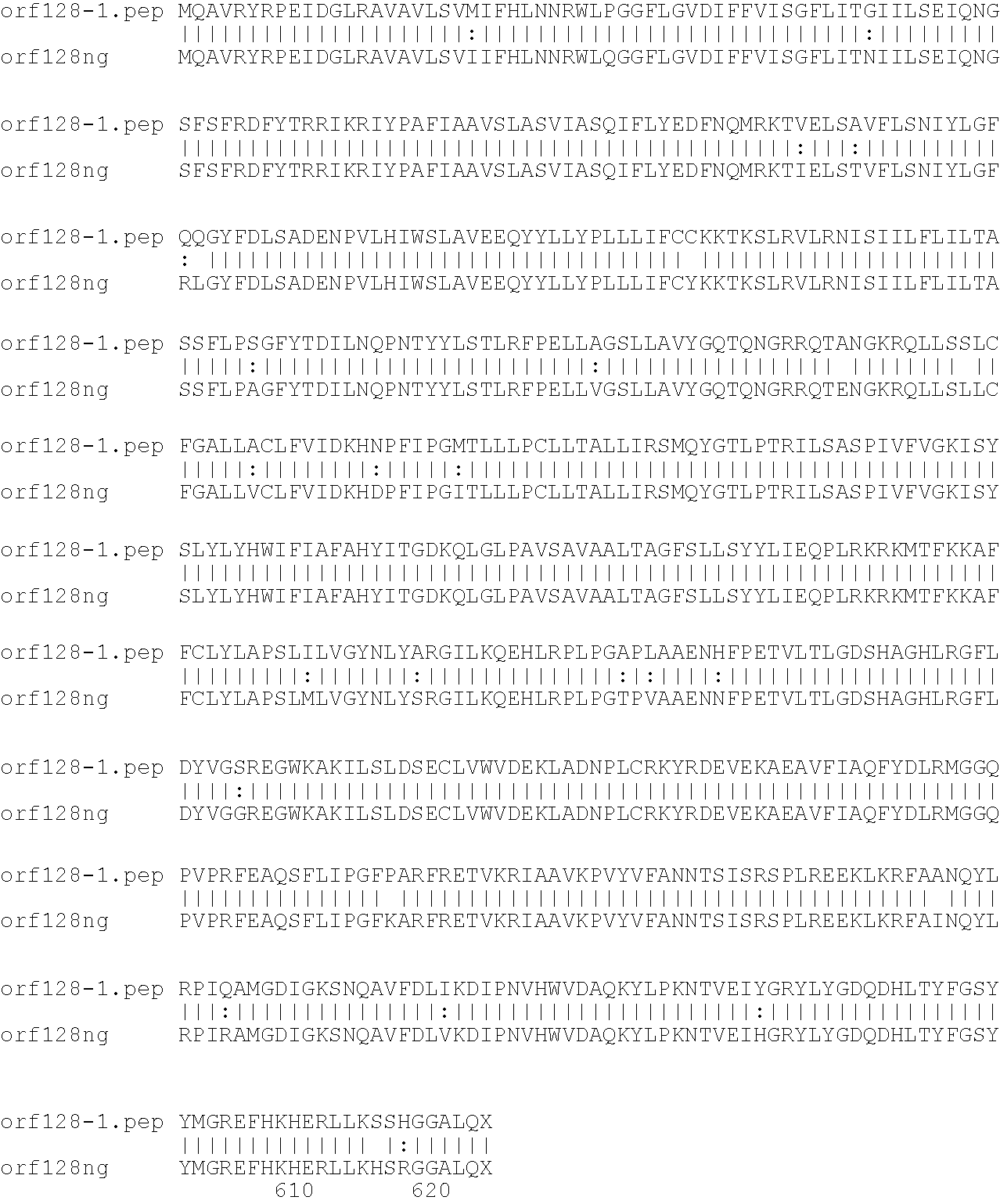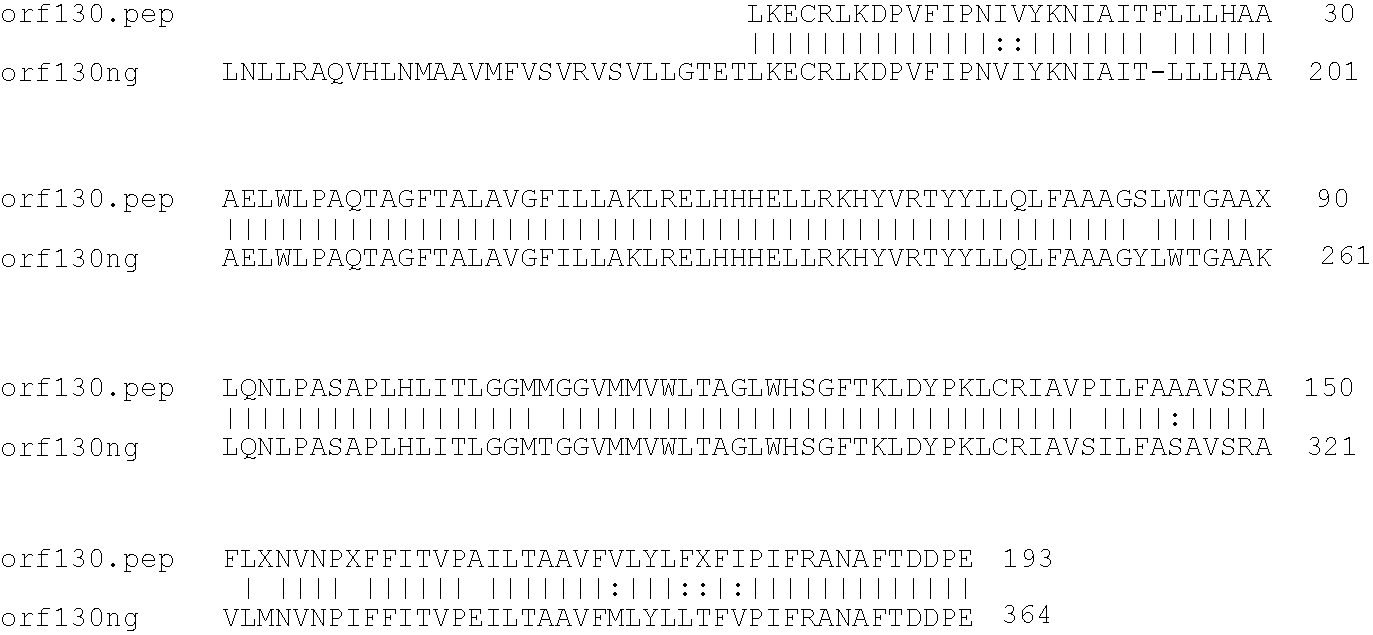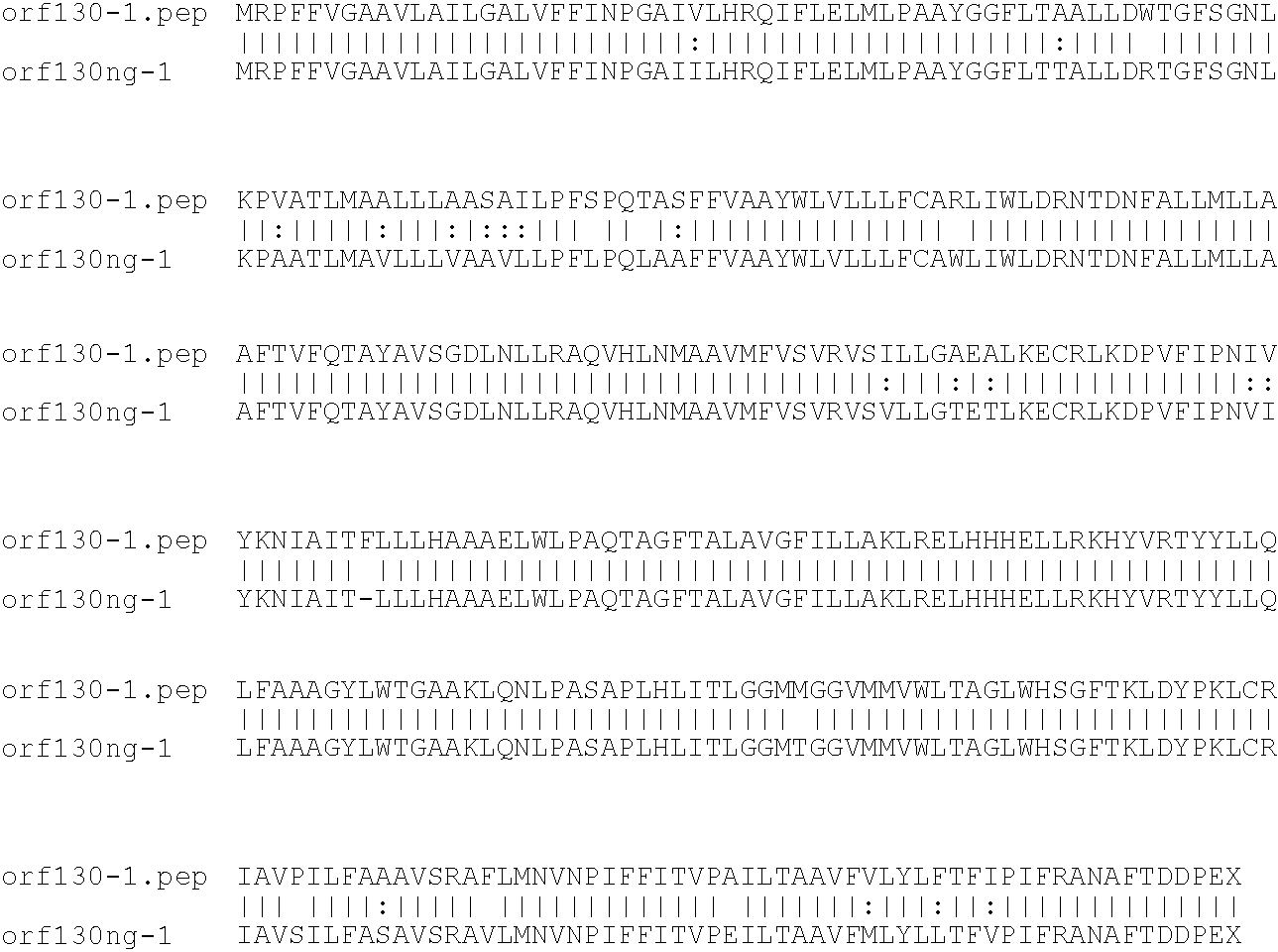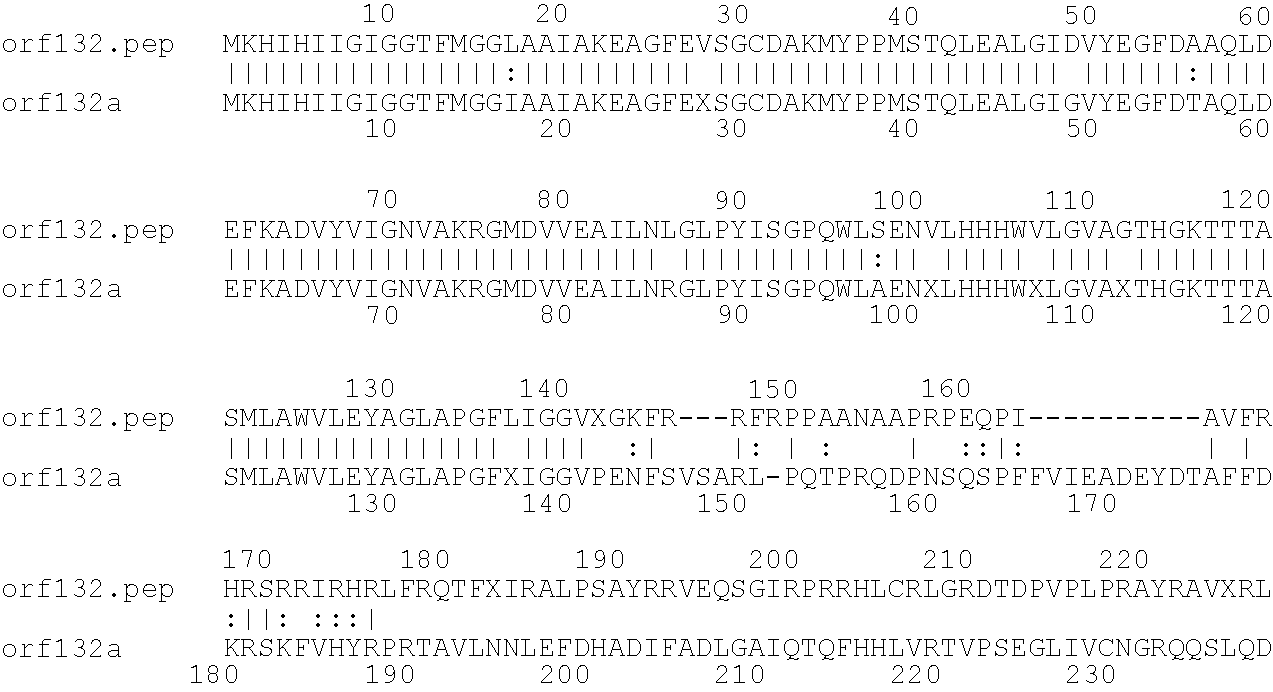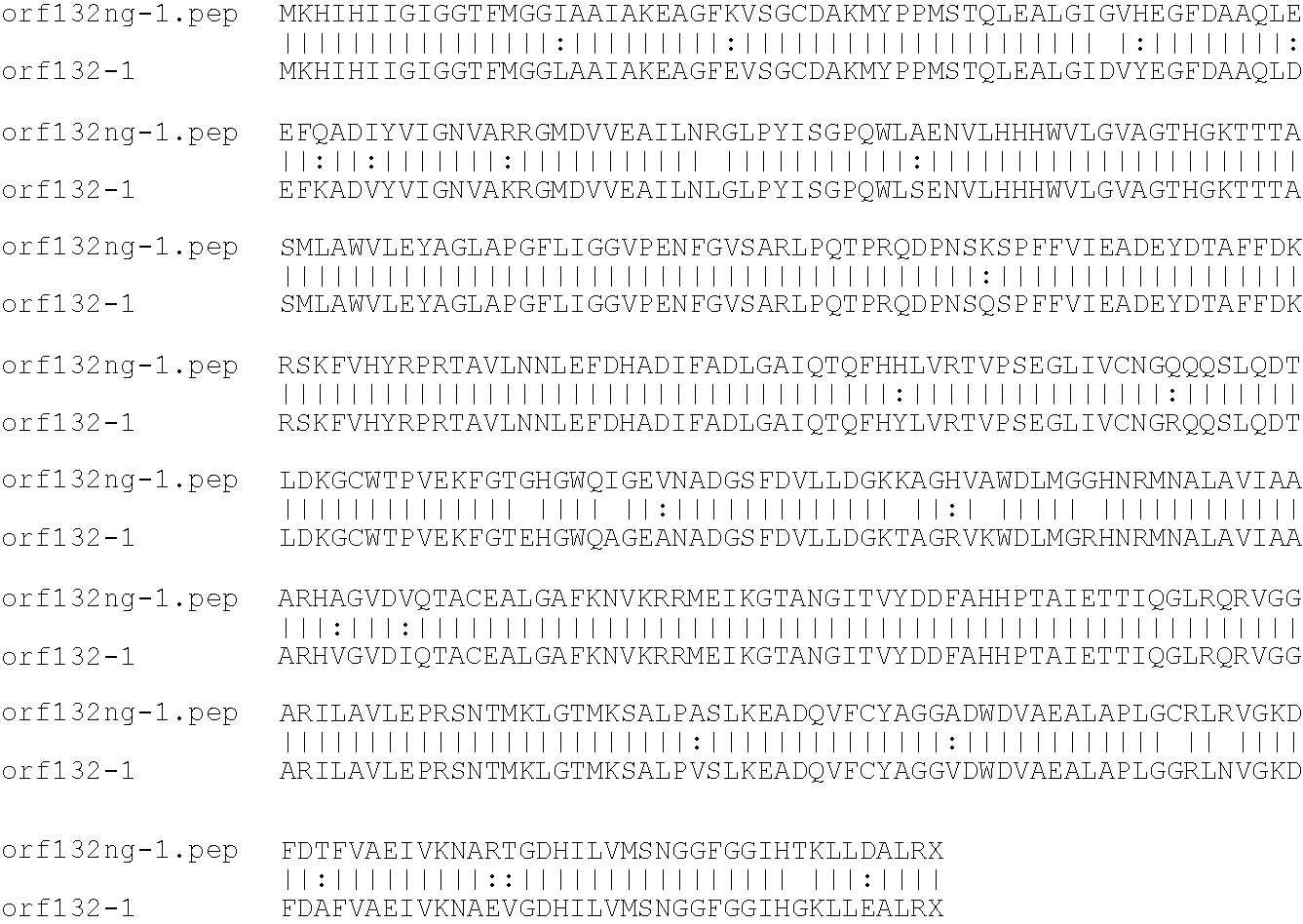[0001]This invention relates to antigens from Neisseria bacteria.
BACKGROUND ART
[0002]Neisseria meningitidis and Neisseria gonorrhoeae are non-motile, gram negative diplococci that are pathogenic in humans. N. meningitidis colonises the pharynx and causes meningitis (and, occasionally, septicaemia in the absence of meningitis); N. gonorrhoeae colonises the genital tract and causes gonorrhea. Although colonising different areas of the body and causing completely different diseases, the two pathogens are closely related, although one feature that clearly differentiates meningococcus from gonococcus is the presence of a polysaccharide capsule that is present in all pathogenic meningococci.
[0003]N. gonorrhoeae caused approximately 800,000 cases per year during the period 1983-1990 in the United States alone (chapter by Meitzner & Cohen, “Vaccines Against Gonococcal Infection”, In: New Generation Vaccines, 2nd edition, ed. Levine, Woodrow, Kaper, & Cobon, Marcel Dekker, New York, 1997, pp. 817-842). The disease causes significant morbidity but limited mortality. Vaccination against N. gonorrhoeae would be highly desirable, but repeated attempts have failed. The main candidate antigens for this vaccine are surface-exposed proteins such as pili, porins, opacity-associated proteins (Opas) and other surface-exposed proteins such as the Lip, Laz, IgA1 protease and transferrin-binding proteins. The lipooligosaccharide (LOS) has also been suggested as vaccine (Meitzner & Cohen, supra).
[0004]N. meningitidis causes both endemic and epidemic disease. In the United States the attack rate is 0.6-1 per 100,000 persons per year, and it can be much greater during outbreaks (see Lieberman et al. (1996) Safety and Immunogenicity of a Serogroups A/C Neisseria meningitidis Oligosaccharide-Protein Conjugate Vaccine in Young Children. JAMA 275(19):1499-1503; Schuchat et al (1997) Bacterial Meningitis in the United States in 1995. N Engl J Med 337(14):970-976). In developing countries, endemic disease rates are much higher and during epidemics incidence rates can reach 500 cases per 100,000 persons per year. Mortality is extremely high, at 10-20% in the United States, and much higher in developing countries. Following the introduction of the conjugate vaccine against Haemophilus influenzae, N. meningitidis is the major cause of bacterial meningitis at all ages in the United States (Schuchat et al (1997) supra).
[0005]Based on the organism's capsular polysaccharide, 12 serogroups of N. meningitidis have been identified. Group A is the pathogen most often implicated in epidemic disease in sub-Saharan Africa. Serogroups B and C are responsible for the vast majority of cases in the United States and in most developed countries. Serogroups W135 and Y are responsible for the rest of the cases in the United States and developed countries. The meningococcal vaccine currently in use is a tetravalent polysaccharide vaccine composed of serogroups A, C, Y and W135. Although efficacious in adolescents and adults, it induces a poor immune response and short duration of protection, and cannot be used in infants [e.g. Morbidity and Mortality weekly report, Vol. 46, No. RR-5 (1997)]. This is because polysaccharides are T-cell independent antigens that induce a weak immune response that cannot be boosted by repeated immunization. Following the success of the vaccination against H. influenzae, conjugate vaccines against serogroups A and C have been developed and are at the final stage of clinical testing (Zollinger W D “New and Improved Vaccines Against Meningococcal Disease” in: New Generation Vaccines, supra, pp. 469-488; Lieberman et al (1996) supra; Costantino et al (1992) Development and phase I clinical testing of a conjugate vaccine against meningococcus A and C. Vaccine 10:691-698).
[0006]Meningococcus B remains a problem, however. This serotype currently is responsible for approximately 50% of total meningitis in the United States, Europe, and South America. The polysaccharide approach cannot be used because the menB capsular polysaccharide is a polymer of α(2-8)-linked N-acetyl neuraminic acid that is also present in mammalian tissue. This results in tolerance to the antigen; indeed, if an immune response were elicited, it would be anti-self, and therefore undesirable. In order to avoid induction of autoimmunity and to induce a protective immune response, the capsular polysaccharide has, for instance, been chemically modified substituting the N-acetyl groups with N-propionyl groups, leaving the specific antigenicity unaltered (Romero & Outschoorn (1994) Current status of Meningococcal group B vaccine candidates: capsular or non-capsular? Clin Microbiol Rev 7(4):559-575).
[0007]Alternative approaches to menB vaccines have used complex mixtures of outer membrane proteins (OMPs), containing either the OMPs alone, or OMPs enriched in porins, or deleted of the class 4 OMPs that are believed to induce antibodies that block bactericidal activity. This approach produces vaccines that are not well characterized. They are able to protect against the homologous strain, but are not effective at large where there are many antigenic variants of the outer membrane proteins. To overcome the antigenic variability, multivalent vaccines containing up to nine different porins have been constructed (e.g. Poolman J T (1992) Development of a meningococcal vaccine. Infect. Agents Dis. 4:13-28). Additional proteins to be used in outer membrane vaccines have been the opa and opc proteins, but none of these approaches have been able to overcome the antigenic variability (e.g. Ala' Aldeen & Borriello (1996) The meningococcal transferrin-binding proteins 1 and 2 are both surface exposed and generate bactericidal antibodies capable of killing homologous and heterologous strains. Vaccine 14(1):49-53).
[0008]A certain amount of sequence data is available for meningococcal and gonoccocal genes and proteins (e.g. EP-A-0467714, WO96/29412), but this is by no means complete. The provision of further sequences could provide an opportunity to identify secreted or surface-exposed proteins that are presumed targets for the immune system and which are not antigenically variable. For instance, some of the identified proteins could be components of efficacious vaccines against meningococcus B, some could be components of vaccines against all meningococcal serotypes, and others could be components of vaccines against all pathogenic Neisseriae.
THE INVENTION
[0009]The invention provides proteins comprising the Neisserial amino acid sequences disclosed in the examples. These sequences relate to N. meningitidis or N. gonorrhoeae.
[0010]It also provides proteins comprising sequences homologous (i.e. having sequence identity) to the Neisserial amino acid sequences disclosed in the examples. Depending on the particular sequence, the degree of identity is preferably greater than 50% (e.g. 65%, 80%, 90%, or more). These homologous proteins include mutants and allelic variants of the sequences disclosed in the examples. Typically, 50% identity or more between two proteins is considered to be an indication of functional equivalence. Identity between the proteins is preferably determined by the Smith-Waterman homology search algorithm as implemented in the MPSRCH program (Oxford Molecular), using an affine gap search with parameters gap open penalty=12 and gap extension penalty=1.
[0011]The invention further provides proteins comprising fragments of the Neisserial amino acid sequences disclosed in the examples. The fragments should comprise at least n consecutive amino acids from the sequences and, depending on the particular sequence, n is 7 or more (e.g. 8, 10, 12, 14, 16, 18, 20 or more). Preferably the fragments comprise an epitope from the sequence.
[0012]The proteins of the invention can, of course, be prepared by various means (e.g. recombinant expression, purification from cell culture, chemical synthesis etc.) and in various forms (e.g. native, fusions etc.). They are preferably prepared in substantially pure or isolated form (i.e. substantially free from other Neisserial or host cell proteins)
[0013]According to a further aspect, the invention provides antibodies which bind to these proteins. These may be polyclonal or monoclonal and may be produced by any suitable means.
[0014]According to a further aspect, the invention provides nucleic acid comprising the Neisserial nucleotide sequences disclosed in the examples. In addition, the invention provides nucleic acid comprising sequences homologous (i.e. having sequence identity) to the Neisserial nucleotide sequences disclosed in the examples.
[0015]Furthermore, the invention provides nucleic acid which can hybridise to the Neisserial nucleic acid disclosed in the examples, preferably under “high stringency” conditions (e.g. 65° C. in a 0.1×SSC, 0.5% SDS solution).
[0016]Nucleic acid comprising fragments of these sequences are also provided. These should comprise at least n consecutive nucleotides from the Neisserial sequences and, depending on the particular sequence, n is 10 or more (eg 12, 14, 15, 18, 20, 25, 30, 35, 40 or more).
[0017]According to a further aspect, the invention provides nucleic acid encoding the proteins and protein fragments of the invention.
[0018]It should also be appreciated that the invention provides nucleic acid comprising sequences complementary to those described above (e.g. for antisense or probing purposes).
[0019]Nucleic acid according to the invention can, of course, be prepared in many ways (e.g. by chemical synthesis, from genomic or cDNA libraries, from the organism itself etc.) and can take various forms (e.g. single stranded, double stranded, vectors, probes etc.).
[0020]In addition, the term “nucleic acid” includes DNA and RNA, and also their analogues, such as those containing modified backbones, and also peptide nucleic acids (PNA) etc.
[0021]According to a further aspect, the invention provides vectors comprising nucleotide sequences of the invention (e.g. expression vectors) and host cells transformed with such vectors.
[0022]According to a further aspect, the invention provides compositions comprising protein, antibody, and/or nucleic acid according to the invention. These compositions may be suitable as vaccines, for instance, or as diagnostic reagents, or as immunogenic compositions.
[0023]The invention also provides nucleic acid, protein, or antibody according to the invention for use as medicaments (e.g. as vaccines) or as diagnostic reagents. It also provides the use of nucleic acid, protein, or antibody according to the invention in the manufacture of: (i) a medicament for treating or preventing infection due to Neisserial bacteria; (ii) a diagnostic reagent for detecting the presence of Neisserial bacteria or of antibodies raised against Neisserial bacteria; and/or (iii) a reagent which can raise antibodies against Neisserial bacteria. Said Neisserial bacteria may be any species or strain (such as N. gonorrhoeae, or any strain of N. meningitidis, such as strain A, strain B or strain C).
[0024]The invention also provides a method of treating a patient, comprising administering to the patient a therapeutically effective amount of nucleic acid, protein, and/or antibody according to the invention.
[0025]According to further aspects, the invention provides various processes.
[0026]A process for producing proteins of the invention is provided, comprising the step of culturing a host cell according to the invention under conditions which induce protein expression.
[0027]A process for producing protein or nucleic acid of the invention is provided, wherein the protein or nucleic acid is synthesised in part or in whole using chemical means.
[0028]A process for detecting polynucleotides of the invention is provided, comprising the steps of: (a) contacting a nucleic probe according to the invention with a biological sample under hybridizing conditions to form duplexes; and (b) detecting said duplexes.
[0029]A process for detecting proteins of the invention is provided, comprising the steps of: (a) contacting an antibody according to the invention with a biological sample under conditions suitable for the formation of an antibody-antigen complexes; and (b) detecting said complexes.
[0030]A summary of standard techniques and procedures which may be employed in order to perform the invention (e.g. to utilise the disclosed sequences for vaccination or diagnostic purposes) follows. This summary is not a limitation on the invention but, rather, gives examples that may be used, but are not required.
General
[0031]The practice of the present invention will employ, unless otherwise indicated, conventional techniques of molecular biology, microbiology, recombinant DNA, and immunology, which are within the skill of the art. Such techniques are explained fully in the literature e.g. Sambrook Molecular Cloning, A Laboratory Manual, Second Edition (1989); DNA Cloning, Volumes I and ii (D. N Glover ed. 1985); Oligonucleotide Synthesis (M. J. Gait ed, 1984); Nucleic Acid Hybridization (B. D. Hames & S. J. Higgins eds. 1984); Transcription and Translation (B. D. Hames & S. J. Higgins eds. 1984); Animal Cell Culture (R. I. Freshney ed. 1986); Immobilized Cells and Enzymes (IRL Press, 1986); B. Perbal, A Practical Guide to Molecular Cloning (1984); the Methods in Enzymology series (Academic Press, Inc.), especially volumes 154 & 155; Gene Transfer Vectors for Mammalian Cells (J. H. Miller and M. P. Calos eds. 1987, Cold Spring Harbor Laboratory); Mayer and Walker, eds. (1987), Immunochemical Methods in Cell and Molecular Biology (Academic Press, London); Scopes, (1987) Protein Purification: Principles and Practice, Second Edition (Springer-Verlag, N.Y.), and Handbook of Experimental Immunology, Volumes I-IV (D. M. Weir and C. C. Blackwell eds 1986).
[0032]Standard abbreviations for nucleotides and amino acids are used in this specification.
[0033]All publications, patents, and patent applications cited herein are incorporated in full by reference. In particular, the contents of UK patent applications 9723516.2, 9724190.5, 9724386.9, 9725158.1, 9726147.3, 9800759.4, and 9819016.8 are incorporated herein.
DEFINITIONS
[0034]A composition containing X is “substantially free of” Y when at least 85% by weight of the total X+Y in the composition is X. Preferably, X comprises at least about 90% by weight of the total of X+Y in the composition, more preferably at least about 95% or even 99% by weight.
[0035]The term “comprising” means “including” as well as “consisting” e.g. a composition “comprising” X may consist exclusively of X or may include something additional to X, such as X+Y.
[0036]The term “heterologous” refers to two biological components that are not found together in nature. The components may be host cells, genes, or regulatory regions, such as promoters. Although the heterologous components are not found together in nature, they can function together, as when a promoter heterologous to a gene is operably linked to the gene. Another example is where a Neisserial sequence is heterologous to a mouse host cell. A further examples would be two epitopes from the same or different proteins which have been assembled in a single protein in an arrangement not found in nature.
[0037]An “origin of replication” is a polynucleotide sequence that initiates and regulates replication of polynucleotides, such as an expression vector. The origin of replication behaves as an autonomous unit of polynucleotide replication within a cell, capable of replication under its own control. An origin of replication may be needed for a vector to replicate in a particular host cell. With certain origins of replication, an expression vector can be reproduced at a high copy number in the presence of the appropriate proteins within the cell. Examples of origins are the autonomously replicating sequences, which are effective in yeast; and the viral T-antigen, effective in COS-7 cells.
[0038]A “mutant” sequence is defined as DNA, RNA or amino acid sequence differing from but having sequence identity with the native or disclosed sequence. Depending on the particular sequence, the degree of sequence identity between the native or disclosed sequence and the mutant sequence is preferably greater than 50% (e.g. 60%, 70%, 80%, 90%, 95%, 99% or more, calculated using the Smith-Waterman algorithm as described above). As used herein, an “allelic variant” of a nucleic acid molecule, or region, for which nucleic acid sequence is provided herein is a nucleic acid molecule, or region, that occurs essentially at the same locus in the genome of another or second isolate, and that, due to natural variation caused by, for example, mutation or recombination, has a similar but not identical nucleic acid sequence. A coding region allelic variant typically encodes a protein having similar activity to that of the protein encoded by the gene to which it is being compared. An allelic variant can also comprise an alteration in the 5′ or 3′ untranslated regions of the gene, such as in regulatory control regions (e.g. see U.S. Pat. No. 5,753,235).
Expression Systems
[0039]The Neisserial nucleotide sequences can be expressed in a variety of different expression systems; for example those used with mammalian cells, baculoviruses, plants, bacteria, and yeast.
[0000]i. Mammalian Systems
[0040]Mammalian expression systems are known in the art. A mammalian promoter is any DNA sequence capable of binding mammalian RNA polymerase and initiating the downstream (3′) transcription of a coding sequence (e.g. structural gene) into mRNA. A promoter will have a transcription initiating region, which is usually placed proximal to the 5′ end of the coding sequence, and a TATA box, usually located 25-30 base pairs (bp) upstream of the transcription initiation site. The TATA box is thought to direct RNA polymerase II to begin RNA synthesis at the correct site. A mammalian promoter will also contain an upstream promoter element, usually located within 100 to 200 bp upstream of the TATA box. An upstream promoter element determines the rate at which transcription is initiated and can act in either orientation [Sambrook et al. (1989) “Expression of Cloned Genes in Mammalian Cells.” In Molecular Cloning: A Laboratory Manual, 2nd ed.].
[0041]Mammalian viral genes are often highly expressed and have a broad host range; therefore sequences encoding mammalian viral genes provide particularly useful promoter sequences. Examples include the SV40 early promoter, mouse mammary tumor virus LTR promoter, adenovirus major late promoter (Ad MLP), and herpes simplex virus promoter. In addition, sequences derived from non-viral genes, such as the murine metallotheionein gene, also provide useful promoter sequences. Expression may be either constitutive or regulated (inducible), depending on the promoter can be induced with glucocorticoid in hormone-responsive cells.
[0042]The presence of an enhancer element (enhancer), combined with the promoter elements described above, will usually increase expression levels. An enhancer is a regulatory DNA sequence that can stimulate transcription up to 1000-fold when linked to homologous or heterologous promoters, with synthesis beginning at the normal RNA start site. Enhancers are also active when they are placed upstream or downstream from the transcription initiation site, in either normal or flipped orientation, or at a distance of more than 1000 nucleotides from the promoter [Maniatis et al. (1987) Science 236:1237; Alberts et al. (1989) Molecular Biology of the Cell, 2nd ed.]. Enhancer elements derived from viruses may be particularly useful, because they usually have a broader host range. Examples include the SV40 early gene enhancer [Dijkema et al (1985) EMBO J. 4:761] and the enhancer/promoters derived from the long terminal repeat (LTR) of the Rous Sarcoma Virus [Gorman et al. (1982b) Proc. Natl. Acad. Sci. 79:6777] and from human cytomegalovirus [Boshart et al. (1985) Cell 41:521]. Additionally, some enhancers are regulatable and become active only in the presence of an inducer, such as a hormone or metal ion [Sassone-Corsi and Borelli (1986) Trends Genet. 2:215; Maniatis et al. (1987) Science 236:1237].
[0043]A DNA molecule may be expressed intracellularly in mammalian cells. A promoter sequence may be directly linked with the DNA molecule, in which case the first amino acid at the N-terminus of the recombinant protein will always be a methionine, which is encoded by the ATG start codon. If desired, the N-terminus may be cleaved from the protein by in vitro incubation with cyanogen bromide.
[0044]Alternatively, foreign proteins can also be secreted from the cell into the growth media by creating chimeric DNA molecules that encode a fusion protein comprised of a leader sequence fragment that provides for secretion of the foreign protein in mammalian cells. Preferably, there are processing sites encoded between the leader fragment and the foreign gene that can be cleaved either in vivo or in vitro. The leader sequence fragment usually encodes a signal peptide comprised of hydrophobic amino acids which direct the secretion of the protein from the cell. The adenovirus triparite leader is an example of a leader sequence that provides for secretion of a foreign protein in mammalian cells.
[0045]Usually, transcription termination and polyadenylation sequences recognized by mammalian cells are regulatory regions located 3′ to the translation stop codon and thus, together with the promoter elements, flank the coding sequence. The 3′ terminus of the mature mRNA is formed by site-specific post-transcriptional cleavage and polyadenylation [Birnstiel et al. (1985) Cell 41:349; Proudfoot and Whitelaw (1988) “Termination and 3′ end processing of eukaryotic RNA. In Transcription and splicing (ed. B. D. Hames and D. M. Glover); Proudfoot (1989) Trends Biochem. Sci. 14: 105]. These sequences direct the transcription of an mRNA which can be translated into the polypeptide encoded by the DNA. Examples of transcription terminater/polyadenylation signals include those derived from SV40 [Sambrook et al (1989) “Expression of cloned genes in cultured mammalian cells.” In Molecular Cloning: A Laboratory Manual].
[0046]Usually, the above described components, comprising a promoter, polyadenylation signal, and transcription termination sequence are put together into expression constructs. Enhancers, introns with functional splice donor and acceptor sites, and leader sequences may also be included in an expression construct, if desired. Expression constructs are often maintained in a replicon, such as an extrachromosomal element (e.g. plasmids) capable of stable maintenance in a host, such as mammalian cells or bacteria. Mammalian replication systems include those derived from animal viruses, which require trans-acting factors to replicate. For example, plasmids containing the replication systems of papovaviruses, such as SV40 [Gluzman (1981) Cell 23:175] or polyomavirus, replicate to extremely high copy number in the presence of the appropriate viral T antigen. Additional examples of mammalian replicons include those derived from bovine papillomavirus and Epstein-Barr virus. Additionally, the replicon may have two replicaton systems, thus allowing it to be maintained, for example, in mammalian cells for expression and in a prokaryotic host for cloning and amplification. Examples of such mammalian-bacteria shuttle vectors include pMT2 [Kaufman et al. (1989) Mol. Cell. Biol. 9:946] and pHEBO [Shimizu et al. (1986) Mol. Cell. Biol. 6:1074].
[0047]The transformation procedure used depends upon the host to be transformed. Methods for introduction of heterologous polynucleotides into mammalian cells are known in the art and include dextran-mediated transfection, calcium phosphate precipitation, polybrene mediated transfection, protoplast fusion, electroporation, encapsulation of the polynucleotide(s) in liposomes, and direct microinjection of the DNA into nuclei.
[0048]Mammalian cell lines available as hosts for expression are known in the art and include many immortalized cell lines available from the American Type Culture Collection (ATCC), including but not limited to, Chinese hamster ovary (CHO) cells, HeLa cells, baby hamster kidney (BHK) cells, monkey kidney cells (COS), human hepatocellular carcinoma cells (e.g. Hep G2), and a number of other cell lines.
[0000]ii. Baculovirus Systems
[0049]The polynucleotide encoding the protein can also be inserted into a suitable insect expression vector, and is operably linked to the control elements within that vector. Vector construction employs techniques which are known in the art. Generally, the components of the expression system include a transfer vector, usually a bacterial plasmid, which contains both a fragment of the baculovirus genome, and a convenient restriction site for insertion of the heterologous gene or genes to be expressed; a wild type baculovirus with a sequence homologous to the baculovirus-specific fragment in the transfer vector (this allows for the homologous recombination of the heterologous gene in to the baculovirus genome); and appropriate insect host cells and growth media.
[0050]After inserting the DNA sequence encoding the protein into the transfer vector, the vector and the wild type viral genome are transfected into an insect host cell where the vector and viral genome are allowed to recombine. The packaged recombinant virus is expressed and recombinant plaques are identified and purified. Materials and methods for baculovirus/insect cell expression systems are commercially available in kit form from, inter alia, Invitrogen, San Diego Calif. (“MaxBac” kit). These techniques are generally known to those skilled in the art and fully described in Summers and Smith, Texas Agricultural Experiment Station Bulletin No. 1555 (1987) (hereinafter “Summers and Smith”).
[0051]Prior to inserting the DNA sequence encoding the protein into the baculovirus genome, the above described components, comprising a promoter, leader (if desired), coding sequence of interest, and transcription termination sequence, are usually assembled into an intermediate transplacement construct (transfer vector). This construct may contain a single gene and operably linked regulatory elements; multiple genes, each with its owned set of operably linked regulatory elements; or multiple genes, regulated by the same set of regulatory elements. Intermediate transplacement constructs are often maintained in a replicon, such as an extrachromosomal element (e.g. plasmids) capable of stable maintenance in a host, such as a bacterium. The replicon will have a replication system, thus allowing it to be maintained in a suitable host for cloning and amplification.
[0052]Currently, the most commonly used transfer vector for introducing foreign genes into AcNPV is pAc373. Many other vectors, known to those of skill in the art, have also been designed. These include, for example, pVL985 (which alters the polyhedrin start codon from ATG to ATT, and which introduces a BamHI cloning site 32 basepairs downstream from the ATT; see Luckow and Summers, Virology (1989) 17:31.
[0053]The plasmid usually also contains the polyhedrin polyadenylation signal (Miller et al. (1988) Ann. Rev. Microbiol., 42:177) and a prokaryotic ampicillin-resistance (amp) gene and origin of replication for selection and propagation in E. coli.
[0054]Baculovirus transfer vectors usually contain a baculovirus promoter. A baculovirus promoter is any DNA sequence capable of binding a baculovirus RNA polymerase and initiating the downstream (5′ to 3′) transcription of a coding sequence (e.g. structural gene) into mRNA. A promoter will have a transcription initiation region which is usually placed proximal to the 5′ end of the coding sequence. This transcription initiation region usually includes an RNA polymerase binding site and a transcription initiation site. A baculovirus transfer vector may also have a second domain called an enhancer, which, if present, is usually distal to the structural gene. Expression may be either regulated or constitutive.
[0055]Structural genes, abundantly transcribed at late times in a viral infection cycle, provide particularly useful promoter sequences. Examples include sequences derived from the gene encoding the viral polyhedron protein, Friesen et al., (1986) “The Regulation of Baculovirus Gene Expression,” in: The Molecular Biology of Baculoviruses (ed. Walter Doerfler); EPO Publ. Nos. 127 839 and 155 476; and the gene encoding the p10 protein, Vlak et al., (1988), J. Gen. Virol. 69:765.
[0056]DNA encoding suitable signal sequences can be derived from genes for secreted insect or baculovirus proteins, such as the baculovirus polyhedrin gene (Carbonell et al. (1988) Gene, 73:409). Alternatively, since the signals for mammalian cell posttranslational modifications (such as signal peptide cleavage, proteolytic cleavage, and phosphorylation) appear to be recognized by insect cells, and the signals required for secretion and nuclear accumulation also appear to be conserved between the invertebrate cells and vertebrate cells, leaders of non-insect origin, such as those derived from genes encoding human α-interferon, Maeda et al., (1985), Nature 315:592; human gastrin-releasing peptide, Lebacq-Verheyden et al., (1988), Molec. Cell. Biol. 8:3129; human IL-2, Smith et al., (1985) Proc. Nat'l Acad. Sci. USA, 82:8404; mouse IL-3, (Miyajima et al., (1987) Gene 58:273; and human glucocerebrosidase, Martin et al. (1988) DNA, 7:99, can also be used to provide for secretion in insects.
[0057]A recombinant polypeptide or polyprotein may be expressed intracellularly or, if it is expressed with the proper regulatory sequences, it can be secreted. Good intracellular expression of nonfused foreign proteins usually requires heterologous genes that ideally have a short leader sequence containing suitable translation initiation signals preceding an ATG start signal. If desired, methionine at the N-terminus may be cleaved from the mature protein by in vitro incubation with cyanogen bromide.
[0058]Alternatively, recombinant polyproteins or proteins which are not naturally secreted can be secreted from the insect cell by creating chimeric DNA molecules that encode a fusion protein comprised of a leader sequence fragment that provides for secretion of the foreign protein in insects. The leader sequence fragment usually encodes a signal peptide comprised of hydrophobic amino acids which direct the translocation of the protein into the endoplasmic reticulum.
[0059]After insertion of the DNA sequence and/or the gene encoding the expression product precursor of the protein, an insect cell host is co-transformed with the heterologous DNA of the transfer vector and the genomic DNA of wild type baculovirus—usually by co-transfection. The promoter and transcription termination sequence of the construct will usually comprise a 2-5 kb section of the baculovirus genome. Methods for introducing heterologous DNA into the desired site in the baculovirus virus are known in the art. (See Summers and Smith supra; Ju et al. (1987); Smith et al., Mol. Cell. Biol. (1983) 3:2156; and Luckow and Summers (1989)). For example, the insertion can be into a gene such as the polyhedrin gene, by homologous double crossover recombination; insertion can also be into a restriction enzyme site engineered into the desired baculovirus gene. Miller et al., (1989), Bioessays 4:91. The DNA sequence, when cloned in place of the polyhedrin gene in the expression vector, is flanked both 5′ and 3′ by polyhedrin-specific sequences and is positioned downstream of the polyhedrin promoter.
[0060]The newly formed baculovirus expression vector is subsequently packaged into an infectious recombinant baculovirus. Homologous recombination occurs at low frequency (between about 1% and about 5%); thus, the majority of the virus produced after cotransfection is still wild-type virus. Therefore, a method is necessary to identify recombinant viruses. An advantage of the expression system is a visual screen allowing recombinant viruses to be distinguished. The polyhedrin protein, which is produced by the native virus, is produced at very high levels in the nuclei of infected cells at late times after viral infection. Accumulated polyhedrin protein forms occlusion bodies that also contain embedded particles. These occlusion bodies, up to 15 μm in size, are highly refractile, giving them a bright shiny appearance that is readily visualized under the light microscope. Cells infected with recombinant viruses lack occlusion bodies. To distinguish recombinant virus from wild-type virus, the transfection supernatant is plaqued onto a monolayer of insect cells by techniques known to those skilled in the art. Namely, the plaques are screened under the light microscope for the presence (indicative of wild-type virus) or absence (indicative of recombinant virus) of occlusion bodies. “Current Protocols in Microbiology” Vol. 2 (Ausubel et al. eds) at 16.8 (Supp. 10, 1990); Summers and Smith, supra; Miller et al. (1989).
[0061]Recombinant baculovirus expression vectors have been developed for infection into several insect cells. For example, recombinant baculoviruses have been developed for, inter alia: Aedes aegypti, Autographa californica, Bombyx mori, Drosophila melanogaster, Spodoptera frugiperda, and Trichoplusia ni (WO 89/046699; Carbonell et al., (1985) J. Virol. 56:153; Wright (1986) Nature 321:718; Smith et al., (1983) Mol. Cell. Biol. 3:2156; and see generally, Fraser, et al. (1989) In Vitro Cell. Dev. Biol. 25:225).
[0062]Cells and cell culture media are commercially available for both direct and fusion expression of heterologous polypeptides in a baculovirus/expression system; cell culture technology is generally known to those skilled in the art. See, eg. Summers and Smith supra.
[0063]The modified insect cells may then be grown in an appropriate nutrient medium, which allows for stable maintenance of the plasmid(s) present in the modified insect host. Where the expression product gene is under inducible control, the host may be grown to high density, and expression induced. Alternatively, where expression is constitutive, the product will be continuously expressed into the medium and the nutrient medium must be continuously circulated, while removing the product of interest and augmenting depleted nutrients. The product may be purified by such techniques as chromatography, e.g. HPLC, affinity chromatography, ion exchange chromatography, etc.; electrophoresis; density gradient centrifugation; solvent extraction, or the like. As appropriate, the product may be further purified, as required, so as to remove substantially any insect proteins which are also secreted in the medium or result from lysis of insect cells, so as to provide a product which is at least substantially free of host debris, e.g. proteins, lipids and polysaccharides.
[0064]In order to obtain protein expression, recombinant host cells derived from the transformants are incubated under conditions which allow expression of the recombinant protein encoding sequence. These conditions will vary, dependent upon the host cell selected. However, the conditions are readily ascertainable to those of ordinary skill in the art, based upon what is known in the art.
[0065]There are many plant cell culture and whole plant genetic expression systems known in the art. Exemplary plant cellular genetic expression systems include those described in patents, such as: U.S. Pat. No. 5,693,506; U.S. Pat. No. 5,659,122; and U.S. Pat. No. 5,608,143. Additional examples of genetic expression in plant cell culture has been described by Zenk, Phytochemistry 30:3861-3863 (1991). Descriptions of plant protein signal peptides may be found in addition to the references described above in Vaulcombe et al., Mol. Gen. Genet. 209:33-40 (1987); Chandler et al., Plant Molecular Biology 3:407-418 (1984); Rogers, J. Biol. Chem. 260:3731-3738 (1985); Rothstein et al., Gene 55:353-356 (1987); Whittier et al., Nucleic Acids Research 15:2515-2535 (1987); Wirsel et al., Molecular Microbiology 3:3-14 (1989); Yu et al., Gene 122:247-253 (1992). A description of the regulation of plant gene expression by the phytohormone, gibberellic acid and secreted enzymes induced by gibberellic acid can be found in R. L. Jones and J. MacMillin, Gibberellins: in: Advanced Plant Physiology, Malcolm B. Wilkins, ed., 1984 Pitman Publishing Limited, London, pp. 21-52. References that describe other metabolically-regulated genes: Sheen, Plant Cell, 2:1027-1038 (1990); Maas et al., EMBO J. 9:3447-3452 (1990); Benkel and Hickey, Proc. Natl. Acad. Sci. 84:1337-1339 (1987)
[0066]Typically, using techniques known in the art, a desired polynucleotide sequence is inserted into an expression cassette comprising genetic regulatory elements designed for operation in plants. The expression cassette is inserted into a desired expression vector with companion sequences upstream and downstream from the expression cassette suitable for expression in a plant host. The companion sequences will be of plasmid or viral origin and provide necessary characteristics to the vector to permit the vectors to move DNA from an original cloning host, such as bacteria, to the desired plant host. The basic bacterial/plant vector construct will preferably provide a broad host range prokaryote replication origin; a prokaryote selectable marker; and, for Agrobacterium transformations, T DNA sequences for Agrobacterium-mediated transfer to plant chromosomes. Where the heterologous gene is not readily amenable to detection, the construct will preferably also have a selectable marker gene suitable for determining if a plant cell has been transformed. A general review of suitable markers, for example for the members of the grass family, is found in Wilmink and Dons, 1993, Plant Mol. Biol. Reptr, 11(2):165-185.
[0067]Sequences suitable for permitting integration of the heterologous sequence into the plant genome are also recommended. These might include transposon sequences and the like for homologous recombination as well as Ti sequences which permit random insertion of a heterologous expression cassette into a plant genome. Suitable prokaryote selectable markers include resistance toward antibiotics such as ampicillin or tetracycline. Other DNA sequences encoding additional functions may also be present in the vector, as is known in the art.
[0068]The nucleic acid molecules of the subject invention may be included into an expression cassette for expression of the protein(s) of interest. Usually, there will be only one expression cassette, although two or more are feasible. The recombinant expression cassette will contain in addition to the heterologous protein encoding sequence the following elements, a promoter region, plant 5′ untranslated sequences, initiation codon depending upon whether or not the structural gene comes equipped with one, and a transcription and translation termination sequence. Unique restriction enzyme sites at the 5′ and 3′ ends of the cassette allow for easy insertion into a pre-existing vector.
[0069]A heterologous coding sequence may be for any protein relating to the present invention. The sequence encoding the protein of interest will encode a signal peptide which allows processing and translocation of the protein, as appropriate, and will usually lack any sequence which might result in the binding of the desired protein of the invention to a membrane. Since, for the most part, the transcriptional initiation region will be for a gene which is expressed and translocated during germination, by employing the signal peptide which provides for translocation, one may also provide for translocation of the protein of interest. In this way, the protein(s) of interest will be translocated from the cells in which they are expressed and may be efficiently harvested. Typically secretion in seeds are across the aleurone or scutellar epithelium layer into the endosperm of the seed. While it is not required that the protein be secreted from the cells in which the protein is produced, this facilitates the isolation and purification of the recombinant protein.
[0070]Since the ultimate expression of the desired gene product will be in a eucaryotic cell it is desirable to determine whether any portion of the cloned gene contains sequences which will be processed out as introns by the host's splicosome machinery. If so, site-directed mutagenesis of the “intron” region may be conducted to prevent losing a portion of the genetic message as a false intron code, Reed and Maniatis, Cell 41:95-105, 1985.
[0071]The vector can be microinjected directly into plant cells by use of micropipettes to mechanically transfer the recombinant DNA. Crossway, Mol. Gen. Genet, 202:179-185, 1985. The genetic material may also be transferred into the plant cell by using polyethylene glycol, Krens, et al., Nature, 296, 72-74, 1982. Another method of introduction of nucleic acid segments is high velocity ballistic penetration by small particles with the nucleic acid either within the matrix of small beads or particles, or on the surface, Klein, et al., Nature, 327, 70-73, 1987 and Knudsen and Muller, 1991, Planta, 185:330-336 teaching particle bombardment of barley endosperm to create transgenic barley. Yet another method of introduction would be fusion of protoplasts with other entities, either minicells, cells, lysosomes or other fusible lipid-surfaced bodies, Fraley, et al., Proc. Natl. Acad. Sci. USA, 79, 1859-1863, 1982.
[0072]The vector may also be introduced into the plant cells by electroporation. (Fromm et al., Proc. Natl. Acad. Sci. USA 82:5824, 1985). In this technique, plant protoplasts are electroporated in the presence of plasmids containing the gene construct. Electrical impulses of high field strength reversibly permeabilize biomembranes allowing the introduction of the plasmids. Electroporated plant protoplasts reform the cell wall, divide, and form plant callus.
[0073]All plants from which protoplasts can be isolated and cultured to give whole regenerated plants can be transformed by the present invention so that whole plants are recovered which contain the transferred gene. It is known that practically all plants can be regenerated from cultured cells or tissues, including but not limited to all major species of sugarcane, sugar beet, cotton, fruit and other trees, legumes and vegetables. Some suitable plants include, for example, species from the genera Fragaria, Lotus, Medicago, Onobrychis, Trifolium, Trigonella, Vigna, Citrus, Linum, Geranium, Manihot, Daucus, Arabidopsis, Brassica, Raphanus, Sinapis, Atropa, Capsicum, Datura, Hyoscyamus, Lycopersion, Nicotiana, Solanum, Petunia, Digitalis, Majorana, Cichorium, Helianthus, Lactuca, Bromus, Asparagus, Antirrhinum, Hererocallis, Nemesia, Pelargonium, Panicum, Pennisetum, Ranunculus, Senecio, Salpiglossis, Cucumis, Browaalia, Glycine, Lolium, Zea, Triticum, Sorghum, and Datura.
[0074]Means for regeneration vary from species to species of plants, but generally a suspension of transformed protoplasts containing copies of the heterologous gene is first provided. Callus tissue is formed and shoots may be induced from callus and subsequently rooted. Alternatively, embryo formation can be induced from the protoplast suspension. These embryos germinate as natural embryos to form plants. The culture media will generally contain various amino acids and hormones, such as auxin and cytokinins. It is also advantageous to add glutamic acid and proline to the medium, especially for such species as corn and alfalfa. Shoots and roots normally develop simultaneously. Efficient regeneration will depend on the medium, on the genotype, and on the history of the culture. If these three variables are controlled, then regeneration is fully reproducible and repeatable.
[0075]In some plant cell culture systems, the desired protein of the invention may be excreted or alternatively, the protein may be extracted from the whole plant. Where the desired protein of the invention is secreted into the medium, it may be collected. Alternatively, the embryos and embryoless-half seeds or other plant tissue may be mechanically disrupted to release any secreted protein between cells and tissues. The mixture may be suspended in a buffer solution to retrieve soluble proteins. Conventional protein isolation and purification methods will be then used to purify the recombinant protein. Parameters of time, temperature pH, oxygen, and volumes will be adjusted through routine methods to optimize expression and recovery of heterologous protein.
[0000]iv. Bacterial Systems
[0076]Bacterial expression techniques are known in the art. A bacterial promoter is any DNA sequence capable of binding bacterial RNA polymerase and initiating the downstream (3′) transcription of a coding sequence (e.g. structural gene) into mRNA. A promoter will have a transcription initiation region which is usually placed proximal to the 5′ end of the coding sequence. This transcription initiation region usually includes an RNA polymerase binding site and a transcription initiation site. A bacterial promoter may also have a second domain called an operator, that may overlap an adjacent RNA polymerase binding site at which RNA synthesis begins. The operator permits negative regulated (inducible) transcription, as a gene repressor protein may bind the operator and thereby inhibit transcription of a specific gene. Constitutive expression may occur in the absence of negative regulatory elements, such as the operator. In addition, positive regulation may be achieved by a gene activator protein binding sequence, which, if present is usually proximal (5′) to the RNA polymerase binding sequence. An example of a gene activator protein is the catabolite activator protein (CAP), which helps initiate transcription of the lac operon in Escherichia coli (E. coli) [Raibaud et al. (1984) Annu. Rev. Genet. 18:173]. Regulated expression may therefore be either positive or negative, thereby either enhancing or reducing transcription.
[0077]Sequences encoding metabolic pathway enzymes provide particularly useful promoter sequences. Examples include promoter sequences derived from sugar metabolizing enzymes, such as galactose, lactose (lac) [Chang et al. (1977) Nature 198:1056], and maltose. Additional examples include promoter sequences derived from biosynthetic enzymes such as tryptophan (trp) [Goeddel et al. (1980) Nuc. Acids Res. 8:4057; Yelverton et al. (1981) Nucl. Acids Res. 9:731; U.S. Pat. No. 4,738,921; EP-A-0036776 and EP-A-0121775]. The g-laotamase (bla) promoter system [Weissmann (1981) “The cloning of interferon and other mistakes.” In Interferon 3 (ed. I. Gresser)], bacteriophage lambda PL [Shimatake et al. (1981) Nature 292:128] and T5 [U.S. Pat. No. 4,689,406] promoter systems also provide useful promoter sequences.
[0078]In addition, synthetic promoters which do not occur in nature also function as bacterial promoters. For example, transcription activation sequences of one bacterial or bacteriophage promoter may be joined with the operon sequences of another bacterial or bacteriophage promoter, creating a synthetic hybrid promoter [U.S. Pat. No. 4,551,433]. For example, the tac promoter is a hybrid trp-lac promoter comprised of both trp promoter and lac operon sequences that is regulated by the lac repressor [Amann et al. (1983) Gene 25:167; de Boer et al. (1983) Proc. Natl. Acad. Sci. 80:21].
[0079]Furthermore, a bacterial promoter can include naturally occurring promoters of non-bacterial origin that have the ability to bind bacterial RNA polymerase and initiate transcription. A naturally occurring promoter of non-bacterial origin can also be coupled with a compatible RNA polymerase to produce high levels of expression of some genes in prokaryotes. The bacteriophage T7 RNA polymerase/promoter system is an example of a coupled promoter system [Studier et al. (1986) J. Mol. Biol. 189:113; Tabor et al. (1985) Proc Natl. Acad. Sci. 82:1074]. In addition, a hybrid promoter can also be comprised of a bacteriophage promoter and an E. coli operator region (EPO-A-0 267 851).
[0080]In addition to a functioning promoter sequence, an efficient ribosome binding site is also useful for the expression of foreign genes in prokaryotes. In E. coli, the ribosome binding site is called the Shine-Dalgarno (SD) sequence and includes an initiation codon (ATG) and a sequence 3-9 nucleotides in length located 3-11 nucleotides upstream of the initiation codon [Shine et al. (1975) Nature 254:34]. The SD sequence is thought to promote binding of mRNA to the ribosome by the pairing of bases between the SD sequence and the 3′ and of E. coli 16S rRNA [Steitz et al. (1979) “Genetic signals and nucleotide sequences in messenger RNA.” In Biological Regulation and Development: Gene Expression (ed. R. F. Goldberger)]. To express eukaryotic genes and prokaryotic genes with weak ribosome-binding site [Sambrook et al. (1989) “Expression of cloned genes in Escherichia coli.” In Molecular Cloning: A Laboratory Manual].
[0081]A DNA molecule may be expressed intracellularly. A promoter sequence may be directly linked with the DNA molecule, in which case the first amino acid at the N-terminus will always be a methionine, which is encoded by the ATG start codon. If desired, methionine at the N-terminus may be cleaved from the protein by in vitro incubation with cyanogen bromide or by either in vivo on in vitro incubation with a bacterial methionine N-terminal peptidase (EPO-A-0 219 237).
[0082]Fusion proteins provide an alternative to direct expression. Usually, a DNA sequence encoding the N-terminal portion of an endogenous bacterial protein, or other stable protein, is fused to the 5′ end of heterologous coding sequences. Upon expression, this construct will provide a fusion of the two amino acid sequences. For example, the bacteriophage lambda cell gene can be linked at the 5′ terminus of a foreign gene and expressed in bacteria. The resulting fusion protein preferably retains a site for a processing enzyme (factor Xa) to cleave the bacteriophage protein from the foreign gene [Nagai et al. (1984) Nature 309:810]. Fusion proteins can also be made with sequences from the lacZ [Jia et al. (1987) Gene 60:197], trpE [Allen et al. (1987) J. Biotechnol. 5:93; Makoff et al. (1989) J. Gen. Microbiol. 135: 11], and Chey [EP-A-0 324 647] genes. The DNA sequence at the junction of the two amino acid sequences may or may not encode a cleavable site. Another example is a ubiquitin fusion protein. Such a fusion protein is made with the ubiquitin region that preferably retains a site for a processing enzyme (e.g. ubiquitin specific processing-protease) to cleave the ubiquitin from the foreign protein. Through this method, native foreign protein can be isolated [Miller et al. (1989) Bio/Technology 7:698].
[0083]Alternatively, foreign proteins can also be secreted from the cell by creating chimeric DNA molecules that encode a fusion protein comprised of a signal peptide sequence fragment that provides for secretion of the foreign protein in bacteria [U.S. Pat. No. 4,336,336]. The signal sequence fragment usually encodes a signal peptide comprised of hydrophobic amino acids which direct the secretion of the protein from the cell. The protein is either secreted into the growth media (gram-positive bacteria) or into the periplasmic space, located between the inner and outer membrane of the cell (gram-negative bacteria). Preferably there are processing sites, which can be cleaved either in vivo or in vitro encoded between the signal peptide fragment and the foreign gene.
[0084]DNA encoding suitable signal sequences can be derived from genes for secreted bacterial proteins, such as the E. coli outer membrane protein gene (ompA) [Masui et al. (1983), in: Experimental Manipulation of Gene Expression; Ghrayeb et al. (1984) EMBO J. 3:2437] and the E. coli alkaline phosphatase signal sequence (phoA) [Oka et al. (1985) Proc. Natl. Acad. Sci. 82:7212]. As an additional example, the signal sequence of the alpha-amylase gene from various Bacillus strains can be used to secrete heterologous proteins from B. subtilis [Palva et al. (1982) Proc. Natl. Acad. Sci. USA 79:5582; EP-A-0 244 042].
[0085]Usually, transcription termination sequences recognized by bacteria are regulatory regions located 3′ to the translation stop codon, and thus together with the promoter flank the coding sequence. These sequences direct the transcription of an mRNA which can be translated into the polypeptide encoded by the DNA. Transcription termination sequences frequently include DNA sequences of about 50 nucleotides capable of forming stem loop structures that aid in terminating transcription. Examples include transcription termination sequences derived from genes with strong promoters, such as the trp gene in E. coli as well as other biosynthetic genes.
[0086]Usually, the above described components, comprising a promoter, signal sequence (if desired), coding sequence of interest, and transcription termination sequence, are put together into expression constructs. Expression constructs are often maintained in a replicon, such as an extrachromosomal element (e.g. plasmids) capable of stable maintenance in a host, such as bacteria. The replicon will have a replication system, thus allowing it to be maintained in a prokaryotic host either for expression or for cloning and amplification. In addition, a replicon may be either a high or low copy number plasmid. A high copy number plasmid will generally have a copy number ranging from about 5 to about 200, and usually about 10 to about 150. A host containing a high copy number plasmid will preferably contain at least about 10, and more preferably at least about 20 plasmids. Either a high or low copy number vector may be selected, depending upon the effect of the vector and the foreign protein on the host.
[0087]Alternatively, the expression constructs can be integrated into the bacterial genome with an integrating vector. Integrating vectors usually contain at least one sequence homologous to the bacterial chromosome that allows the vector to integrate. Integrations appear to result from recombinations between homologous DNA in the vector and the bacterial chromosome. For example, integrating vectors constructed with DNA from various Bacillus strains integrate into the Bacillus chromosome (EP-A-0 127 328). Integrating vectors may also be comprised of bacteriophage or transposon sequences.
[0088]Usually, extrachromosomal and integrating expression constructs may contain selectable markers to allow for the selection of bacterial strains that have been transformed. Selectable markers can be expressed in the bacterial host and may include genes which render bacteria resistant to drugs such as ampicillin, chloramphenicol, erythromycin, kanamycin (neomycin), and tetracycline [Davies et al. (1978) Annu. Rev. Microbiol. 32:469]. Selectable markers may also include biosynthetic genes, such as those in the histidine, tryptophan, and leucine biosynthetic pathways.
[0089]Alternatively, some of the above described components can be put together in transformation vectors. Transformation vectors are usually comprised of a selectable market that is either maintained in a replicon or developed into an integrating vector, as described above.
[0090]Expression and transformation vectors, either extra-chromosomal replicons or integrating vectors, have been developed for transformation into many bacteria. For example, expression vectors have been developed for, inter alia, the following bacteria: Bacillus subtilis [Palva et al. (1982) Proc. Natl. Acad. Sci. USA 79:5582; EP-A-0 036 259 and EP-A-0 063 953; WO 84/04541], Escherichia coli [Shimatake et al. (1981) Nature 292:128; Amann et al. (1985) Gene 40:183; Studier et al. (1986) J. Mol. Biol. 189:113; EP-A-0 036 776, EP-A-0 136 829 and EP-A-0 136 907], Streptococcus cremoris [Powell et al. (1988) Appl. Environ. Microbiol. 54:655]; Streptococcus lividans [Powell et al. (1988) Appl. Environ. Microbiol. 54:655], Streptomyces lividans [U.S. Pat. No. 4,745,056].
[0091]Methods of introducing exogenous DNA into bacterial hosts are well-known in the art, and usually include either the transformation of bacteria treated with CaCl2or other agents, such as divalent cations and DMSO. DNA can also be introduced into bacterial cells by electroporation. Transformation procedures usually vary with the bacterial species to be transformed. See e.g. [Masson et al. (1989) FEMS Microbiol. Lett. 60:273; Palva et al. (1982) Proc. Natl. Acad. Sci. USA 79:5582; EP-A-0 036 259 and EP-A-0 063 953; WO 84/04541, Bacillus], [Miller et al. (1988) Proc. Natl. Acad. Sci. 85:856; Wang et al. (1990) J. Bacteriol. 172:949, Campylobacter], [Cohen et al. (1973) Proc. Natl. Acad. Sci. 69:2110; Dower et al. (1988) Nucleic Acids Res. 16:6127; Kushner (1978) “An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In Genetic Engineering: Proceedings of theInternational Symposium on Genetic Engineering (eds. H. W. Boyer and S, Nicosia); Mandel et al. (1970) J. Mol. Biol. 53:159; Taketo (1988) Biochim. Biophys. Acta 949:318; Escherichia], [Chassy et al. (1987) FEMS Microbiol. Lett. 44:173 Lactobacillus]; [Fiedler et al. (1988) Anal. Biochem 170:38, Pseudomonas]; [Augustin et al. (1990) FEMS Microbiol. Lett. 66:203, Staphylococcus], [Barany et al. (1980) J. Bacteriol. 144:698; Harlander (1987) “Transformation of Streptococcus lactis by electroporation, in: Streptococcal Genetics (ed. J. Ferretti and R. Curtiss III); Perry et al. (1981) Infect. Immun. 32:1295; Powell et al. (1988) Appl. Environ. Microbiol. 54:655; Somkuti et al. (1987) Proc. 4th Evr. Cong. Biotechnology 1:412, Streptococcus].
[0000]v. Yeast Expression
[0092]Yeast expression systems are also known to one of ordinary skill in the art. A yeast promoter is any DNA sequence capable of binding yeast RNA polymerase and initiating the downstream (3′) transcription of a coding sequence (e.g. structural gene) into mRNA. A promoter will have a transcription initiation region which is usually placed proximal to the 5′ end of the coding sequence. This transcription initiation region usually includes an RNA polymerase binding site (the “TATA Box”) and a transcription initiation site. A yeast promoter may also have a second domain called an upstream activator sequence (UAS), which, if present, is usually distal to the structural gene. The UAS permits regulated (inducible) expression. Constitutive expression occurs in the absence of a UAS. Regulated expression may be either positive or negative, thereby either enhancing or reducing transcription.
[0093]Yeast is a fermenting organism with an active metabolic pathway, therefore sequences encoding enzymes in the metabolic pathway provide particularly useful promoter sequences. Examples include alcohol dehydrogenase (ADH) (EP-A-0 284 044), enolase, glucokinase, glucose-6-phosphate isomerase, glyceraldehyde-3-phosphate-dehydrogenase (GAP or GAPDH), hexokinase, phosphofructokinase, 3-phosphoglycerate mutase, and pyruvate kinase (PyK) (EPO-A-0 329 203). The yeast PHO5 gene, encoding acid phosphatase, also provides useful promoter sequences [Myanohara et al. (1983) Proc. Natl. Acad. Sci. USA 80:1].
[0094]In addition, synthetic promoters which do not occur in nature also function as yeast promoters. For example, UAS sequences of one yeast promoter may be joined with the transcription activation region of another yeast promoter, creating a synthetic hybrid promoter. Examples of such hybrid promoters include the ADH regulatory sequence linked to the GAP transcription activation region (U.S. Pat. Nos. 4,876,197 and 4,880,734). Other examples of hybrid promoters include promoters which consist of the regulatory sequences of either the ADH2, GAL4, GAL10, OR PHO5 genes, combined with the transcriptional activation region of a glycolytic enzyme gene such as GAP or PyK (EP-A-0 164 556). Furthermore, a yeast promoter can include naturally occurring promoters of non-yeast origin that have the ability to bind yeast RNA polymerase and initiate transcription. Examples of such promoters include, inter alia, [Cohen et al. (1980) Proc. Natl. Acad. Sci. USA 77:1078; Henikoff et al. (1981) Nature 283:835; Hollenberg et al. (1981) Curr. Topics Microbiol. Immunol. 96:119; Hollenberg et al. (1979) “The Expression of Bacterial Antibiotic Resistance Genes in the Yeast Saccharomyces cerevisiae,” in: Plasmids of Medical, Environmental and Commercial Importance (eds. K. N. Timmis and A. Puhler); Mercerau-Puigalon et al. (1980) Gene 11:163; Panthier et al. (1980) Curr. Genet. 2:109;].
[0095]A DNA molecule may be expressed intracellularly in yeast. A promoter sequence may be directly linked with the DNA molecule, in which case the first amino acid at the N-terminus of the recombinant protein will always be a methionine, which is encoded by the ATG start codon. If desired, methionine at the N-terminus may be cleaved from the protein by in vitro incubation with cyanogen bromide.
[0096]Fusion proteins provide an alternative for yeast expression systems, as well as in mammalian, baculovirus, and bacterial expression systems. Usually, a DNA sequence encoding the N-terminal portion of an endogenous yeast protein, or other stable protein, is fused to the 5′ end of heterologous coding sequences. Upon expression, this construct will provide a fusion of the two amino acid sequences. For example, the yeast or human superoxide dismutase (SOD) gene, can be linked at the 5′ terminus of a foreign gene and expressed in yeast. The DNA sequence at the junction of the two amino acid sequences may or may not encode a cleavable site. See eg. EP-A-0 196 056. Another example is a ubiquitin fusion protein. Such a fusion protein is made with the ubiquitin region that preferably retains a site for a processing enzyme (e.g. ubiquitin-specific processing protease) to cleave the ubiquitin from the foreign protein. Through this method, therefore, native foreign protein can be isolated (e.g. WO88/024066).
[0097]Alternatively, foreign proteins can also be secreted from the cell into the growth media by creating chimeric DNA molecules that encode a fusion protein comprised of a leader sequence fragment that provide for secretion in yeast of the foreign protein. Preferably, there are processing sites encoded between the leader fragment and the foreign gene that can be cleaved either in vivo or in vitro. The leader sequence fragment usually encodes a signal peptide comprised of hydrophobic amino acids which direct the secretion of the protein from the cell.
[0098]DNA encoding suitable signal sequences can be derived from genes for secreted yeast proteins, such as the yeast invertase gene (EP-A-0 012 873; JPO. 62,096,086) and the A-factor gene (U.S. Pat. No. 4,588,684). Alternatively, leaders of non-yeast origin, such as an interferon leader, exist that also provide for secretion in yeast (EP-A-0 060 057).
[0099]A preferred class of secretion leaders are those that employ a fragment of the yeast alpha-factor gene, which contains both a “pre” signal sequence, and a “pro” region. The types of alpha-factor fragments that can be employed include the full-length pre-pro alpha factor leader (about 83 amino acid residues) as well as truncated alpha-factor leaders (usually about 25 to about 50 amino acid residues) (U.S. Pat. Nos. 4,546,083 and 4,870,008; EP-A-0 324 274). Additional leaders employing an alpha-factor leader fragment that provides for secretion include hybrid alpha-factor leaders made with a presequence of a first yeast, but a pro-region from a second yeast alphafactor. (e.g. see WO 89/02463.)
[0100]Usually, transcription termination sequences recognized by yeast are regulatory regions located 3′ to the translation stop codon, and thus together with the promoter flank the coding sequence. These sequences direct the transcription of an mRNA which can be translated into the polypeptide encoded by the DNA. Examples of transcription terminator sequence and other yeast-recognized termination sequences, such as those coding for glycolytic enzymes.
[0101]Usually, the above described components, comprising a promoter, leader (if desired), coding sequence of interest, and transcription termination sequence, are put together into expression constructs. Expression constructs are often maintained in a replicon, such as an extrachromosomal element (e.g. plasmids) capable of stable maintenance in a host, such as yeast or bacteria. The replicon may have two replication systems, thus allowing it to be maintained, for example, in yeast for expression and in a prokaryotic host for cloning and amplification. Examples of such yeast-bacteria shuttle vectors include YEp24 [Botstein et al. (1979) Gene 8:17-24], pCl/1 [Brake et al. (1984) Proc. Natl. Acad. Sci. USA 81:4642-4646], and YRp17 [Stinchcomb et al. (1982) J. Mol. Biol. 158:157]. In addition, a replicon may be either a high or low copy number plasmid. A high copy number plasmid will generally have a copy number ranging from about 5 to about 200, and usually about 10 to about 150. A host containing a high copy number plasmid will preferably have at least about 10, and more preferably at least about 20. Enter a high or low copy number vector may be selected, depending upon the effect of the vector and the foreign protein on the host. See e.g. Brake et al., supra.
[0102]Alternatively, the expression constructs can be integrated into the yeast genome with an integrating vector. Integrating vectors usually contain at least one sequence homologous to a yeast chromosome that allows the vector to integrate, and preferably contain two homologous sequences flanking the expression construct. Integrations appear to result from recombinations between homologous DNA in the vector and the yeast chromosome [Orr-Weaver et al. (1983) Methods in Enzymol. 101:228-245]. An integrating vector may be directed to a specific locus in yeast by selecting the appropriate homologous sequence for inclusion in the vector. See Orr-Weaver et al., supra. One or more expression construct may integrate, possibly affecting levels of recombinant protein produced [Rine et al. (1983) Proc. Natl. Acad. Sci. USA 80:6750]. The chromosomal sequences included in the vector can occur either as a single segment in the vector, which results in the integration of the entire vector, or two segments homologous to adjacent segments in the chromosome and flanking the expression construct in the vector, which can result in the stable integration of only the expression construct.
[0103]Usually, extrachromosomal and integrating expression constructs may contain selectable markers to allow for the selection of yeast strains that have been transformed. Selectable markers may include biosynthetic genes that can be expressed in the yeast host, such as ADE2, HIS4, LEU2, TRP1, and ALG7, and the G418 resistance gene, which confer resistance in yeast cells to tunicamycin and G418, respectively. In addition, a suitable selectable marker may also provide yeast with the ability to grow in the presence of toxic compounds, such as metal. For example, the presence of CUP1 allows yeast to grow in the presence of copper ions [Butt et al. (1987) Microbiol, Rev. 51:351].
[0104]Alternatively, some of the above described components can be put together into transformation vectors. Transformation vectors are usually comprised of a selectable marker that is either maintained in a replicon or developed into an integrating vector, as described above.
[0105]Expression and transformation vectors, either extrachromosomal replicons or integrating vectors, have been developed for transformation into many yeasts. For example, expression vectors have been developed for, inter alia, the following yeasts: Candida albicans [Kurtz, et al. (1986) Mol. Cell. Biol. 6:142], Candida maltosa [Kunze, et al. (1985) J. Basic Microbiol. 25:141]. Hansenula polymorpha [Gleeson, et al. (1986) J. Gen. Microbiol. 132:3459; Roggenkamp et al. (1986) Mol. Gen. Genet. 202:302], Kluyveromyces fragilis [Das, et al. (1984) J. Bacteriol. 158:1165], Kluyveromyces lactis [De Louvencourt et al. (1983) J. Bacteriol. 154:737; Van den Berg et al. (1990) Bio/Technology 8:135], Pichia guillerimondii [Kunze et al. (1985) J. Basic Microbiol. 25:141], Pichia pastoris [Cregg, et al. (1985) Mol. Cell. Biol. 5:3376; U.S. Pat. Nos. 4,837,148 and 4,929,555], Saccharomyces cerevisiae [Hinnen et al. (1978) Proc. Natl. Acad. Sci. USA 75:1929; Ito et al. (1983) J. Bacteriol. 153:163], Schizosaccharomyces pombe [Beach and Nurse (1981) Nature 300:706], and Yarrowia lipolytica [Davidow, et al. (1985) Curr. Genet. 10:380471 Gaillardin, et al. (1985) Curr. Genet. 10:49].
[0106]Methods of introducing exogenous DNA into yeast hosts are well-known in the art, and usually include either the transformation of spheroplasts or of intact yeast cells treated with alkali cations. Transformation procedures usually vary with the yeast species to be transformed. See eg. [Kurtz et al. (1986) Mol. Cell. Biol. 6:142; Kunze et al. (1985) J. Basic Microbiol. 25:141; Candida]; [Gleeson et al. (1986) J. Gen. Microbiol. 132:3459; Roggenkamp et al. (1986) Mol. Gen. Genet. 202:302; Hansenula]; [Das et al. (1984) J. Bacteriol. 158:1165; De Louvencourt et al. (1983) J. Bacteriol. 154:1165; Van den Berg et al. (1990) Bio/Technology 8:135; Kluyveromyces]; [Cregg et al. (1985) Mol. Cell. Biol. 5:3376; Kunze et al. (1985) J. Basic Microbiol. 25:141; U.S. Pat. Nos. 4,837,148 and 4,929,555; Pichia]; [Hinnen et al. (1978) Proc. Natl. Acad. Sci. USA 75; 1929; Ito et al. (1983) J. Bacteriol. 153:163 Saccharomyces]; [Beach and Nurse (1981) Nature 300:706; Schizosaccharomyces]; [Davidow et al. (1985) Curr. Genet. 10:39; Gaillardin et al. (1985) Curr. Genet. 10:49; Yarrowia].
Antibodies
[0107]As used herein, the term “antibody” refers to a polypeptide or group of polypeptides composed of at least one antibody combining site. An “antibody combining site” is the three-dimensional binding space with an internal surface shape and charge distribution complementary to the features of an epitope of an antigen, which allows a binding of the antibody with the antigen. “Antibody” includes, for example, vertebrate antibodies, hybrid antibodies, chimeric antibodies, humanised antibodies, altered antibodies, univalent antibodies, Fab proteins, and single domain antibodies.
[0108]Antibodies against the proteins of the invention are useful for affinity chromatography, immunoassays, and distinguishing/identifying Neisserial proteins.
[0109]Antibodies to the proteins of the invention, both polyclonal and monoclonal, may be prepared by conventional methods. In general, the protein is first used to immunize a suitable animal, preferably a mouse, rat, rabbit or goat. Rabbits and goats are preferred for the preparation of polyclonal sera due to the volume of serum obtainable, and the availability of labeled anti-rabbit and anti-goat antibodies. Immunization is generally performed by mixing or emulsifying the protein in saline, preferably in an adjuvant such as Freund's complete adjuvant, and injecting the mixture or emulsion parenterally (generally subcutaneously or intramuscularly). A dose of 50-200 μg/injection is typically sufficient. Immunization is generally boosted 2-6 weeks later with one or more injections of the protein in saline, preferably using Freund's incomplete adjuvant. One may alternatively generate antibodies by in vitro immunization using methods known in the art, which for the purposes of this invention is considered equivalent to in vivo immunization. Polyclonal antisera is obtained by bleeding the immunized animal into a glass or plastic container, incubating the blood at 25° C. for one hour, followed by incubating at 4° C. for 2-18 hours. The serum is recovered by centrifugation (e.g. 1,000 g for 10 minutes). About 20-50 ml per bleed may be obtained from rabbits.
[0110]Monoclonal antibodies are prepared using the standard method of Kohler & Milstein [Nature (1975) 256:495-96], or a modification thereof. Typically, a mouse or rat is immunized as described above. However, rather than bleeding the animal to extract serum, the spleen (and optionally several large lymph nodes) is removed and dissociated into single cells. If desired, the spleen cells may be screened (after removal of nonspecifically adherent cells) by applying a cell suspension to a plate or well coated with the protein antigen. B-cells expressing membrane-bound immunoglobulin specific for the antigen bind to the plate, and are not rinsed away with the rest of the suspension. Resulting B-cells, or all dissociated spleen cells, are then induced to fuse with myeloma cells to form hybridomas, and are cultured in a selective medium (e.g. hypoxanthine, aminopterin, thymidine medium, “HAT”). The resulting hybridomas are plated by limiting dilution, and are assayed for the production of antibodies which bind specifically to the immunizing antigen (and which do not bind to unrelated antigens). The selected MAb-secreting hybridomas are then cultured either in vitro (e.g. in tissue culture bottles or hollow fiber reactors), or in vivo (as ascites in mice).
[0111]If desired, the antibodies (whether polyclonal or monoclonal) may be labeled using conventional techniques. Suitable labels include fluorophores, chromophores, radioactive atoms (particularly32P and125I), electron-dense reagents, enzymes, and ligands having specific binding partners. Enzymes are typically detected by their activity. For example, horseradish peroxidase is usually detected by its ability to convert 3,3′,5,5′-tetramethylbenzidine (TMB) to a blue pigment, quantifiable with a spectrophotometer. “Specific binding partner” refers to a protein capable of binding a ligand molecule with high specificity, as for example in the case of an antigen and a monoclonal antibody specific therefor. Other specific binding partners include biotin and avidin or streptavidin, IgG and protein A, and the numerous receptor-ligand couples known in the art. It should be understood that the above description is not meant to categorize the various labels into distinct classes, as the same label may serve in several different modes. For example,125I may serve as a radioactive label or as an electron-dense reagent. HRP may serve as enzyme or as antigen for a MAb. Further, one may combine various labels for desired effect. For example, MAbs and avidin also require labels in the practice of this invention: thus, one might label a MAb with biotin, and detect its presence with avidin labeled with125I, or with an anti-biotin MAb labeled with HRP. Other permutations and possibilities will be readily apparent to those of ordinary skill in the art, and are considered as equivalents within the scope of the instant invention.
Pharmaceutical Compositions
[0112]Pharmaceutical compositions can comprise either polypeptides, antibodies, or nucleic acid of the invention. The pharmaceutical compositions will comprise a therapeutically effective amount of either polypeptides, antibodies, or polynucleotides of the claimed invention.
[0113]The term “therapeutically effective amount” as used herein refers to an amount of a therapeutic agent to treat, ameliorate, or prevent a desired disease or condition, or to exhibit a detectable therapeutic or preventative effect. The effect can be detected by, for example, chemical markers or antigen levels. Therapeutic effects also include reduction in physical symptoms, such as decreased body temperature. The precise effective amount for a subject will depend upon the subject's size and health, the nature and extent of the condition, and the therapeutics or combination of therapeutics selected for administration. Thus, it is not useful to specify an exact effective amount in advance. However, the effective amount for a given situation can be determined by routine experimentation and is within the judgement of the clinician.
[0114]For purposes of the present invention, an effective dose will be from about 0.01 mg/kg to 50 mg/kg or 0.05 mg/kg to about 10 mg/kg of the DNA constructs in the individual to which it is administered.
[0115]A pharmaceutical composition can also contain a pharmaceutically acceptable carrier. The term “pharmaceutically acceptable carrier” refers to a carrier for administration of a therapeutic agent, such as antibodies or a polypeptide, genes, and other therapeutic agents. The term refers to any pharmaceutical carrier that does not itself induce the production of antibodies harmful to the individual receiving the composition, and which may be administered without undue toxicity. Suitable carriers may be large, slowly metabolized macromolecules such as proteins, polysaccharides, polylactic acids, polyglycolic acids, polymeric amino acids, amino acid copolymers, and inactive virus particles. Such carriers are well known to those of ordinary skill in the art.
[0116]Pharmaceutically acceptable salts can be used therein, for example, mineral acid salts such as hydrochlorides, hydrobromides, phosphates, sulfates, and the like; and the salts of organic acids such as acetates, propionates, malonates, benzoates, and the like. A thorough discussion of pharmaceutically acceptable excipients is available in Remington's Pharmaceutical Sciences (Mack Pub. Co., N.J. 1991).
[0117]Pharmaceutically acceptable carriers in therapeutic compositions may contain liquids such as water, saline, glycerol and ethanol. Additionally, auxiliary substances, such as wetting or emulsifying agents, pH buffering substances, and the like, may be present in such vehicles. Typically, the therapeutic compositions are prepared as injectables, either as liquid solutions or suspensions; solid forms suitable for solution in, or suspension in, liquid vehicles prior to injection may also be prepared. Liposomes are included within the definition of a pharmaceutically acceptable carrier.
Delivery Methods
[0118]Once formulated, the compositions of the invention can be administered directly to the subject. The subjects to be treated can be animals; in particular, human subjects can be treated.
[0119]Direct delivery of the compositions will generally be accomplished by injection, either subcutaneously, intraperitoneally, intravenously or intramuscularly or delivered to the interstitial space of a tissue. The compositions can also be administered into a lesion. Other modes of administration include oral and pulmonary administration, suppositories, and transdermal or transcutaneous applications (e.g. see WO98/20734), needles, and gene guns or hyposprays. Dosage treatment may be a single dose schedule or a multiple dose schedule.
Vaccines
[0120]Vaccines according to the invention may either be prophylactic (i.e. to prevent infection) or therapeutic (i.e. to treat disease after infection).
[0121]Such vaccines comprise immunising antigen(s), immunogen(s), polypeptide(s), protein(s) or nucleic acid, usually in combination with “pharmaceutically acceptable carriers,” which include any carrier that does not itself induce the production of antibodies harmful to the individual receiving the composition. Suitable carriers are typically large, slowly metabolized macromolecules such as proteins, polysaccharides, polylactic acids, polyglycolic acids, polymeric amino acids, amino acid copolymers, lipid aggregates (such as oil droplets or liposomes), and inactive virus particles. Such carriers are well known to those of ordinary skill in the art. Additionally, these carriers may function as immunostimulating agents (“adjuvants”). Furthermore, the antigen or immunogen may be conjugated to a bacterial toxoid, such as a toxoid from diphtheria, tetanus, cholera, H. pylori, etc. pathogens.
[0122]Preferred adjuvants to enhance effectiveness of the composition include, but are not limited to: (1) aluminum salts (alum), such as aluminum hydroxide, aluminum phosphate, aluminum sulfate, etc; (2) oil-in-water emulsion formulations (with or without other specific immunostimulating agents such as muramyl peptides (see below) or bacterial cell wall components), such as for example (a) MF59™ (WO 90/14837; Chapter 10 in Vaccine design: the subunit and adjuvant approach, eds. Powell & Newman, Plenum Press 1995), containing 5% Squalene, 0.5% Tween 80, and 0.5% Span 85 (optionally containing various amounts of MTP-PE (see below), although not required) formulated into submicron particles using a microfluidizer such as Model 110Y microfluidizer (Microfluidics, Newton, Mass.), (b) SAF, containing 10% Squalane, 0.4% Tween 80, 5% pluronic-blocked polymer L 121, and thr-MDP (see below) either microfluidized into a submicron emulsion or vortexed to generate a larger particle size emulsion, and (c) Ribi™ adjuvant system (RAS), (Ribi Immunochem, Hamilton, Mont.) containing 2% Squalene, 0.2% Tween 80, and one or more bacterial cell wall components from the group consisting of monophosphorylipid A (MPL), trehalose dimycolate (TDM), and cell wall skeleton (CWS), preferably MPL+CWS (Detox™); (3) saponin adjuvants, such as Stimulon™ (Cambridge Bioscience, Worcester, Mass.) may be used or particles generated therefrom such as ISCOMs (immunostimulating complexes); (4) Complete Freund's Adjuvant (CFA) and Incomplete Freund's Adjuvant (IFA); (5) cytokines, such as interleukins (e.g. IL-1, IL-2, IL-4, IL-5, IL-6, IL-7, IL-12, etc.), interferons (e.g. gamma interferon), macrophage colony stimulating factor (M-CSF), tumor necrosis factor (TNF), etc; and (6) other substances that act as immunostimulating agents to enhance the effectiveness of the composition. Alum and MF59™ are preferred.
[0123]As mentioned above, muramyl peptides include, but are not limited to, N-acetyl-muramyl-L-threonyl-D-isoglutamine (thr-MDP), N-acetyl-normuramyl-L-alanyl-D-isoglutamine (nor-MDP), N-acetylmuramyl-L-alanyl-D-isoglutaminyl-L-alanine-2-(1′-2′-dipalmitoyl-sn-glycero-3-hydroxyphosphoryloxy)-ethylamine (MTP-PE), etc.
[0124]The immunogenic compositions (e.g. the immunising antigen/immunogen/polypeptide/protein/nucleic acid, pharmaceutically acceptable carrier, and adjuvant) typically will contain diluents, such as water, saline, glycerol, ethanol, etc. Additionally, auxiliary substances, such as wetting or emulsifying agents, pH buffering substances, and the like, may be present in such vehicles.
[0125]Typically, the immunogenic compositions are prepared as injectables, either as liquid solutions or suspensions; solid forms suitable for solution in, or suspension in, liquid vehicles prior to injection may also be prepared. The preparation also may be emulsified or encapsulated in liposomes for enhanced adjuvant effect, as discussed above under pharmaceutically acceptable carriers.
[0126]Immunogenic compositions used as vaccines comprise an immunologically effective amount of the antigenic or immunogenic polypeptides, as well as any other of the above-mentioned components, as needed. By “immunologically effective amount”, it is meant that the administration of that amount to an individual, either in a single dose or as part of a series, is effective for treatment or prevention. This amount varies depending upon the health and physical condition of the individual to be treated, the taxonomic group of individual to be treated (e.g. nonhuman primate, primate, etc.), the capacity of the individual's immune system to synthesize antibodies, the degree of protection desired, the formulation of the vaccine, the treating doctor's assessment of the medical situation, and other relevant factors. It is expected that the amount will fall in a relatively broad range that can be determined through routine trials.
[0127]The immunogenic compositions are conventionally administered parenterally, e.g. by injection, either subcutaneously, intramuscularly, or transdermally/transcutaneously (e.g. WO98/20734). Additional formulations suitable for other modes of administration include oral and pulmonary formulations, suppositories, and transdermal applications. Dosage treatment may be a single dose schedule or a multiple dose schedule. The vaccine may be administered in conjunction with other immunoregulatory agents.
[0128]As an alternative to protein-based vaccines, DNA vaccination may be employed [e.g. Robinson & Torres (1997) Seminars in Immunology 9:271-283; Donnelly et al. (1997) Annu Rev Immunol 15:617-648; see later herein].
Gene Delivery Vehicles
[0129]Gene therapy vehicles for delivery of constructs including a coding sequence of a therapeutic of the invention, to be delivered to the mammal for expression in the mammal, can be administered either locally or systemically. These constructs can utilize viral or non-viral vector approaches in in vivo or ex vivo modality. Expression of such coding sequence can be induced using endogenous mammalian or heterologous promoters. Expression of the coding sequence in vivo can be either constitutive or regulated.
[0130]The invention includes gene delivery vehicles capable of expressing the contemplated nucleic acid sequences. The gene delivery vehicle is preferably a viral vector and, more preferably, a retroviral, adenoviral, adeno-associated viral (AAV), herpes viral, or alphavirus vector. The viral vector can also be an astrovirus, coronavirus, orthomyxovirus, papovavirus, paramyxovirus, parvovirus, picornavirus, poxvirus, or togavirus viral vector. See generally, Jolly (1994) Cancer Gene Therapy 1:51-64; Kimura (1994) Human Gene Therapy 5:845-852; Connelly (1995) Human Gene Therapy 6:185-193; and Kaplitt (1994) Nature Genetics 6:148-153.
[0131]Retroviral vectors are well known in the art and we contemplate that any retroviral gene therapy vector is employable in the invention, including B, C and D type retroviruses, xenotropic retroviruses (for example, NZB-X1, NZB-X2 and NZB9-1 (see O'Neill (1985) J. Virol. 53:160) polytropic retroviruses e.g. MCF and MCF-M LV (see Kelly (1983) J. Virol. 45:291), spumaviruses and lentiviruses. See RNA Tumor Viruses, Second Edition, Cold Spring Harbor Laboratory, 1985.
[0132]Portions of the retroviral gene therapy vector may be derived from different retroviruses. For example, retrovector LTRs may be derived from a Murine Sarcoma Virus, a tRNA binding site from a Rous Sarcoma Virus, a packaging signal from a Murine Leukemia Virus, and an origin of second strand synthesis from an Avian Leukosis Virus.
[0133]These recombinant retroviral vectors may be used to generate transduction competent retroviral vector particles by introducing them into appropriate packaging cell lines (see U.S. Pat. No. 5,591,624). Retrovirus vectors can be constructed for site-specific integration into host cell DNA by incorporation of a chimeric integrase enzyme into the retroviral particle (see WO96/37626). It is preferable that the recombinant viral vector is a replication defective recombinant virus.
[0134]Packaging cell lines suitable for use with the above-described retrovirus vectors are well known in the art, are readily prepared (see WO95/30763 and WO92/05266), and can be used to create producer cell lines (also termed vector cell lines or “VCLs”) for the production of recombinant vector particles. Preferably, the packaging cell lines are made from human parent cells (e.g. HT1080 cells) or mink parent cell lines, which eliminates inactivation in human serum.
[0135]Preferred retroviruses for the construction of retroviral gene therapy vectors include Avian Leukosis Virus, Bovine Leukemia, Virus, Murine Leukemia Virus, Mink-Cell Focus-Inducing Virus, Murine Sarcoma Virus, Reticuloendotheliosis Virus and Rous Sarcoma Virus. Particularly preferred Murine Leukemia Viruses include 4070A and 1504A (Hartley and Rowe (1976) J Virol 19:19-25), Abelson (ATCC No. VR-999), Friend (ATCC No. VR-245), Graffi, Gross (ATCC Nol VR-590), Kirsten, Harvey Sarcoma Virus and Rauscher (ATCC No. VR-998) and Moloney Murine Leukemia Virus (ATCC No. VR-190). Such retroviruses may be obtained from depositories or collections such as the American Type Culture Collection (“ATCC”) in Rockville, Md. or isolated from known sources using commonly available techniques.
[0136]Exemplary known retroviral gene therapy vectors employable in this invention include those described in patent applications GB2200651, EP0415731, EP0345242, EP0334301, WO89/02468; WO89/05349, WO89/09271, WO90/02806, WO90/07936, WO94/03622, WO93/25698, WO93/25234, WO93/11230, WO93/10218, WO91/02805, WO91/02825, WO95/07994, U.S. Pat. No. 5,219,740, U.S. Pat. No. 4,405,712, U.S. Pat. No. 4,861,719, U.S. Pat. No. 4,980,289, U.S. Pat. No. 4,777,127, U.S. Pat. No. 5,591,624. See also Vile (1993) Cancer Res 53:3860-3864; Vile (1993) Cancer Res 53:962-967; Ram (1993) Cancer Res 53 (1993) 83-88; Takamiya (1992) J Neurosci Res 33:493-503; Baba (1993) J Neurosurg 79:729-735; Mann (1983) Cell 33:153; Cane (1984) Proc Natl Acad Sci 81:6349; and Miller (1990) Human Gene Therapy 1.
[0137]Human adenoviral gene therapy vectors are also known in the art and employable in this invention. See, for example, Berkner (1988) Biotechniques 6:616 and Rosenfeld (1991) Science 252:431, and WO93/07283, WO93/06223, and WO93/07282. Exemplary known adenoviral gene therapy vectors employable in this invention include those described in the above referenced documents and in WO94/12649, WO93/03769, WO93/19191, WO94/28938, WO95/11984, WO95/00655, WO95/27071, WO95/29993, WO95/34671, WO96/05320, WO94/08026, WO94/11506, WO93/06223, WO94/24299, WO95/14102, WO95/24297, WO95/02697, WO94/28152, WO94/24299, WO95/09241, WO95/25807, WO95/05835, WO94/18922 and WO95/09654. Alternatively, administration of DNA linked to killed adenovirus as described in Curiel (1992) Hum. Gene Ther. 3:147-154 may be employed. The gene delivery vehicles of the invention also include adenovirus associated virus (AAV) vectors. Leading and preferred examples of such vectors for use in this invention are the AAV-2 based vectors disclosed in Srivastava, WO93/09239. Most preferred AAV vectors comprise the two AAV inverted terminal repeats in which the native D-sequences are modified by substitution of nucleotides, such that at least 5 native nucleotides and up to 18 native nucleotides, preferably at least 10 native nucleotides up to 18 native nucleotides, most preferably 10 native nucleotides are retained and the remaining nucleotides of the D-sequence are deleted or replaced with non-native nucleotides. The native D-sequences of the AAV inverted terminal repeats are sequences of 20 consecutive nucleotides in each AAV inverted terminal repeat (i.e. there is one sequence at each end) which are not involved in HP formation. The non-native replacement nucleotide may be any nucleotide other than the nucleotide found in the native D-sequence in the same position. Other employable exemplary AAV vectors are pWP-19, pWN-1, both of which are disclosed in Nahreini (1993) Gene 124:257-262. Another example of such an AAV vector is psub201 (see Samulski (1987) J. Virol. 61:3096). Another exemplary AAV vector is the Double-D ITR vector. Construction of the Double-D ITR vector is disclosed in U.S. Pat. No. 5,478,745. Still other vectors are those disclosed in Carter U.S. Pat. No. 4,797,368 and Muzyczka U.S. Pat. No. 5,139,941, Chartejee U.S. Pat. No. 5,474,935, and Kotin WO94/288157. Yet a further example of an AAV vector employable in this invention is SSV9AFABTKneo, which contains the AFP enhancer and albumin promoter and directs expression predominantly in the liver. Its structure and construction are disclosed in Su (1996) Human Gene Therapy 7:463-470. Additional AAV gene therapy vectors are described in U.S. Pat. No. 5,354,678, U.S. Pat. No. 5,173,414, U.S. Pat. No. 5,139,941, and U.S. Pat. No. 5,252,479.
[0138]The gene therapy vectors of the invention also include herpes vectors. Leading and preferred examples are herpes simplex virus vectors containing a sequence encoding a thymidine kinase polypeptide such as those disclosed in U.S. Pat. No. 5,288,641 and EP0176170 (Roizman). Additional exemplary herpes simplex virus vectors include HFEM/ICP6-LacZ disclosed in WO95/04139 (Wistar Institute), pHSVlac described in Geller (1988) Science 241:1667-1669 and in WO90/09441 and WO92/07945, HSV Us3::pgC-lacZ described in Fink (1992) Human Gene Therapy 3:11-19 and HSV 7134, 2 RH 105 and GAL4 described in EP 0453242 (Breakefield), and those deposited with the ATCC as accession numbers ATCC VR-977 and ATCC VR-260.
[0139]Also contemplated are alpha virus gene therapy vectors that can be employed in this invention. Preferred alpha virus vectors are Sindbis viruses vectors. Togaviruses, Semliki Forest virus (ATCC VR-67; ATCC VR-1247), Middleberg virus (ATCC VR-370), Ross River virus (ATCC VR-373; ATCC VR-1246), Venezuelan equine encephalitis virus (ATCC VR923; ATCC VR-1250; ATCC VR-1249; ATCC VR-532), and those described in U.S. Pat. Nos. 5,091,309, 5, 217,879, and WO92/10578. More particularly, those alpha virus vectors described in U.S. Ser. No. 08/405,627, filed Mar. 15, 1995, WO94/21792, WO92/10578, WO95/07994, U.S. Pat. No. 5,091,309 and U.S. Pat. No. 5,217,879 are employable. Such alpha viruses may be obtained from depositories or collections such as the ATCC in Rockville, Md. or isolated from known sources using commonly available techniques. Preferably, alphavirus vectors with reduced cytotoxicity are used (see U.S. Ser. No. 08/679,640).
[0140]DNA vector systems such as eukarytic layered expression systems are also useful for expressing the nucleic acids of the invention. See WO95/07994 for a detailed description of eukaryotic layered expression systems. Preferably, the eukaryotic layered expression systems of the invention are derived from alphavirus vectors and most preferably from Sindbis viral vectors.
[0141]Other viral vectors suitable for use in the present invention include those derived from poliovirus, for example ATCC VR-58 and those described in Evans, Nature 339 (1989) 385 and Sabin (1973) J. Biol. Standardization 1:115; rhinovirus, for example ATCC VR-1110 and those described in Arnold (1990) J Cell Biochem L401; pox viruses such as canary pox virus or vaccinia virus, for example ATCC VR-111 and ATCC VR-2010 and those described in Fisher-Hoch (1989) Proc Natl Acad Sci 86:317; Flexner (1989) Ann NY Acad Sci 569:86, Flexner (1990) Vaccine 8:17; in U.S. Pat. No. 4,603,112 and U.S. Pat. No. 4,769,330 and WO89/01973; SV40 virus, for example ATCC VR-305 and those described in Mulligan (1979) Nature 277:108 and Madzak (1992) J Gen Virol 73:1533; influenza virus, for example ATCC VR-797 and recombinant influenza viruses made employing reverse genetics techniques as described in U.S. Pat. No. 5,166,057 and in Enami (1990) Proc Natl Acad Sci 87:3802-3805; Enami & Palese (1991) J Virol 65:2711-2713 and Luytjes (1989) Cell 59:110, (see also McMichael (1983) NEJ Med 309:13, and Yap (1978) Nature 273:238 and Nature (1979) 277:108); human immunodeficiency virus as described in EP-0386882 and in Buchschacher (1992) J. Virol. 66:2731; measles virus, for example ATCC VR-67 and VR-1247 and those described in EP-0440219; Aura virus, for example ATCC VR-368; Bebaru virus, for example ATCC VR-600 and ATCC VR-1240; Cabassou virus, for example ATCC VR-922; Chikungunya virus, for example ATCC VR-64 and ATCC VR-1241; Fort Morgan Virus, for example ATCC VR-924; Getah virus, for example ATCC VR-369 and ATCC VR-1243; Kyzylagach virus, for example ATCC VR-927; Mayaro virus, for example ATCC VR-66; Mucambo virus, for example ATCC VR-580 and ATCC VR-1244; Ndumu virus, for example ATCC VR-371; Pixunavirus, for example ATCC VR-372 and ATCC VR-1245; Tonate virus, for example ATCC VR-925; Triniti virus, for example ATCC VR-469; Una virus, for example ATCC VR-374; Whataroa virus, for example ATCC VR-926; Y-62-33 virus, for example ATCC VR-375; O'Nyong virus, Eastern encephalitis virus, for example ATCC VR-65 and ATCC VR-1242; Western encephalitis virus, for example ATCC VR-70, ATCC VR-1251, ATCC VR-622 and ATCC VR-1252; and coronavirus, for example ATCC VR-740 and those described in Hamre (1966) Proc Soc Exp Biol Med 121:190.
[0142]Delivery of the compositions of this invention into cells is not limited to the above mentioned viral vectors. Other delivery methods and media may be employed such as, for example, nucleic acid expression vectors, polycationic condensed DNA linked or unlinked to killed adenovirus alone, for example see U.S. Ser. No. 08/366,787, filed Dec. 30, 1994 and Curiel (1992) Hum Gene Ther 3:147-154 ligand linked DNA, for example see Wu (1989) J Biol Chem 264:16985-16987, eucaryotic cell delivery vehicles cells, for example see U.S. Ser. No. 08/240,030, filed May 9, 1994, and U.S. Ser. No. 08/404,796, deposition of photopolymerized hydrogel materials, hand-held gene transfer particle gun, as described in U.S. Pat. No. 5,149,655, ionizing radiation as described in U.S. Pat. No. 5,206,152 and in WO92/11033, nucleic charge neutralization or fusion with cell membranes. Additional approaches are described in Philip (1994) Mol Cell Biol 14:2411-2418 and in Woffendin (1994) Proc Natl Acad Sci 91:1581-1585.
[0143]Particle mediated gene transfer may be employed, for example see U.S. Ser. No. 60/023,867. Briefly, the sequence can be inserted into conventional vectors that contain conventional control sequences for high level expression, and then incubated with synthetic gene transfer molecules such as polymeric DNA-binding cations like polylysine, protamine, and albumin, linked to cell targeting ligands such as asialoorosomucoid, as described in Wu & Wu (1987) J. Biol. Chem. 262:4429-4432, insulin as described in Hucked (1990) Biochem Pharmacol 40:253-263, galactose as described in Plank (1992) Bioconjugate Chem 3:533-539, lactose or transferrin.
[0144]Naked DNA may also be employed. Exemplary naked DNA introduction methods are described in WO 90/11092 and U.S. Pat. No. 5,580,859. Uptake efficiency may be improved using biodegradable latex beads. DNA coated latex beads are efficiently transported into cells after endocytosis initiation by the beads. The method may be improved further by treatment of the beads to increase hydrophobicity and thereby facilitate disruption of the endosome and release of the DNA into the cytoplasm.
[0145]Liposomes that can act as gene delivery vehicles are described in U.S. Pat. No. 5,422,120, WO95/13796, WO94/23697, WO91/14445 and EP-524,968. As described in U.S. Ser. No. 60/023,867, on non-viral delivery, the nucleic acid sequences encoding a polypeptide can be inserted into conventional vectors that contain conventional control sequences for high level expression, and then be incubated with synthetic gene transfer molecules such as polymeric DNA-binding cations like polylysine, protamine, and albumin, linked to cell targeting ligands such as asialoorosomucoid, insulin, galactose, lactose, or transferrin. Other delivery systems include the use of liposomes to encapsulate DNA comprising the gene under the control of a variety of tissue-specific or ubiquitously-active promoters. Further non-viral delivery suitable for use includes mechanical delivery systems such as the approach described in Woffendin et al (1994) Proc. Natl. Acad. Sci. USA 91(24):11581-11585. Moreover, the coding sequence and the product of expression of such can be delivered through deposition of photopolymerized hydrogel materials. Other conventional methods for gene delivery that can be used for delivery of the coding sequence include, for example, use of hand-held gene transfer particle gun, as described in U.S. Pat. No. 5,149,655; use of ionizing radiation for activating transferred gene, as described in U.S. Pat. No. 5,206,152 and WO92/11033
[0146]Exemplary liposome and polycationic gene delivery vehicles are those described in U.S. Pat. Nos. 5,422,120 and 4,762,915; in WO 95/13796; WO94/23697; and WO91/14445; in EP-0524968; and in Stryer, Biochemistry, pages 236-240 (1975) W.H. Freeman, San Francisco; Szoka (1980) Biochem Biophys Acta 600:1; Bayer (1979) Biochen Biophys Acta 550:464; Rivnay (1987) Meth Enzymol 149:119; Wang (1987) Proc Natl Acad Sci 84:7851; Plant (1989) Anal Biochem 176:420.
[0147]A polynucleotide composition can comprises therapeutically effective amount of a gene therapy vehicle, as the term is defined above. For purposes of the present invention, an effective dose will be from about 0.01 mg/kg to 50 mg/kg or 0.05 mg/kg to about 10 mg/kg of the DNA constructs in the individual to which it is administered.
Delivery Methods
[0148]Once formulated, the polynucleotide compositions of the invention can be administered (1) directly to the subject; (2) delivered ex vivo, to cells derived from the subject; or (3) in vitro for expression of recombinant proteins. The subjects to be treated can be mammals or birds. Also, human subjects can be treated.
[0149]Direct delivery of the compositions will generally be accomplished by injection, either subcutaneously, intraperitoneally, intravenously or intramuscularly or delivered to the interstitial space of a tissue. The compositions can also be administered into a lesion. Other modes of administration include oral and pulmonary administration, suppositories, and transdermal or transcutaneous applications (e.g. see WO98/20734), needles, and gene guns or hyposprays. Dosage treatment may be a single dose schedule or a multiple dose schedule.
[0150]Methods for the ex vivo delivery and reimplantation of transformed cells into a subject are known in the art and described in e.g. WO93/14778. Examples of cells useful in ex vivo applications include, for example, stem cells, particularly hematopoetic, lymph cells, macrophages, dendritic cells, or tumor cells.
[0151]Generally, delivery of nucleic acids for both ex vivo and in vitro applications can be accomplished by the following procedures, for example, dextran-mediated transfection, calcium phosphate precipitation, polybrene mediated transfection, protoplast fusion, electroporation, encapsulation of the polynucleotide(s) in liposomes, and direct microinjection of the DNA into nuclei, all well known in the art.
Polynucleotide and Polypeptide Pharmaceutical Compositions
[0152]In addition to the pharmaceutically acceptable carriers and salts described above, the following additional agents can be used with polynucleotide and/or polypeptide compositions.
A. Polypeptides
[0153]One example are polypeptides which include, without limitation: asioloorosomucoid (ASOR); transferrin; asialoglycoproteins; antibodies; antibody fragments; ferritin; interleukins; interferons, granulocyte, macrophage colony stimulating factor (GM-CSF), granulocyte colony stimulating factor (G-CSF), macrophage colony stimulating factor (M-CSF), stem cell factor and erythropoietin. Viral antigens, such as envelope proteins, can also be used. Also, proteins from other invasive organisms, such as the 17 amino acid peptide from the circumsporozoite protein of plasmodium falciparum known as RII.
B. Hormones, Vitamins, etc.
[0154]Other groups that can be included are, for example: hormones, steroids, androgens, estrogens, thyroid hormone, or vitamins, folic acid.
C. Polyalkylenes, Polysaccharides, etc.
[0155]Also, polyalkylene glycol can be included with the desired polynucleotides/polypeptides. In a preferred embodiment, the polyalkylene glycol is polyethlylene glycol. In addition, mono-, di-, or polysaccarides can be included. In a preferred embodiment of this aspect, the polysaccharide is dextran or DEAE-dextran. Also, chitosan and poly(lactide-co-glycolide)
D. Lipids, and Liposomes
[0156]The desired polynucleotide/polypeptide can also be encapsulated in lipids or packaged in liposomes prior to delivery to the subject or to cells derived therefrom.
[0157]Lipid encapsulation is generally accomplished using liposomes which are able to stably bind or entrap and retain nucleic acid. The ratio of condensed polynucleotide to lipid preparation can vary but will generally be around 1:1 (mg DNA: micromoles lipid), or more of lipid. For a review of the use of liposomes as carriers for delivery of nucleic acids, see, Hug and Sleight (1991) Biochim. Biophys. Acta. 1097:1-17; Straubinger (1983) Meth. Enzymol. 101:512-527.
[0158]Liposomal preparations for use in the present invention include cationic (positively charged), anionic (negatively charged) and neutral preparations. Cationic liposomes have been shown to mediate intracellular delivery of plasmid DNA (Felgner (1987) Proc. Natl. Acad. Sci. USA 84:7413-7416); mRNA (Malone (1989) Proc. Natl. Acad. Sci. USA 86:6077-6081); and purified transcription factors (Debs (1990) J. Biol. Chem. 265:10189-10192), in functional form.
[0159]Cationic liposomes are readily available. For example, N[1-2,3-dioleyloxy)propyl]-N,N,N-triethylammonium (DOTMA) liposomes are available under the trademark Lipofectin, from GIBCO BRL, Grand Island, N.Y. (See, also, Felgner supra). Other commercially available liposomes include transfectace (DDAB/DOPE) and DOTAP/DOPE (Boerhinger). Other cationic liposomes can be prepared from readily available materials using techniques well known in the art. See, e.g. Szoka (1978) Proc. Natl. Acad. Sci. USA 75:4194-4198; WO90/11092 for a description of the synthesis of DOTAP (1,2-bis(oleoyloxy)-3-(trimethylammonio)propane) liposomes.
[0160]Similarly, anionic and neutral liposomes are readily available, such as from Avanti Polar Lipids (Biriningham, Ala.), or can be easily prepared using readily available materials. Such materials include phosphatidyl choline, cholesterol, phosphatidyl ethanolamine, dioleoylphosphatidyl choline (DOPC), dioleoylphosphatidyl glycerol (DOPG), dioleoylphoshatidyl ethanolamine (DOPE), among others. These materials can also be mixed with the DOTMA and DOTAP starting materials in appropriate ratios. Methods for making liposomes using these materials are well known in the art.
[0161]The liposomes can comprise multilammelar vesicles (MLVs), small unilamellar vesicles (SUVs), or large unilamellar vesicles (LUVs). The various liposome-nucleic acid complexes are prepared using methods known in the art. See eg. Straubinger (1983) Meth. Immunol. 101:512-527; Szoka (1978) Proc. Natl. Acad. Sci. USA 75:4194-4198; Papahadjopoulos (1975) Biochim. Biophys. Acta 394:483; Wilson (1979) Cell 17:77); Deamer & Bangham (1976) Biochim. Biophys. Acta 443:629; Ostro (1977) Biochem. Biophys. Res. Commun. 76:836; Fraley (1979) Proc. Natl. Acad. Sci. USA 76:3348); Enoch & Strittmatter (1979) Proc. Natl. Acad. Sci. USA 76:145; Fraley (1980) J. Biol. Chem. (1980) 255:10431; Szoka & Papahadjopoulos (1978) Proc. Natl. Acad. Sci. USA 75:145; and Schaefer-Ridder (1982) Science 215:166.
E. Lipoproteins
[0162]In addition, lipoproteins can be included with the polynucleotide/polypeptide to be delivered. Examples of lipoproteins to be utilized include: chylomicrons, HDL, IDL, LDL, and VLDL. Mutants, fragments, or fusions of these proteins can also be used. Also, modifications of naturally occurring lipoproteins can be used, such as acetylated LDL. These lipoproteins can target the delivery of polynucleotides to cells expressing lipoprotein receptors. Preferably, if lipoproteins are including with the polynucleotide to be delivered, no other targeting ligand is included in the composition.
[0163]Naturally occurring lipoproteins comprise a lipid and a protein portion. The protein portion are known as apoproteins. At the present, apoproteins A, B, C, D, and E have been isolated and identified. At least two of these contain several proteins, designated by Roman numerals, AI, AII, AIV; CI, CII, CIII.
[0164]A lipoprotein can comprise more than one apoprotein. For example, naturally occurring chylomicrons comprises of A, B, C, and E, over time these lipoproteins lose A and acquire C and E apoproteins. VLDL comprises A, B, C, and E apoproteins, LDL comprises apoprotein B; and HDL comprises apoproteins A, C, and E.
[0165]The amino acid of these apoproteins are known and are described in, for example, Breslow (1985) Annu Rev. Biochem 54:699; Law (1986) Adv. Exp Med. Biol. 151:162; Chen (1986) J Biol Chem 261:12918; Kane (1980) Proc Natl Acad Sci USA 77:2465; and Utennann (1984) Hum Genet. 65:232.
[0166]Lipoproteins contain a variety of lipids including, triglycerides, cholesterol (free and esters), and phopholipids. The composition of the lipids varies in naturally occurring lipoproteins. For example, chylomicrons comprise mainly triglycerides. A more detailed description of the lipid content of naturally occurring lipoproteins can be found, for example, in Meth. Enzymol. 128 (1986). The composition of the lipids are chosen to aid in conformation of the apoprotein for receptor binding activity. The composition of lipids can also be chosen to facilitate hydrophobic interaction and association with the polynucleotide binding molecule.
[0167]Naturally occurring lipoproteins can be isolated from serum by ultracentrifugation, for instance. Such methods are described in Meth. Enzymol. (supra); Pitas (1980) J. Biochem. 255:5454-5460 and Mahey (1979) J Clin. Invest 64:743-750. Lipoproteins can also be produced by in vitro or recombinant methods by expression of the apoprotein genes in a desired host cell. See, for example, Atkinson (1986) Annu Rev Biophys Chem 15:403 and Radding (1958) Biochim Biophys Acta 30: 443. Lipoproteins can also be purchased from commercial suppliers, such as Biomedical Techniologies, Inc., Stoughton, Mass., USA. Further description of lipoproteins can be found in Zuckermann et al. PCT/US97/14465.
F. Polycationic Agents
[0168]Polycationic agents can be included, with or without lipoprotein, in a composition with the desired polynucleotide/polypeptide to be delivered.
[0169]Polycationic agents, typically, exhibit a net positive charge at physiological relevant pH and are capable of neutralizing the electrical charge of nucleic acids to facilitate delivery to a desired location. These agents have both in vitro, ex vivo, and in vivo applications. Polycationic agents can be used to deliver nucleic acids to a living subject either intramuscularly, subcutaneously, etc.
[0170]The following are examples of useful polypeptides as polycationic agents: polylysine, polyarginine, polyornithine, and protamine. Other examples include histones, protamines, human serum albumin, DNA binding proteins, non-histone chromosomal proteins, coat proteins from DNA viruses, such as (X174, transcriptional factors also contain domains that bind DNA and therefore may be useful as nucleic aid condensing agents. Briefly, transcriptional factors such as C/CEBP, c-jun, c-fos, AP-1, AP-2, AP-3, CPF, Prot-1, Sp-1, Oct-1, Oct-2, CREP, and TFIID contain basic domains that bind DNA sequences.
[0171]Organic polycationic agents include: spermine, spermidine, and purtrescine.
[0172]The dimensions and of the physical properties of a polycationic agent can be extrapolated from the list above, to construct other polypeptide polycationic agents or to produce synthetic polycationic agents.
[0173]Synthetic polycationic agents which are useful include, for example, DEAE-dextran, polybrene. Lipofectin™, and lipofectAMINE™ are monomers that form polycationic complexes when combined with polynucleotides/polypeptides.
Immunodiagnostic Assays
[0174]Neisserial antigens of the invention can be used in immunoassays to detect antibody levels (or, conversely, anti-Neisserial antibodies can be used to detect antigen levels). Immunoassays based on well defined, recombinant antigens can be developed to replace invasive diagnostics methods. Antibodies to Neisserial proteins within biological samples, including for example, blood or serum samples, can be detected. Design of the immunoassays is subject to a great deal of variation, and a variety of these are known in the art. Protocols for the immunoassay may be based, for example, upon competition, or direct reaction, or sandwich type assays. Protocols may also, for example, use solid supports, or may be by immunoprecipitation. Most assays involve the use of labeled antibody or polypeptide; the labels may be, for example, fluorescent, chemiluminescent, radioactive, or dye molecules. Assays which amplify the signals from the probe are also known; examples of which are assays which utilize biotin and avidin, and enzyme-labeled and mediated immunoassays, such as ELISA assays.
[0175]Kits suitable for immunodiagnosis and containing the appropriate labeled reagents are constructed by packaging the appropriate materials, including the compositions of the invention, in suitable containers, along with the remaining reagents and materials (for example, suitable buffers, salt solutions, etc.) required for the conduct of the assay, as well as suitable set of assay instructions.
Nucleic Acid Hybridisation
[0176]“Hybridization” refers to the association of two nucleic acid sequences to one another by hydrogen bonding. Typically, one sequence will be fixed to a solid support and the other will be free in solution. Then, the two sequences will be placed in contact with one another under conditions that favor hydrogen bonding. Factors that affect this bonding include: the type and volume of solvent; reaction temperature; time of hybridization; agitation; agents to block the non-specific attachment of the liquid phase sequence to the solid support (Denhardt's reagent or BLOTTO); concentration of the sequences; use of compounds to increase the rate of association of sequences (dextran sulfate or polyethylene glycol); and the stringency of the washing conditions following hybridization. See Sambrook et al. [supra] Volume 2, chapter 9, pages 9.47 to 9.57.
[0177]“Stringency” refers to conditions in a hybridization reaction that favor association of very similar sequences over sequences that differ. For example, the combination of temperature and salt concentration should be chosen that is approximately 120 to 200° C. below the calculated Tm of the hybrid under study. The temperature and salt conditions can often be determined empirically in preliminary experiments in which samples of genomic DNA immobilized on filters are hybridized to the sequence of interest and then washed under conditions of different stringencies. See Sambrook et al. at page 9.50.
[0178]Variables to consider when performing, for example, a Southern blot are (1) the complexity of the DNA being blotted and (2) the homology between the probe and the sequences being detected. The total amount of the fragment(s) to be studied can vary a magnitude of 10, from 0.1 to 1 μg for a plasmid or phage digest to 10−9to 10−8g for a single copy gene in a highly complex eukaryotic genome. For lower complexity polynucleotides, substantially shorter blotting, hybridization, and exposure times, a smaller amount of starting polynucleotides, and lower specific activity of probes can be used. For example, a single-copy yeast gene can be detected with an exposure time of only 1 hour starting with 1 μg of yeast DNA, blotting for two hours, and hybridizing for 4-8 hours with a probe of 108cpm/μg. For a single-copy mammalian gene a conservative approach would start with 10 μg of DNA, blot overnight, and hybridize overnight in the presence of 10% dextran sulfate using a probe of greater than 108cpm/μg, resulting in an exposure time of ˜24 hours.
[0179]Several factors can affect the melting temperature (Tm) of a DNA-DNA hybrid between the probe and the fragment of interest, and consequently, the appropriate conditions for hybridization and washing. In many cases the probe is not 100% homologous to the fragment. Other commonly encountered variables include the length and total G+C content of the hybridizing sequences and the ionic strength and formamide content of the hybridization buffer. The effects of all of these factors can be approximated by a single equation:
[0000]
Tm=81+16.6(log10Ci)+0.4[%(G+C)]−0.6(% formamide)−600/n−1.5(% mismatch).
[0000]where Ci is the salt concentration (monovalent ions) and n is the length of the hybrid in base pairs (slightly modified from Meinkoth & Wahl (1984) Anal. Biochem. 138: 267-284).
[0180]In designing a hybridization experiment, some factors affecting nucleic acid hybridization can be conveniently altered. The temperature of the hybridization and washes and the salt concentration during the washes are the simplest to adjust. As the temperature of the hybridization increases (i.e. stringency), it becomes less likely for hybridization to occur between strands that are nonhomologous, and as a result, background decreases. If the radiolabeled probe is not completely homologous with the immobilized fragment (as is frequently the case in gene family and interspecies hybridization experiments), the hybridization temperature must be reduced, and background will increase. The temperature of the washes affects the intensity of the hybridizing band and the degree of background in a similar manner. The stringency of the washes is also increased with decreasing salt concentrations.
[0181]In general, convenient hybridization temperatures in the presence of 50% formamide are 42° C. for a probe with is 95% to 100% homologous to the target fragment, 37° C. for 90% to 95% homology, and 32° C. for 85% to 90% homology. For lower homologies, formamide content should be lowered and temperature adjusted accordingly, using the equation above. If the homology between the probe and the target fragment are not known, the simplest approach is to start with both hybridization and wash conditions which are nonstringent. If non-specific bands or high background are observed after autoradiography, the filter can be washed at high stringency and reexposed. If the time required for exposure makes this approach impractical, several hybridization and/or washing stringencies should be tested in parallel.
Nucleic Acid Probe Assays
[0182]Methods such as PCR, branched DNA probe assays, or blotting techniques utilizing nucleic acid probes according to the invention can determine the presence of cDNA or mRNA. A probe is said to “hybridize” with a sequence of the invention if it can form a duplex or double stranded complex, which is stable enough to be detected.
[0183]The nucleic acid probes will hybridize to the Neisserial nucleotide sequences of the invention (including both sense and antisense strands). Though many different nucleotide sequences will encode the amino acid sequence, the native Neisserial sequence is preferred because it is the actual sequence present in cells. mRNA represents a coding sequence and so a probe should be complementary to the coding sequence; single-stranded cDNA is complementary to mRNA, and so a cDNA probe should be complementary to the non-coding sequence.
[0184]The probe sequence need not be identical to the Neisserial sequence (or its complement)—some variation in the sequence and length can lead to increased assay sensitivity if the nucleic acid probe can form a duplex with target nucleotides, which can be detected. Also, the nucleic acid probe can include additional nucleotides to stabilize the formed duplex. Additional Neisserial sequence may also be helpful as a label to detect the formed duplex. For example, a non-complementary nucleotide sequence may be attached to the 5′ end of the probe, with the remainder of the probe sequence being complementary to a Neisserial sequence. Alternatively, non-complementary bases or longer sequences can be interspersed into the probe, provided that the probe sequence has sufficient complementarity with the a Neisserial sequence in order to hybridize therewith and thereby form a duplex which can be detected.
[0185]The exact length and sequence of the probe will depend on the hybridization conditions, such as temperature, salt condition and the like. For example, for diagnostic applications, depending on the complexity of the analyte sequence, the nucleic acid probe typically contains at least 10-20 nucleotides, preferably 15-25, and more preferably at least 30 nucleotides, although it may be shorter than this. Short primers generally require cooler temperatures to form sufficiently stable hybrid complexes with the template.
[0186]Probes may be produced by synthetic procedures, such as the triester method of Matteucci et al. [J. Am. Chem. Soc. (1981) 103:3185], or according to Urdea et al. [Proc. Natl. Acad. Sci. USA (1983) 80:7461], or using commercially available automated oligonucleotide synthesizers.
[0187]The chemical nature of the probe can be selected according to preference. For certain applications, DNA or RNA are appropriate. For other applications, modifications may be incorporated e.g. backbone modifications, such as phosphorothioates or methylphosphonates, can be used to increase in vivo half-life, alter RNA affinity, increase nuclease resistance etc. [e.g. see Agrawal & Iyer (1995) Curr Opin Biotechnol 6:12-19; Agrawal (1996) TIBTECH 14:376-387]; analogues such as peptide nucleic acids may also be used [e.g. see Corey (1997) TIBTECH 15:224-229; Buchardt et al. (1993) TIBTECH 11:384-386].
[0188]Alternatively, the polymerase chain reaction (PCR) is another well-known means for detecting small amounts of target nucleic acids. The assay is described in: Mullis et al. [Meth. Enzymol. (1987) 155: 335-350]; U.S. Pat. Nos. 4,683,195 and 4,683,202. Two “primer” nucleotides hybridize with the target nucleic acids and are used to prime the reaction. The primers can comprise sequence that does not hybridize to the sequence of the amplification target (or its complement) to aid with duplex stability or, for example, to incorporate a convenient restriction site. Typically, such sequence will flank the desired Neisserial sequence.
[0189]A thermostable polymerase creates copies of target nucleic acids from the primers using the original target nucleic acids as a template. After a threshold amount of target nucleic acids are generated by the polymerase, they can be detected by more traditional methods, such as Southern blots. When using the Southern blot method, the labelled probe will hybridize to the Neisserial sequence (or its complement).
[0190]Also, mRNA or cDNA can be detected by traditional blotting techniques described in Sambrook et al [supra]. mRNA, or cDNA generated from mRNA using a polymerase enzyme, can be purified and separated using gel electrophoresis. The nucleic acids on the gel are then blotted onto a solid support, such as nitrocellulose. The solid support is exposed to a labelled probe and then washed to remove any unhybridized probe. Next, the duplexes containing the labeled probe are detected. Typically, the probe is labelled with a radioactive moiety.
BRIEF DESCRIPTION OF THE DRAWINGS
[0191]FIGS. 1-20 show biochemical data obtained in the Examples, and also sequence analysis, for ORFs 37, 5, 2, 15, 22, 28, 32, 4, 61, 76, 89, 97, 106, 138, 23, 25, 27, 79, 85 and 132. M1 and M2 are molecular weight markers. Arrows indicate the position of the main recombinant product or, in Western blots, the position of the main N. meningitidis immunoreactive band. TP indicates N. meningitidis total protein extract; OMV indicates N. meningitidis outer membrane vesicle preparation. In bactericidal assay results: a diamond (♦) shows preimmune data; a triangle (▴) shows GST control data; a circle () shows data with recombinant N. meningitidis protein. Computer analyses show a hydrophilicity plot (upper), an antigenic index plot (middle), and an AMPHI analysis (lower). The AMPHI program has been used to predict T-cell epitopes [Gao et al. (1989) J. Immunol. 143:3007; Roberts et al. (1996) AIDS Res Hum Retrovir 12:593; Quakyi et al. (1992) Scand J Immunol suppl. 11:9) and is available in the Protean package of DNASTAR, Inc. (1228 South Park Street, Madison, Wis. 53715 USA).
EXAMPLES
[0192]The examples describe nucleic acid sequences which have been identified in N. meningitidis, along with their putative translation products, and also those of N. gonorrhoeae. Not all of the nucleic acid sequences are complete i.e. they encode less than the full-length wild-type protein.
[0193]The examples are generally in the following format:
- a nucleotide sequence which has been identified in N. meningitidis (strain B)
- the putative translation product of this sequence
- a computer analysis of the translation product based on database comparisons
- corresponding gene and protein sequences identified in N. meningitidis (strain A) and in N. gonorrhoeae
- a description of the characteristics of the proteins which indicates that they might be suitably antigenic
- results of biochemical analysis (expression, purification, ELISA, FACS etc.)
[0200]The examples typically include details of sequence identity between species and strains. Proteins that are similar in sequence are generally similar in both structure and function, and the sequence identity often indicates a common evolutionary origin. Comparison with sequences of proteins of known function is widely used as a guide for the assignment of putative protein function to a new sequence and has proved particularly useful in whole-genome analyses.
[0201]Sequence comparisons were performed at NCBI (http://www.ncbi.nlm.nih.gov) using the algorithms BLAST, BLAST2, BLASTn, BLASTp, tBLASTn, BLASTx, & tBLASTx [e.g. see also Altschul et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25:2289-3402]. Searches were performed against the following databases: non-redundant GenBank+EMBL+DDBJ+PDB sequences and non-redundant GenBank CDS translations+PDB+SwissProt+SPupdate+PIR sequences.
[0202]To compare Meningococcal and Gonococcal sequences, the tBLASTx algorithm was used, as implemented at http://www.genome.ou.edu/gono_blast.html. The FASTA algorithm was also used to compare the ORFs (from GCG Wisconsin Package, version 9.0).
[0203]Dots within nucleotide sequences (e.g. position 495 in SEQ ID 11) represent nucleotides which have been arbitrarily introduced in order to maintain a reading frame. In the same way, double-underlined nucleotides were removed. Lower case letters (e.g. position 496 in SEQ ID 11) represent ambiguities which arose during alignment of independent sequencing reactions (some of the nucleotide sequences in the examples are derived from combining the results of two or more experiments).
[0204]Nucleotide sequences were scanned in all six reading frames to predict the presence of hydrophobic domains using an algorithm based on the statistical studies of Esposti et al. [Critical evaluation of the hydropathy of membrane proteins (1990) Eur J Biochem 190:207-219]. These domains represent potential transmembrane regions or hydrophobic leader sequences.
[0205]Open reading frames were predicted from fragmented nucleotide sequences using the program ORFFINDER(NCBI).
[0206]Underlined amino acid sequences indicate possible transmembrane domains or leader sequences in the ORFs, as predicted by the PSORT algorithm (http://www.psort.nibb.acjp). Functional domains were also predicted using the MOTIFS program (GCG Wisconsin & PROSITE).
[0207]Various tests can be used to assess the in vivo immunogencity of the proteins identified in the examples. For example, the proteins can be expressed recombinantly and used to screen patient sera by immunoblot. A positive reaction between the protein and patient serum indicates that the patient has previously mounted an immune response to the protein in question i.e. the protein is an immunogen. This method can also be used to identify immunodominant proteins.
[0208]The recombinant protein can also be conveniently used to prepare antibodies e.g. in a mouse. These can be used for direct confirmation that a protein is located on the cell-surface. Labelled antibody (e.g. fluorescent labelling for FACS) can be incubated with intact bacteria and the presence of label on the bacterial surface confirms the location of the protein.
[0209]In particular, the following methods (A) to (S) were used to express, purify and biochemically characterise the proteins of the invention:
A) Chromosomal DNA Preparation
[0210]N. meningitidis strain 2996 was grown to exponential phase in 100 ml of GC medium, harvested by centrifugation, and resuspended in 5 ml buffer (20% Sucrose, 50 mM Tris-HCl, 50 mM EDTA, pH8). After 10 minutes incubation on ice, the bacteria were lysed by adding 10 ml lysis solution (50 mM NaCl, 1% Na-Sarkosyl, 50 μg/ml Proteinase K), and the suspension was incubated at 37° C. for 2 hours. Two phenol extractions (equilibrated to pH 8) and one ChCl3/isoamylalcohol (24:1) extraction were performed. DNA was precipitated by addition of 0.3M sodium acetate and 2 volumes ethanol, and was collected by centrifugation. The pellet was washed once with 70% ethanol and redissolved in 4 ml buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8). The DNA concentration was measured by reading the OD at 260 nm.
B) Oligonucleotide Design
[0211]Synthetic oligonucleotide primers were designed on the basis of the coding sequence of each ORF, using (a) the meningococcus B sequence when available, or (b) the gonococcus/meningococcus A sequence, adapted to the codon preference usage of meningococcus as necessary. Any predicted signal peptides were omitted, by deducing the 5′-end amplification primer sequence immediately downstream from the predicted leader sequence.
[0212]For most ORFs, the 5′ primers included two restriction enzyme recognition sites (BamHI-NdeI, BamHI-NheI, or EcoRI-NheI, depending on the gene's own restriction pattern); the 3′ primers included a XhoI restriction site. This procedure was established in order to direct the cloning of each amplification product (corresponding to each ORF) into two different expression systems: pGEX-KG (using either BamHI-XhoI or EcoRI-XhoI), and pET21b+ (using either NdeI-XhoI or NheI-XhoI).
[0000]| 5′-end primer tail: | CGCGGATCCCATATG | (BamHI-NdeI) |
|
| CGCGGATCCGCTAGC | (BamHI-NheI) |
|
| CCGGAATTCTAGCTAGC | (EcoRI-NheI) |
|
| 3′-end primer tail: | CCCGCTCGAG | (XhoI) |
[0213]For ORFs 5, 15, 17, 19, 20, 22, 27, 28, 65 & 89, two different amplifications were performed to clone each ORF in the two expression systems. Two different 5′ primers were used for each ORF; the same 3′ XhoI primer was used as before:
[0000]| 5′-end primer tail: | GGAATTCCATATGGCCATGG | (NdeI) |
|
| 5′-end primer tail: | CGGGATCC | (BamHI) |
[0214]ORF 76 was cloned in the pTRC expression vector and expressed as an amino-terminus His-tag fusion. In this particular case, the predicted signal peptide was included in the final product. NheI-BamHI restriction sites were incorporated using primers:
[0000]| 5′-end primer tail: | GATCAGCTAGCCATATG | (NheI) |
|
| 3′-end primer tail: | CGGGATCC | (BamHI) |
[0215]As well as containing the restriction enzyme recognition sequences, the primers included nucleotides which hybridized to the sequence to be amplified. The number of hybridizing nucleotides depended on the melting temperature of the whole primer, and was determined for each primer using the formulae:
[0000]
Tm=4(G+C)+2(A+T) (tail excluded)
[0000]
Tm=64.9+0.41(% GC)−600/N (whole primer)
[0216]The average melting temperature of the selected oligos were 65-70° C. for the whole oligo and 50-55° C. for the hybridising region alone.
[0217]Table I (page 487) shows the forward and reverse primers used for each amplification. In certain cases, it will be noted that the sequence of the primer does not exactly match the sequence in the ORF. When initial amplifications were performed, the complete 5′ and/or 3′ sequence was not known for some meningococcal ORFs, although the corresponding sequences had been identified in gonococcus. For amplification, the gonococcal sequences could thus be used as the basis for primer design, altered to take account of codon preference. In particular, the following codons were changed: ATA→ATT; TCG→TCT; CAG→CAA; AAG→AAA; GAG→GAA; CGA→CGC; CGG→CGC; GGG→GGC. Italicised nucleotides in Table I indicate such a change. It will be appreciated that, once the complete sequence has been identified, this approach is generally no longer necessary.
[0218]Oligos were synthesized by a Perkin Elmer 394 DNA/RNA Synthesizer, eluted from the columns in 2 ml NH4OH, and deprotected by 5 hours incubation at 56° C. The oligos were precipitated by addition of 0.3M Na-Acetate and 2 volumes ethanol. The samples were then centrifuged and the pellets resuspended in either 100 μl or 1 ml of water. OD260was determined using a Perkin Elmer Lambda Bio spectophotometer and the concentration was determined and adjusted to 2-10 pmol/μl.
C) Amplification
[0219]The standard PCR protocol was as follows: 50-200ng of genomic DNA were used as a template in the presence of 20-40 μM of each oligo, 400-800 μM dNTPs solution, 1×PCR buffer (including 1.5 mM MgCl2), 2.5 units TaqI DNA polymerase (using Perkin-Elmer AmpliTaQ, GIBCO Platinum, Pwo DNA polymerase, or Tahara Shuzo Taq polymerase).
[0220]In some cases, PCR was optimsed by the addition of 10 μl DMSO or 50 μl 2M betaine.
[0221]After a hot start (adding the polymerase during a preliminary 3 minute incubation of the whole mix at 95° C.), each sample underwent a double-step amplification: the first 5 cycles were performed using as the hybridization temperature the one of the oligos excluding the restriction enzymes tail, followed by 30 cycles performed according to the hybridization temperature of the whole length oligos. The cycles were followed by a final 10 minute extension step at 72° C.
[0222]The standard cycles were as follows:
[0000] |
| First 5 cycles | 30 seconds | 30 seconds | 30-60 seconds |
| 95° C. | 50-55° C. | 72° C. |
| Last 30 cycles | 30 seconds | 30 seconds | 30-60 seconds |
| 95° C. | 65-70° C. | 72° C. |
|
[0223]The elongation time varied according to the length of the ORF to be amplified.
[0224]The amplifications were performed using either a 9600 or a 2400 Perkin Elmer GeneAmp PCR System. To check the results, 1/10 of the amplification volume was loaded onto a 1-1.5% agarose gel and the size of each amplified fragment compared with a DNA molecular weight marker.
[0225]The amplified DNA was either loaded directly on a 1% agarose gel or first precipitated with ethanol and resuspended in a suitable volume to be loaded on a 1% agarose gel. The DNA fragment corresponding to the right size band was then eluted and purified from gel, using the Qiagen Gel Extraction Kit, following the instructions of the manufacturer. The final volume of the DNA fragment was 30 μl or 50 μl of either water or 10 mM Tris, pH 8.5.
D) Digestion of PCR Fragments
[0226]The purified DNA corresponding to the amplified fragment was split into 2 aliquots and double-digested with:
- NdeI/XhoI or NheI/XhoI for cloning into pET-21b+ and further expression of the protein as a C-terminus His-tag fusion
- BamHI/XhoI or EcoRI/XhoI for cloning into pGEX-KG and further expression of the protein as N-terminus GST fusion.
- For ORF 76, NheI/BamHI for cloning into pTRC-HisA vector and further expression of the protein as N-terminus His-tag fusion.
- EcoRI/PstI, EcoRI/SalI, SalI/PstI for cloning into pGex-His and further expression of the protein as N-terminus His-tag fusion
[0231]Each purified DNA fragment was incubated (37° C. for 3 hours to overnight) with 20 units of each restriction enzyme (New England Biolabs) in a either 30 or 40 μl final volume in the presence of the appropriate buffer. The digestion product was then purified using the QIAquick PCR purification kit, following the manufacturer's instructions, and eluted in a final volume of 30 or 50 μl of either water or 10 mM Tris-HCl, pH 8.5. The final DNA concentration was determined by 1% agarose gel electrophoresis in the presence of titrated molecular weight marker.
[0000]E) Digestion of the Cloning Vectors (pET22B, pGEX-KG, pTRC-His A, and pGex-His)
[0232]10 μg plasmid was double-digested with 50 units of each restriction enzyme in 200 μl reaction volume in the presence of appropriate buffer by overnight incubation at 37° C. After loading the whole digestion on a 1% agarose gel, the band corresponding to the digested vector was purified from the gel using the Qiagen QIAquick Gel Extraction Kit and the DNA was eluted in 50 μl of 10 mM Tris-HCl, pH 8.5. The DNA concentration was evaluated by measuring OD260of the sample, and adjusted to 50 μg/μl. 1 μl of plasmid was used for each cloning procedure.
[0233]The vector pGEX-His is a modified pGEX-2T vector carrying a region encoding six histidine residues upstream to the thrombin cleavage site and containing the multiple cloning site of the vector pTRC99 (Pharmacia).
F) Cloning
[0234]The fragments corresponding to each ORF, previously digested and purified, were ligated in both pET22b and pGEX-KG. In a final volume of 20 μl, a molar ratio of 3:1 fragment/vector was ligated using 0.5 μl of NEB T4 DNA ligase (400 units/μl), in the presence of the buffer supplied by the manufacturer. The reaction was incubated at room temperature for 3 hours. In some experiments, ligation was performed using the Boheringer “Rapid Ligation Kit”, following the manufacturer's instructions.
[0235]In order to introduce the recombinant plasmid in a suitable strain, 100 μl E. coli DH5 competent cells were incubated with the ligase reaction solution for 40 minutes on ice, then at 37° C. for 3 minutes, then, after adding 800 μl LB broth, again at 37° C. for 20 minutes. The cells were then centrifuged at maximum speed in an Eppendorf microfuge and resuspended in approximately 200 μl of the supernatant. The suspension was then plated on LB ampicillin (100 mg/ml).
[0236]The screening of the recombinant clones was performed by growing 5 randomly-chosen colonies overnight at 37° C. in either 2 ml (pGEX or pTC clones) or 5 ml (pET clones) LB broth+100 μg/ml ampicillin. The cells were then pelletted and the DNA extracted using the Qiagen QIAprep Spin Miniprep Kit, following the manufacturer's instructions, to a final volume of 30 μl. 5 μl of each individual miniprep (approximately 1 g) were digested with either NdeI/XhoI or BamHI/XhoI and the whole digestion loaded onto a 1-1.5% agarose gel (depending on the expected insert size), in parallel with the molecular weight marker (IKb DNA Ladder, GIBCO). The screening of the positive clones was made on the base of the correct insert size.
[0237]For the cloning of ORFs 110, 111, 113, 115, 119, 122, 125 & 130, the double-digested PCR product was ligated into double-digested vector using EcoRI-PstI cloning sites or, for ORFs 115 & 127, EcoRI-SalI or, for ORF 122, SalI-PstI. After cloning, the recombinant plasmids were introduced in the E. coli host W3110. Individual clones were grown overnight at 37° C. in L-broth with 50 μl/ml ampicillin.
G) Expression
[0238]Each ORF cloned into the expression vector was transformed into the strain suitable for expression of the recombinant protein product. 1 μl of each construct was used to transform 30 μl of E. coli BL21 (pGEX vector), E. coli TOP 10 (PTRC vector) or E. coli BL21-DE3 (PET vector), as described above. In the case of the pGEX-His vector, the same E. coli strain (W3110) was used for initial cloning and expression. Single recombinant colonies were inoculated into 2 ml LB+Amp (100 μg/ml), incubated at 37° C. overnight, then diluted 1:30 in 20 ml of LB+Amp (100 μg/ml) in 100 ml flasks, making sure that the OD600ranged between 0.1 and 0.15. The flasks were incubated at 30° C. into gyratory water bath shakers until OD indicated exponential growth suitable for induction of expression (0.4-0.8 OD for pET and pTRC vectors; 0.8-1 OD for pGEX and pGEX-His vectors). For the pET, pTRC and pGEX-His vectors, the protein expression was induced by addition of 1 mM IPTG, whereas in the case of pGEX system the final concentration of IPTG was 0.2 mM. After 3 hours incubation at 30° C., the final concentration of the sample was checked by OD. In order to check expression, 1 ml of each sample was removed, centrifuged in a microfuge, the pellet resuspended in PBS, and analysed by 12% SDS-PAGE with Coomassie Blue staining. The whole sample was centrifuged at 6000 g and the pellet resuspended in PBS for further use.
H) GST-Fusion Proteins Large-Scale Purification.
[0239]A single colony was grown overnight at 37° C. on LB+Amp agar plate. The bacteria were inoculated into 20 ml of LB+Amp liquid colture in a water bath shaker and grown overnight. Bacteria were diluted 1:30 into 600 ml of fresh medium and allowed to grow at the optimal temperature (20-37° C.) to OD5500.8-1. Protein expression was induced with 0.2 mM IPTG followed by three hours incubation. The culture was centrifuged at 800 rpm at 4° C. The supernatant was discarded and the bacterial pellet was resuspended in 7.5 ml cold PBS. The cells were disrupted by sonication on ice for 30 sec at 40 W using a Branson sonifier B-15, frozen and thawed twice and centrifuged again. The supernatant was collected and mixed with 150 μl Glutatione-Sepharose 4B resin (Pharmacia) (previously washed with PBS) and incubated at room temperature for 30 minutes. The sample was centrifuged at 700 g for 5 minutes at 4° C. The resin was washed twice with 10 ml cold PBS for 10 minutes, resuspended in 1 ml cold PBS, and loaded on a disposable column. The resin was washed twice with 2 ml cold PBS until the flow-through reached OD280of 0.02-0.06. The GST-fusion protein was eluted by addition of 700 μl cold Glutathione elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl) and fractions collected until the OD280was 0.1. 21 μl of each fraction were loaded on a 12% SDS gel using either Biorad SDS-PAGE Molecular weight standard broad range (M1) (200, 116.25, 97.4, 66.2, 45,31, 21.5, 14.4, 6.5 kDa) or Amersham Rainbow Marker (M2) (220, 66, 46, 30, 21.5, 14.3 kDa) as standards. As the MW of GST is 26 kDa, this value must be added to the MW of each GST-fusion protein.
I) His-Fusion Solubility Analysis (ORFs 111-129)
[0240]To analyse the solubility of the His-fusion expression products, pellets of 3 ml cultures were resuspended in buffer M1 [500 μl PBS pH 7.2]. 25 μl lysozyme (10 mg/ml) was added and the bacteria were incubated for 15 min at 4° C. The pellets were sonicated for 30 sec at 40 W using a Branson sonifier B-15, frozen and thawed twice and then separated again into pellet and supernatant by a centrifugation step. The supernatant was collected and the pellet was resuspended in buffer M2 [8M urea, 0.5M NaCl, 20 mM imidazole and 0.1 M NaH2PO4] and incubated for 3 to 4 hours at 4° C. After centrifugation, the supernatant was collected and the pellet was resuspended in buffer M3 [6M guanidinium-HCl, 0.5M NaCl, 20 mM imidazole and 0.1 M NaH2PO4] overnight at 4° C. The supernatants from all steps were analysed by SDS-PAGE.
[0241]The proteins expressed from ORFs 113, 119 and 120 were found to be soluble in PBS, whereas ORFs 111, 122, 126 and 129 need urea and ORFs 125 and 127 need guanidium-HCl for their solubilization.
J) His-Fusion Large-Scale Purification.
[0242]A single colony was grown overnight at 37° C. on a LB+Amp agar plate. The bacteria were inoculated into 20 ml of LB+Amp liquid culture and incubated overnight in a water bath shaker. Bacteria were diluted 1:30 into 600 ml fresh medium and allowed to grow at the optimal temperature (20-37° C.) to OD5500.6-0.8. Protein expression was induced by addition of 1 mM IPTG and the culture further incubated for three hours. The culture was centrifuged at 800 rpm at 4° C., the supernatant was discarded and the bacterial pellet was resuspended in 7.5 ml of either (i) cold buffer A (300 mM NaCl, 50 mM phosphate buffer, 10 mM imidazole, pH 8) for soluble proteins or (ii) buffer B (urea 8M, 10 mM Tris-HCl, 100 mM phosphate buffer, pH 8.8) for insoluble proteins. The cells were disrupted by sonication on ice for 30 sec at 40 W using a Branson sonifier B-15, frozen and thawed two times and centrifuged again.
[0243]For insoluble proteins, the supernatant was stored at −20° C., while the pellets were resuspended in 2 ml buffer C (6M guanidine hydrochloride, 100 mM phosphate buffer, 10 mM Tris-HCl, pH 7.5) and treated in a homogenizer for 10 cycles. The product was centrifuged at 13000 rpm for 40 minutes.
[0244]Supernatants were collected and mixed with 150 μl Ni2+-resin (Pharmacia) (previously washed with either buffer A or buffer B, as appropriate) and incubated at room temperature with gentle agitation for 30 minutes. The sample was centrifuged at 700 g for 5 minutes at 4° C. The resin was washed twice with 10 ml buffer A or B for 10 minutes, resuspended in 1 ml buffer A or B and loaded on a disposable column. The resin was washed at either (i) 4° C. with 2 ml cold buffer A or (ii) room temperature with 2 ml buffer B, until the flow-through reached OD280of 0.02-0.06.
[0245]The resin was washed with either (i) 2 ml cold 20 mM imidazole buffer (300 mM NaCl, 50 mM phosphate buffer, 20 mM imidazole, pH 8) or (ii) buffer D (urea 8M, 10 mM Tris-HCl, 100 mM phosphate buffer, pH 6.3) until the flow-through reached the O.D280of 0.02-0.06. The His-fusion protein was eluted by addition of 700 μl of either (i) cold elution buffer A (300 mM NaCl, 50 mM phosphate buffer, 250 mM imidazole, pH 8) or (ii) elution buffer B (urea 8M, 10 mM Tris-HCl, 100 mM phosphate buffer, pH 4.5) and fractions collected until the O.D280was 0.1. 21 μl of each fraction were loaded on a 12% SDS gel.
K) His-Fusion Proteins Renaturation
[0246]10% glycerol was added to the denatured proteins. The proteins were then diluted to 20 μg/ml using dialysis buffer I (10% glycerol, 0.5M arginine, 50 mM phosphate buffer, 5 mM reduced glutathione, 0.5 mM oxidised glutathione, 2M urea, pH 8.8) and dialysed against the same buffer at 4° C. for 12-14 hours. The protein was further dialysed against dialysis buffer II (10% glycerol, 0.5M arginine, 50 mM phosphate buffer, 5 mM reduced glutathione, 0.5 mM oxidised glutathione, pH 8.8) for 12-14 hours at 4° C. Protein concentration was evaluated using the formula:
[0000]
Protein (mg/ml)=(1.55×OD280)−(0.76×OD260)
L) His-Fusion Large-Scale Purification (ORFs 111-129)
[0247]500 ml of bacterial cultures were induced and the fusion proteins were obtained soluble in buffer M1, M2 or M3 using the procedure described above. The crude extract of the bacteria was loaded onto a Ni-NTA superflow column (Quiagen) equilibrated with buffer M1, M2 or M3 depending on the solubilization buffer of the fusion proteins. Unbound material was eluted by washing the column with the same buffer. The specific protein was eluted with the corresponding buffer containing 500 mM imidazole and dialysed against the corresponding buffer without imidazole. After each run the columns were sanitized by washing with at least two column volumes of 0.5 M sodium hydroxide and reequilibrated before the next use.
M) Mice Immunisations
[0248]20 μg of each purified protein were used to immunise mice intraperitoneally. In the case of ORFs 2, 4, 15, 22, 27, 28, 37, 76, 89 and 97, Balb-C mice were immunised with Al(OH)3as adjuvant on days 1, 21 and 42, and immune response was monitored in samples taken on day 56. For ORFs 44, 106 and 132, CD1 mice were immunised using the same protocol. For ORFs 25 and 40, CD1 mice were immunised using Freund's adjuvant, rather than AL(OH)3, and the same immunisation protocol was used, except that the immune response was measured on day 42, rather than 56. Similarly, for ORFs 23, 32,38 and 79, CD1 mice were immunised with Freund's adjuvant, but the immune response was measured on day 49.
N) ELISA Assay (Sera Analysis)
[0249]The acapsulated MenB M7 strain was plated on chocolate agar plates and incubated overnight at 37° C. Bacterial colonies were collected from the agar plates using a sterile dracon swab and inoculated into 7 ml of Mueller-Hinton Broth (Difco) containing 0.25% Glucose. Bacterial growth was monitored every 30 minutes by following OD620. The bacteria were let to grow until the OD reached the value of 0.3-0.4. The culture was centrifuged for 10 minutes at 10000 rpm. The supernatant was discarded and bacteria were washed once with PBS, resuspended in PBS containing 0.025% formaldehyde, and incubated for 2 hours at room temperature and then overnight at 4° C. with stirring. 100 μl bacterial cells were added to each well of a 96 well Greiner plate and incubated overnight at 4° C. The wells were then washed three times with PBT washing buffer (0.1% Tween-20 in PBS). 200 μl of saturation buffer (2.7% Polyvinylpyrrolidone 10 in water) was added to each well and the plates incubated for 2 hours at 37° C. Wells were washed three times with PBT. 200 μl of diluted sera (Dilution buffer: 1% BSA, 0.1% Tween-20, 0.1% NaN3in PBS) were added to each well and the plates incubated for 90 minutes at 37° C. Wells were washed three times with PBT. 100 μl of HRP-conjugated rabbit anti-mouse (Dako) serum diluted 1:2000 in dilution buffer were added to each well and the plates were incubated for 90 minutes at 37° C. Wells were washed three times with PBT buffer. 100 μl of substrate buffer for HRP (25 ml of citrate buffer pH5, 10 mg of O-phenildiamine and 10 μl of H2O) were added to each well and the plates were left at room temperature for 20 minutes. 100 μl H2SO4was added to each well and OD490was followed. The ELISA was considered positive when OD490was 2.5 times the respective pre-immune sera.
O) FACScan Bacteria Binding Assay Procedure.
[0250]The acapsulated MenB M7 strain was plated on chocolate agar plates and incubated overnight at 37° C. Bacterial colonies were collected from the agar plates using a sterile dracon swab and inoculated into 4 tubes containing 8 ml each Mueller-Hinton Broth (Difco) containing 0.25% glucose. Bacterial growth was monitored every 30 minutes by following OD620. The bacteria were let to grow until the OD reached the value of 0.35-0.5. The culture was centrifuged for 10 minutes at 4000 rpm. The supernatant was discarded and the pellet was resuspended in blocking buffer (1% BSA, 0.4% NaN3) and centrifuged for 5 minutes at 400 rpm. Cells were resuspended in blocking buffer to reach OD620of 0.07. 100 μl bacterial cells were added to each well of a Costar 96 well plate. 100 μl of diluted (1:200) sera (in blocking buffer) were added to each well and plates incubated for 2 hours at 4° C. Cells were centrifuged for 5 minutes at 400 rpm, the supernatant aspirated and cells washed by addition of 200 μl/well of blocking buffer in each well. 100 μl of R-Phicoerytrin conjugated F(ab)2goat anti-mouse, diluted 1:100, was added to each well and plates incubated for 1 hour at 4° C. Cells were spun down by centrifugation at 400 rpm for 5 minutes and washed by addition of 200 μl/well of blocking buffer. The supernatant was aspirated and cells resuspended in 200 μl/well of PBS, 0.25% formaldehyde. Samples were transferred to FACScan tubes and read. The condition for FACScan setting were: FL1 on, FL2 and FL3 off; FSC-H threshold:92; FSC PMT Voltage: E 02; SSC PMT: 474; Amp. Gains 7.1; FL-2 PMT: 539; compensation values: 0.
P) OMV Preparations
[0251]Bacteria were grown overnight on 5 GC plates, harvested with a loop and resuspended in 10 ml 20 mM Tris-HCl. Heat inactivation was performed at 56° C. for 30 minutes and the bacteria disrupted by sonication for 10 minutes on ice (50% duty cycle, 50% output). Unbroken cells were removed by centrifugation at 5000 g for 10 minutes and the total cell envelope fraction recovered by centrifugation at 50000 g at 4° C. for 75 minutes. To extract cytoplasmic membrane proteins from the crude outer membranes, the whole fraction was resuspended in 2% sarkosyl (Sigma) and incubated at room temperature for 20 minutes. The suspension was centrifuged at 10000 g for 10 minutes to remove aggregates, and the supernatant further ultracentrifuged at 50000 g for 75 minutes to pellet the outer membranes. The outer membranes were resuspended in 10 mM Tris-HCl, pH8 and the protein concentration measured by the Bio-Rad Protein assay, using BSA as a standard.
Q) Whole Extracts Preparation
[0252]Bacteria were grown overnight on a GC plate, harvested with a loop and resuspended in 1 ml of 20 mM Tris-HCl. Heat inactivation was performed at 56° C. for 30 minutes.
R) Western Blotting
[0253]Purified proteins (500ng/lane), outer membrane vesicles (5 μg) and total cell extracts (25 μg) derived from MenB strain 2996 were loaded on 15% SDS-PAGE and transferred to a nitrocellulose membrane. The transfer was performed for 2 hours at 150 mA at 4° C., in transferring buffer (0.3% Tris base, 1.44% glycine, 20% methanol). The membrane was saturated by overnight incubation at 4° C. in saturation buffer (10% skimmed milk, 0.1% Triton X100 in PBS). The membrane was washed twice with washing buffer (3% skimmed milk, 0.1% Triton X100 in PBS) and incubated for 2 hours at 37° C. with mice sera diluted 1:200 in washing buffer. The membrane was washed twice and incubated for 90 minutes with a 1:2000 dilution of horseradish peroxidase labelled anti-mouse Ig. The membrane was washed twice with 0.1% Triton X100 in PBS and developed with the Opti-4CN Substrate Kit (Bio-Rad). The reaction was stopped by adding water.
[0254]S) Bactericidal Assay
[0255]MC58 strain was grown overnight at 37° C. on chocolate agar plates. 5-7 colonies were collected and used to inoculate 7 ml Mueller-Hinton broth. The suspension was incubated at 37° C. on a nutator and let to grow until OD620was 0.5-0.8. The culture was aliquoted into sterile 1.5 ml Eppendorf tubes and centrifuged for 20 minutes at maximum speed in a microfuge. The pellet was washed once in Gey's buffer (Gibco) and resuspended in the same buffer to an OD620of 0.5, diluted 1:20000 in Gey's buffer and stored at 25° C.
[0256]50 μl of Gey's buffer/1% BSA was added to each well of a 96-well tissue culture plate. 25 μl of diluted mice sera (1:100 in Gey's buffer/0.2% BSA) were added to each well and the plate incubated at 4° C. 25 μl of the previously described bacterial suspension were added to each well. 25 μl of either heat-inactivated (56° C. waterbath for 30 minutes) or normal baby rabbit complement were added to each well. Immediately after the addition of the baby rabbit complement, 22 μl of each sample/well were plated on Mueller-Hinton agar plates (time 0). The 96-well plate was incubated for 1 hour at 37° C. with rotation and then 22 μl of each sample/well were plated on Mueller-Hinton agar plates (time 1). After overnight incubation the colonies corresponding to time 0 and time 1 hour were counted.
[0257]Table II (page 493) gives a summary of the cloning, expression and purification results.
Example 1
[0258]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 1>:
[0000]| 1 | ATGAAACAGA CAGTCAA.AT GCTTGCCGCC GCCCTGATTG CCTTGGGCTT | |
|
| 51 | GAACCGACCG GTGTGGNCGG ATGACGTATC GGATTTTCGG GAAAACTTGC |
|
| 101 | A.GCGGCAGC ACAGGGAAAT GCAGCAGCCC AATACAATTT GGGCGCAATG |
|
| 151 | TAT.TACAAA GGACGCGCGT GCGCCGGGAT GATGCTGAAG CGGTCAGATG |
|
| 201 | GTATCGGCAG CCGGCGGAAC AGGGGTTAGC CCAAGCCCAA TACAATTTGG |
|
| 251 | GCTGGATGTA TGCCAACGGG CGCGC.GTGC GCCAAGATGA TACCGAAGCG |
|
| 301 | GTCAGATGGT ATCGGCAGGC GGCAGCGCAG GGGGTTGTCC AAGCCCAATA |
|
| 351 | CAATTTGGGC GTGATATATG CCGAAGGACG TGGAGTGCGC CAAGACGATG |
|
| 401 | TCGAAGCGGT CAGATGGTTT CGGCAGGCGG CAGCGCAGGG GGTAGCCCAA |
|
| 451 | GCCCAAAACA ATTTGGGCGT GATGTATGCC GAAAGANCGC GCGTGCGCCA |
|
| 501 | AGACCG... |
[0259]This corresponds to the amino acid sequence <SEQ ID 2; ORF37>:
[0000]| 1 | MKQTVXMLAA ALIALGLNRP VWXDDVSDFR ENLXAAAQGN AAAQYNLGAM | |
|
| 51 | YXQRTRVRRD DAEAVRWYRQ PAEQGLAQAQ YNLGWMYANG RXVRQDDTEA |
|
| 101 | VRWYRQAAAQ GVVQAQYNLG VIYAEGRGVR QDDVEAVRWF RQAAAQGVAQ |
|
| 151 | AQNNLGVMYA ERXRVRQD... |
[0260]Further work revealed the complete nucleotide sequence <SEQ ID 3>:
[0000]| 1 | ATGAAACAGA CAGTCAAATG GCTTGCCGCC GCCCTGATTG CCTTGGGCTT | |
|
| 51 | GAACCGAGCG GTGTGGGCGG ATGACGTATC GGATTTTCGG GAAAACTTGC |
|
| 101 | AGGCGGCAGC ACAGGGAAAT GCAGCAGCCC AATACAATTT GGGCGCAATG |
|
| 151 | TATTACAAAG GACGCGGCGT GCGCCGGGAT GATGCTGAAG CGGTCAGATG |
|
| 201 | GTATCGGCAG GCGGCGGAAC AGGGGTTAGC CCAAGCCCAA TACAATTTGG |
|
| 251 | GCTGGATGTA TGCCAACGGG CGCGGCGTGC GCCAAGATGA TACCGAAGCG |
|
| 301 | GTCAGATGGT ATCGGCAGGC GGCAGCGCAG GGGGTTGTCC AAGCCCAATA |
|
| 351 | CAATTTGGGC GTGATATATG CCGAAGGACG TGGAGTGCGC CAAGACGATG |
|
| 401 | TCGAAGCGGT CAGATGGTTT CGGCAGGCGG CAGCGCAGGG GGTAGCCCAA |
|
| 451 | GCCCAAAACA ATTTGGGCGT GATGTATGCC GAAAGACGCG GCGTGCGCCA |
|
| 501 | AGACCGCGCC CTTGCACAAG AATGGTTTGG CAAGGCTTGT CAAAACGGAG |
|
| 551 | ACCAAGACGG CTGCGACAAT GACCAACGCC TGAAGGCGGG TTATTGA |
[0261]This corresponds to the amino acid sequence <SEQ ID 4; ORF37-1>:
[0000]| 1 | MKQTVKWLAA ALIALGLNRA VWADDVSDFR ENLQAAAQGN AAAQYNLGAM | |
|
| 51 | YYKGRGVRRD DAEAVRWYRQ AAEQGLAQAQ YNLGWMYANG RGVRQDDTEA |
|
| 101 | VRWYRQAAAQ GVVQAQYNLG VIYAEGRGVR QDDVEAVRWF RQAAAQGVAQ |
|
| 151 | AQNNLGVMYA ERRGVRQDRA LAQEWFGKAC QNGDQDGCDN DQRLKAGY* |
[0262]Further work identified the corresponding gene in strain A of N. meningitidis <SEQ ID 5>:
[0000]| 1 | ATGAAACAGA CAGTCAAATG GCTTGCCGCC GCCCTGATTG CCTTGGGCTT | |
|
| 51 | GAACCAAGCG GTGTGGGCGG ATGACGTATC GGATTTTCGG GAAAACTTGC |
|
| 101 | AGGCGGCAGC ACAGGGAAAT GCAGCAGCCC AAAACAATTT GGGCGTGATG |
|
| 151 | TATGCCGAAA GACGCGGCGT GCGCCAAGAC CGCGCCCTTG CACAAGAATG |
|
| 201 | GCTTGGCAAG GCTTGTCAAA ACGGATACCA AGACAGCTGC GACAATGACC |
|
| 251 | AACGCCTGAA AGCGGGTTAT TGA |
[0263]This encodes a protein having amino acid sequence <SEQ ID 6; ORF37a>:
[0000]| 1 | MKQTVKWLAA ALIALGLNQA VWADDVSDFR ENLQAAAQGN AAAQNNLGVM | |
|
| 51 | YAERRGVRQD RALAQEWLGK ACQNGYQDSC DNDQRLKAGY * |
[0264]The originally-identified partial strain B sequence (ORF37) shows 68.0% identity over a 75aa overlap with ORF37a:
[0000]
[0265]Further work identified the corresponding gene in N. gonorrhoeae <SEQ ID 7>:
[0000]| 1 | ATGAAACAGA CAGTCAAATG GCTTGCCGCC GCCCTGATTG CCTTGGGCTT | |
|
| 51 | GAACCAAGCG GTGTGGGCGG GTGACGTATC GGATTTTCGG GAAAACTTGC |
|
| 101 | AGgcggcaGA ACaggGAAAT GCAGCAGCCC AATTCAATTT GGGCGTGATG |
|
| 151 | TATGAAAATG GACAAGGAGT TCGTCAAGAT TATGTACAGG CAGTGCAGTG |
|
| 201 | GTATCGCAAG GCTTCAGAAC AAGGGGATGC CCAAGCCCAA TACAATTTGG |
|
| 251 | GCTTGATGTA TTACGATGGA CGCGGCGTGC GCCAAGACCT TGCGCTCGCT |
|
| 301 | CAACAATGGC TTGGCAAGGC TTGTCAAAAC GGAGACCAAA ACAGCTGCGA |
|
| 351 | CAATGACCAA CGCCTGAAGG CGGGTTATTA A |
[0266]This encodes a protein having amino acid sequence <SEQ ID 8; ORF37ng>:
[0000]| 1 | MKQTVKWLAA ALIALGLNQA VWAGDVSDFR ENLQAAEQGN AAAQFNLGVM | |
|
| 51 | YENGQGVRQD YVQAVQWYRK ASEQGDAQAQ YNLGLMYYDG RGVRQDLALA |
|
| 101 | QQWLGKACQN GDQNSCDNDQ RLKAGY* |
[0267]The originally-identified partial strain B sequence (ORF37) shows 64.9% identity over a 111aa overlap with ORF37ng:
[0000]
[0268]The complete strain B sequence (ORF37-1) and ORF37ng show 51.5% identity in 198 aa overlap:
[0000]
[0269]Computer analysis of these amino acid sequences indicates a putative leader sequence, and it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0270]ORF37-1 (11 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 1A shows the results of affinity purification of the GST-fusion protein, and FIG. 1B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for ELISA (positive result), FACS analysis (FIG. 1C), and a bactericidal assay (FIG. 1D). These experiments confirm that ORF37-1 is a surface-exposed protein, and that it is a useful immunogen.
[0271]FIG. 1E shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF37-1.
Example 2
[0272]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 9>:
[0000] | TTCGGCGA CATCGGCGGT TTGAAGGTCA ATGCCCCCGT |
| |
| CAAATCCGCA GGCGTATTGG TCGGGCGCGT CGGCGCTATC |
| |
| GGACTTGACC CGAAATCCTA TCAGGCGAGG GTGCGCCTCG |
| |
| ATTTGGACGG CAAGTATCAG TTCAGCAGCG ACGTTTCCGC |
| |
| GCAAATCCTG ACTTCsGGAC TTTTGGGCGA GCAGTACATC |
| |
| GGGCTGCAGC AGGGCGGCGA CACGGAAAAC CTTGCTGCCG |
| |
| GCGACACCAT CTCCGTAACC AGTTCTGCAA TGGTTCTGGA |
| |
| AAACCTTATC GGCAAATTCA TGACGAGTTT TGCCGAGAAA |
| |
| AATGCCGACG GCGGCAATGC GGAAAAAGCC GCCGAATAA |
[0273]This corresponds to the amino acid sequence <SEQ ID 10>:
[0000]| 1 | FGDIGGLKVN APVKSAGVLV GRVGAIGLDP KSYQARVRLD LDGKYQFSSD | |
|
| 51 | VSAQILTSGL LGEQYIGLQQ GGDTENLAAG DTISVTSSAM VLENLIGKFM |
|
| 101 | TSFAEKNADG GNAEKAAE* |
[0274]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Hypothetical H. influenzae Protein (ybrd.haein; Accession Number p45029)
[0275]SEQ ID 9 and ybrd.haein show 48.4% aa identity in 122 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0276]SEQ ID 9 shows 99.2% identity over a 118aa overlap with a predicted ORF from N. gonorrhoeae.
[0000]
[0277]The complete yrbd H. influenzae sequence has a leader sequence and it is expected that the full-length homologous N. meningitidis protein will also have one. This suggests that it is either a membrane protein, a secreted protein, or a surface protein and that the protein, or one of its epitopes, could be a useful antigen for vaccines or diagnostics.
Example 3
[0278]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 11>:
[0000]| 1 | ..ATTTTGATAT ACCTCATCCG CAAGAATCTA GGTTCGCCCG TCTTCTTCTT | |
|
| 51 | TCAGGAACGC CCCGGAAAGG ACGGAAAACC TTTTAAAATG GTCAAATTCC |
|
| 101 | GTTCCATGCG CGACGGCTTG TATTCAGACG GCATTCCGCT GCCCGACGGA |
|
| 151 | GAACGCCTGA CACCGTTCGG CAAAAAACTG CGTGCCGcCA GTwTGGACGA |
|
| 201 | ACTGCCTGAA TTATGGAATA TCTTAAAAGG CGAGATGAGC CTGGTCGGCC |
|
| 251 | CCCGCCCGCT GCTGATGCAA TATCTGCCGC TGTACGACAA CTTCCAAAAC |
|
| 301 | CGCCGCCACG AAATGAAACC CGGCATTACC GGCTGGGCGC AGGTCAACGG |
|
| 351 | GCGCAACGCg CTTTCGTGGG ACGAAAAATT CGCCTGCGAT GTTTGGTATA |
|
| 401 | TCGACCACTT CAGCCTGTGC CTCGACATCA AAATCCTACT GCTGACGGTT |
|
| 451 | AAAAAAGTAT TAATCAAGGA AGGGATTTCC GCACAGGGCG AACA.aCCAT |
|
| 501 | GCCCCCTTTC ACAGGAAAAC GCAAACTCGC CGTCGTCGGT GCGGGCGGAC |
|
| 551 | ACGGAAAAGT CGTTGCCGAC CTTGCCGCCG CACTCGGCCG GTACAGGGAA |
|
| 601 | ATCGTTTTTC TGGACGACCG CGCACAAGGC AGCGTCAACG GCTTTTCCGT |
|
| 651 | CATCGGCACG ACGCTGCTGC TTGAAAACAG TTTATCGCCC GAACAATACG |
|
| 701 | ACGTCGCCGT CGCCGTCGGC AACAACCGCA TCCGCCGCCA AATCGCCGAA |
|
| 751 | AAAGCCGCCG CGCTCGGCTT CGCCCTGCCC GTACTGGTTC ATCCGGACGC |
|
| 801 | GACCGTCTCG CCTTCTGCAA CAGTCGGACA AGGCAGCGTC GTTATGGCGA |
|
| 851 | AAGCGGTCG.. |
[0279]This corresponds to the amino acid sequence <SEQ ID 12; ORF3>:
[0000]| 1 | ..ILIYLIRKNL GSPVFFFQER PGKDGKPFKM VKFRSMRDGL YSDGIPLPDG | |
|
| 51 | ERLTPFGKKL RAASXDELPE LWNILKGEMS LVGPRPLLMQ YLPLYDNFQN |
|
| 101 | RRHEMKPGIT GWAQVNGRNA LSWDEKFACD VWYIDHFSLC LDIKILLLTV |
|
| 151 | KKVLIKEGIS AQGEXTMPPF TGKRKLAVVG AGGHGKVVAD LAAALGRYRE |
|
| 201 | IVFLDDRAQG SVNGFSVIGT TLLLENSLSP EQYDVAVAVG NNRIRRQIAE |
|
| 251 | KAAALGFALP VLVHPDATVS PSATVGQGSV VMAKAV.. |
[0280]Further sequence analysis revealed the complete nucleotide sequence <SEQ ID 13>:
[0000]| 1 | ATGAGTAAAT TCTTCAAACG CCTGTTTGAC ATTGTTGCCT CCGCCTCGGG | |
|
| 51 | ACTGATTTTC CTCTCGCCAG TATTTTTGAT TTTGATATAC CTCATCCGCA |
|
| 101 | AGAATCTAGG TTCGCCCGTC TTCTTCTTTC AGGAACGCCC CGGAAAGGAC |
|
| 151 | GGAAAACCTT TTAAAATGGT CAAATTCCGT TCCATGCGCG ACGCGCTTGA |
|
| 201 | TTCAGACGGC ATTCCGCTGC CCGACGGAGA ACGCCTGACA CCGTTCGGCA |
|
| 251 | AAAAACTGCG TGCCGCCAGT TTGGACGAAC TGCCTGAATT ATGGAATATC |
|
| 301 | TTAAAAGGCG AGATGAGCCT GGTCGGCCCC CGCCCGCTGC TGATGCAATA |
|
| 351 | TCTGCCGCTG TACGACAACT TCCAAAACCG CCGCCACGAA ATGAAACCCG |
|
| 401 | GCATTACCGG CTGGGCGCAG GTCAACGGGC GCAACGCGCT TTCGTGGGAC |
|
| 451 | GAAAAATTCG CCTGCGATGT TTGGTATATC GACCACTTCA GCCTGTGCCT |
|
| 501 | CGACATCAAA ATCCTACTGC TGACGGTTAA AAAAGTATTA ATCAAGGAAG |
|
| 551 | GGATTTCCGC ACAGGGCGAA GCCACCATGC CCCCTTTCAC AGGAAAACGC |
|
| 601 | AAACTCGCCG TCGTCGGTGC GGGCGGACAC GGAAAAGTCG TTGCCGACCT |
|
| 651 | TGCCGCCGCA CTCGGCCGGT ACAGGGAAAT CGTTTTTCTG GACGACCGCG |
|
| 701 | CACAAGGCAG CGTCAACGGC TTTTCCGTCA TCGGCACGAC GCTGCTGCTT |
|
| 751 | GAAAACAGTT TATCGCCCGA ACAATACGAC GTCGCCGTCG CCGTCGGCAA |
|
| 801 | CAACCGCATC CGCCGCCAAA TCGCCGAAAA AGCCGCCGCG CTCGGCTTCG |
|
| 851 | CCCTGCCCGT TCTGGTTCAT CCGGACGCGA CCGTCTCGCC TTCTGCAACA |
|
| 901 | GTCGGACAAG GCAGCGTCGT TATGGCGAAA GCCGTCGTAC AGGCAGGCAG |
|
| 951 | CGTATTGAAA GACGGCGTGA TTGTGAACAC TGCCGCCACC GTCGATCACG |
|
| 1001 | ACTGCCTGCT TAACGCTTTC GTCCACATCA GCCCAGGCGC GCACCTGTCG |
|
| 1051 | GGCAACACGC ATATCGGCGA AGAAAGCTGG ATAGGCACGG GCGCGTGCAG |
|
| 1101 | CCGCCAGCAG ATCCGTATCG GCAGCCGCGC AACCATTGGA GCGGGCGCAG |
|
| 1151 | TCGTCGTACG CGACGTTTCA GACGGCATGA CCGTCGCGGG CAATCCGGCA |
|
| 1201 | AAGCCGCTGC CGCGCAAAAA CCCCGAGACC TCGACAGCAT AA |
[0281]This corresponds to the amino acid sequence <SEQ ID 14; ORF3-1>:
[0000]| 1 | MSKFFKRLFD IVASASGLIF LSPVFLILIY LIRKNLGSPV FFFQERPGKD | |
|
| 51 | GKPFKMVKFR SMRDALDSDG IPLPDGERLT PFGKKLRAAS LDELPELWNI |
|
| 101 | LKGEMSLVGP RPLLMQYLPL YDNFQNRRHE MKPGITGWAQ VNGRNALSWD |
|
| 151 | EKFACDVWYI DHFSLCLDIK ILLLTVKKVL IKEGISAQGE ATMPPFTGKR |
|
| 201 | KLAVVGAGGH GKVVADLAAA LGRYREIVFL DDRAQGSVNG FSVIGTTLLL |
|
| 251 | ENSLSPEQYD VAVAVGNNRI RRQIAEKAAA DGFALPVLVH PDATVSPSAT |
|
| 301 | VGQGSVVMAK AVVQAGSVLK DGVIVNTAAT VDHDCLLNAF VHISPGAHLS |
|
| 351 | GNTHIGEESW IGTGACSRQQ IRIGSRATIG AGAVVVRDVS DGMTVAGNPA |
|
| 401 | KPLPRKNPET STA* |
[0282]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0283]ORF3 shows 93.0% identity over a 286aa overlap with an ORF (ORF3a) from strain A of N. meningitidis.
[0000]
[0284]The complete length ORF3a nucleotide sequence <SEQ ID 15> is:
[0000]| 1 | ATGAGTAAAT TCTTCAAACG CCTGTTTGAC ATTGTTGCCT CCGCCTCGGG | |
|
| 51 | ACTGATTTTC CTCTCGCCAG TATTTTTGAT TTTGATATAC CTCATCCGCA |
|
| 101 | AGAATCTGGG TTCGCCCGTC TTCTTCTTTC AGGAACGCCC CGGAAAGGAC |
|
| 151 | GGAAAACCTT TTAAAATGGT CAAATTCCGT TCCATGCACG ACGCGCTTGA |
|
| 201 | TTCAGACGGC ATTCTGCTGC CCGACGGAGA ACGCCTGACA CCGTTCGGCA |
|
| 251 | AAAAACTGCG TGCCGCCAGT TTGGACGAAC TGCCCGAACT GTGGAACGTC |
|
| 301 | CTCAAAGGCG ACATGAGCCT GGTCGGCCCC CGCCCGCTGC TGATGCAATA |
|
| 351 | TCTGCCGCTG TACGACAACT TCCAAAACCG CCGCCACGAA ATGAAACCGG |
|
| 401 | GCATTACCGG CTGGGCGCAG GTCAACGGGC GCAACGCGCT TTCGTGGGAC |
|
| 451 | GAACGCTTCG CATGCGACAT CTGGTATATC GACCACTTCA GCCTGTGCCT |
|
| 501 | CGAGATCAAA ATCCTACTGC TGACGGTTAA AAAAGTATTA ATCAAAGAAG |
|
| 551 | GGATTTCCGC ACAGGGCGAA GCCACCATGC CCCCTTTCAC AGGAAAACGC |
|
| 601 | AAACTTGCCG TCGTCGGTGC GGGCGGACAC GGCAAAGTCG TTGCCGAGCT |
|
| 651 | TGCCGCCGCA CTCGGCACAT ACGGCGAAAT CGTTTTTCTG GACGACCGCG |
|
| 701 | TCCAAGGCAG CGTCAACGGC TTCCCCGTCA TCGGCACGAC GCTGCTGCTT |
|
| 751 | GAAAACAGTT TATCGCCCGA ACAATTCGAC ATCGCCGTCG CCGTCGGCAA |
|
| 801 | CAACCGCATC CGCCGCCAAA TCGCCGAAAA AGCCGCCGCG CTCGGCTTCG |
|
| 851 | CCCTGCCCGT CCTGATTCAT CCGGACTCGA CCGTCTCGCC TTCTGCAACA |
|
| 901 | GTCGGACAAG GCGGCGTCGT TATGGCGAAA GCCGTCGTAC AGGCTGACAG |
|
| 951 | CGTATTGAAA GACGGCGTAA TTGTGAACAC TGCCGCCACC GTCGATCACG |
|
| 1001 | ATTGCCTGCT TGATGCTTTC GTCCACATCA GCCCGGGCGC GCACCTGTCG |
|
| 1051 | GGCAACACGC GTATCGGCGA AGAAAGCTGG ATAGGCACAG GCGCGTGCAG |
|
| 1101 | CCGCCAGCAG ATCCGTATCG GCAGCCGCGC AACCATTGGA GCGGGCGCAG |
|
| 1151 | TCGTCGTGCG CGACGTTTCA GACGGCATGA CCGTCGCGGG CAACCCGGCA |
|
| 1201 | AAACCATTGG CAGGCAAAAA TACCGAGACC CTGCGGTCGT AA |
[0285]This is predicted to encode a protein having amino acid sequence <SEQ ID 16>:
[0000]| 1 | MSKFFKRLFD IVASASGLIF LSPVFLILIY LIRKNLGSPV FFFQERPGKD | |
|
| 51 | GKPFKMVKFR SMHDALDSDG ILLPDGERLT PFGKKLRAAS LDELPELWNV |
|
| 101 | LKGDMSLVGP RPLLMQYLPL YDNFQNRRHE MKPGITGWAQ VNGRNALSWD |
|
| 151 | ERFACDIWYI DHFSLCLDIK ILLLTVKKVL IKEGISAQGE ATMPPFTGKR |
|
| 201 | KLAVVGAGGH GKVVAELAAA LGTYGEIVFL DDRVQGSVNG FPVIGTTLLL |
|
| 251 | ENSLSPEQFD IAVAVGNNRI RRQIAEKAAA LGFALPVLIH PDSTVSPSAT |
|
| 301 | VGQGGVVMAK AVVQADSVLK DGVIVNTAAT VDHDCLLDAF VHISPGAHLS |
|
| 351 | GNTRIGEESW IGTGACSRQQ IRIGSRATIG AGAVVVRDVS DGMTVAGNPA |
|
| 401 | KPLAGKNTET LRS* |
[0286]Two transmembrane domains are underlined.
[0287]ORF3-1 shows 94.6% identity in 410 aa overlap with ORF3a:
[0000]
[0000]Homology with Hypothetical Protein Encoded by yvfc Gene (Accession Z71928) of B. subtilis
[0288]ORF3 and YVFC proteins show 55% aa identity in 170 aa overlap (BLASTp):
[0000]| ORF3 | 3 | IYLIRKNLGSPVFFFQERPGKDGKPFKMVKFRSMRDGLYSDGIPLPDGERLTPFGKKLRA | 62 | |
| | I ++R +GSPVFF Q RPG GKPF + KFR+M D S G LPD RLT G+ +R | |
| yvfc | 27 | IAVVRLKIGSPVFFKQVRPGLHGKPFTLYKFRTMTDERDSKGNLLPDEVRLTKTGRLIRK | 86 |
|
| ORF3 | 63 | ASXDELPELWNILKGEMSLVGPRPLLMQYLPLYDNFQNRRHEMKPGITGWAQVNGRNALS | 122 |
| | S DELP+L N+LKG++SLVGPRPLLM YLPLY Q RRHE+KPGITGWAQ+NGRNA+S | |
| yvfc | 87 | LSIDELPQLLNVLKGDLSLVGPRPLLMDYLPLYTEKQARRHEVKPGITGWAQINGRNAIS | 146 |
|
| ORF3 | 123 | WDEKFACDVWYIDHFSLCLDXXXXXXXXXXXXXXEGISAQGEXTMPPFTG | 172 |
| | W++KF DVWY+D++S LD EGI T FTG | |
| yvfc | 147 | WEKKFELDVWYVDNWSFFLDLKILCLTVRKVLVSEGIQQTNHVTAERFTG | 196 |
Homology with a Predicted ORF from
N. gonorrhoeae [0289]ORF3 shows 86.3% identity over a 286aa overlap with a predicted ORF (ORF3.ng) from N. gonorrhoeae:
[0000]
[0290]The complete length ORF3ng nucleotide sequence <SEQ ID 17> is:
[0000]| 1 | ATGAGTAAAG CCGTCAAACG CCTGTTCGAC ATCATCGCAT CCGCATCGGG | |
|
| 51 | GCTGATTGTC CTGTCGCCCG TGTTTTTGGT TTTAATATAC CTCATCCGCA |
|
| 101 | AAAACTTAGG TTCGCCCGTC TTCTTCattC GGGAACGCCc cgGAAAGGAc |
|
| 151 | ggaaaacCTT TTAAAATGGT CAAATTCCGT TCCAtgcgcg acgcgcttGA |
|
| 201 | TTCAGACGGC ATTCCGCTGC CCGATAGCGA ACGCCTGACC GATTTCGGCA |
|
| 251 | AAAAATTACG CGCCACCAGT TTGGACGAAC TTCCTGAATT ATGGAATGTC |
|
| 301 | CTCAAAGGCG AGATGAGCCT GGTCGGCCCC CGCCCGCTTT TGATGCAGTA |
|
| 351 | TCTGCCGCTT TACAACAAAT TTCAAAACCG CCGCCACGAA ATGAAACCGG |
|
| 401 | GCATTACCGG CTGGGCGCAG GTCAACGGGC GCAACGCGCT TTCGTGGGAC |
|
| 451 | GAAAAGTTCT CCTGCGATGT TTGGTACACC GACAATTTCA GCTTTTGGCT |
|
| 501 | GGATATGAAA ATCCTGTTTC TGACAGTCAA AAAAGTCTTG ATTAAAGAAG |
|
| 551 | GCATTTCGGC GCAAGGGGAA GCCACCATGC CCCCTTTCGC GGGGAATCGC |
|
| 601 | AAACTCGCCG TTATCGGCGC GGGCGGACAC GGCAAAGTCG TTGCCGAGCT |
|
| 651 | TGCCGCCGCA CTCGGCACAT ACGGCGAAAT CGTTTTTCTG GACGACCGCA |
|
| 701 | CCCAAGGCAG CGTCAACGGC TTCCCCGTCA TCGGCACGAC GCTGCTGCTT |
|
| 751 | GAAAACAGTT TATCGCCCGA ACAATTCGAC ATCACCGTCG CCGTCGGCAA |
|
| 801 | CAACCGCATC CGCCGCCAAA TCACCGAAAA CGCCGCCGCG CTCGGCTTCA |
|
| 851 | AACTGCCCGT TCTGATTCAT CCCGACGCGA CCGTCTCGCC TTCTGCAATA |
|
| 901 | ATCGGACAAG GCAGCGTCGT AATGGCGAAA GCCGTCGTAC AGGCCGGCAG |
|
| 951 | CGTATTGAAA GACGGCGTGA TTGTGAACAC TGCCGCCACC GTCGATCACG |
|
| 1001 | ACTGCCTGCT TGACGCTTTC GtccaCATCA GCCCGGGCGC GCACCTGTCG |
|
| 1051 | GGCAACACGC GTATCGGCGA AGAAAGCCGG ATAGGCACGG GCGCGTGCAG |
|
| 1101 | CCGCCAGCAG ACAACCGTCG GCAGCGGGGT TACCgccgGT GCAGGGgcGG |
|
| 1151 | TTATCGTATG CGACATCCCG GACGGCATGA CCGTCGCGGG CAACCCGGCA |
|
| 1201 | AAGCCCCTTA CGGGCAAAAA CCCCAAGACC GGGACGGCAT AA |
[0291]This encodes a protein having amino acid sequence <SEQ ID 18>:
[0000]| 1 | MSKAVKRLFD IIASASGLIV LSPVFLVLIY LIRKNLGSPV FFIRERPGKD |
|
| 51 | GKPFKMVKFR SMRDALDSDG IPLPDSERLT DFGKKLRATS LDELPELWNV |
|
| 101 | LKGEMSLVGP RPLLMQYLPL YNKFQNRRHE MKPGITGWAQ VNGRNALSWD |
|
| 151 | EKFSCDVWYT DNFSFWLDMK ILFLTVKKVL IKEGISAQGE ATMPPFAGNR |
|
| 201 | KLAVIGAGGH GKVVAELAAA LGTYGEIVFL DDRTQGSVNG FPVIGTTLLL |
|
| 251 | ENSLSPEQFD ITVAVGNNRI RRQITENAAA LGFKLPVLIH PDATVSPSAI |
|
| 301 | IGQGSVVMAK AVVQAGSVLK DGVIVNTAAT VDHDCLLDAF VHISPGAHLS |
|
| 351 | GNTRIGEESR IGTGACSRQQ TTVGSGVTAG AGAVIVCDIP DGMTVAGNPA |
|
| 401 | KPLTGKNPKT GTA* |
[0292]This protein shows 86.9% identity in 413 aa overlap with ORF3-1:
[0000]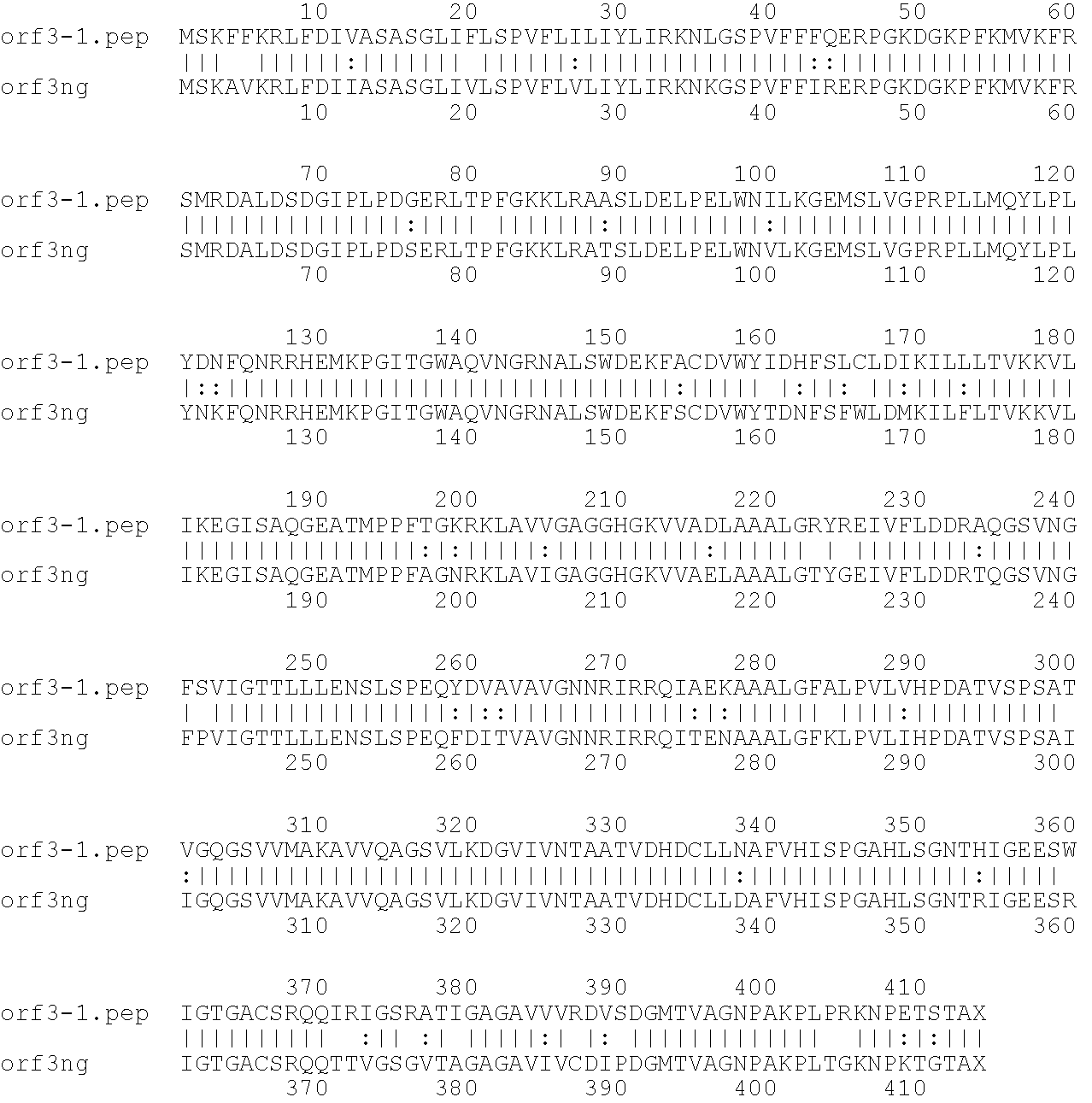
[0293]In addition, ORF3ng shows significant homology with a hypothetical protein from B. subtilis:
[0000]| gnl|PID|e238668 (Z71928) hypothetical protein [Bacillus subtilis] | |
| >gi|1945702|gnl|PID|e313004 (Z94043) hypothetical protein [Bacillus subtilis] |
| >gi|2635938|gnl|PID|e1186113 (Z99121) similar to capsular polysaccharide |
| biosynthesis [Bacillus subtilis]Length = 202 |
| Score = 235 bits (594), Expect = 3e−61 |
| Identities = 114/195 (58%), Positives = 142/195 (72%) |
|
| Query: | 5 | VKRLFDIIASASGLIVLSPVFLVLIYLIRKNLGSPVFFIRERPGKDGKPFKMVKFRSMRD | 64 | |
| | +KRLFD+ A+ L S + L I ++R +GSPVFF + RPG GKPF + KFR+M D | |
| Sbjct: | 3 | LKRLFDLTAAIFLLCCTSVIILFTIAVVRLKIGSPVFFKQVRPGLHGKPFTLYKFRTMTD | 62 |
|
| Query: | 65 | ALDSDGIPLPDSERLTDFGKKLRATSLDELPELWNVLKGEMSLVGPRPLLMQYLPLYNKF | 124 |
| | DS G LPD RLT G+ +R S+DELP+L NVLKG++SLVGPRPLLM YLPLY + | |
| Sbjct: | 63 | ERDSKGNLLPDEVRLTKTGRLIRKLSIDELPQLLNVLKGDLSLVGPRPLLMDYLPLYTEK | 122 |
|
| Query: | 125 | QNRRHEMKPGITGWAQVNGRNALSWDEKFSCDVWYTDNFSFWLDMKILFLTVKKVLIKEG | 184 |
| | Q RRHE+KPGITGWAQ+NGRNA+SW++KF DVWY DN+SF+LD+KIL LTV+KVL+ EG | |
| Sbjct: | 123 | QARRHEVKPGITGWAQINGRNAISWEKKFELDVWYVDNWSFFLDLKILCLTVRKVLVSEG | 182 |
|
| Query: | 185 | ISAQGEATMPPFAGN | 199 |
| | I T F G+ | |
| Sbjct: | 183 | IQQTNHVTAERFTGS | 197 |
[0294]The hypothetical product of yvfc gene shows similarity to EXOY of R. meliloti, an exopolysaccharide production protein. Based on this and on the two predicted transmembrane regions in the homologous N. gonorrhoeae sequence, it is predicted that these proteins, or their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 4
[0295]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 19>:
[0000]| 1 | ..AACCATATGG CGATTGTCAT CGACGAATAC GGCGGCACAT CCGGCTTGGT | |
|
| 51 | CACCTTTGAA GACATCATCG AGCAAATCGT CGGCGAAATC GAAGACGAGT |
|
| 101 | TTGACGAAGA CGATAGCGCC GACAATATCC ATGCCGTTTC TTCAGACACG |
|
| 151 | TGGCGCATCC ATGCAGCTAC CGAAATCGAA GACATCAACA CCTTCTTCGG |
|
| 201 | CACGGAATAC AGCATCGAAG AAGCCGACAC CATT.GGCGG CCTGGTCATT |
|
| 251 | CAAGAGTTGG GACATCTGCC CGTGCGCGGC GAAAAAGTCC TTATCGGCGG |
|
| 301 | TTTGCAGTTC ACCGTCGCAC GCGCCGACAA CCGCCGCCTG CATACGCTGA |
|
| 351 | TGGCGACCCG CGTGAAGTAA GC........ .....ACCGC CGTTTCTGCA |
|
| 401 | CAGTTTAG |
[0296]This corresponds to amino acid sequence <SEQ ID 20; ORF5>:
[0000]| 1 | ..NHMAIVIDEY GGTSGLVTFE DIIEQIVGEI EDEFDEDDSA DNIHAVSSDT | |
|
| 51 | WRIHAATEIE DINTFFGTEY SIEEADTIXR PGHSRVGTSA RARRKSPYRR |
|
| 101 | FAVHRRTRRQ PPPAYADGDP REVS....XR RFCTV* |
[0297]Further sequence analysis revealed the complete DNA sequence to be <SEQ ID 21>:
[0000]| 1 | ATGGACGGCG CACAACCGAA AACGAATTTT TTTGAACGCC TGATTGCCCG | |
|
| 51 | ACTCGCCCGC GAACCCGATT CCGCCGAAGA CGTATTAAAC CTGCTTCGGC |
|
| 101 | AGGCGCACGA GCAGGAAGTT TTTGATGCGG ATACGCTTTT AAGATTGGAA |
|
| 151 | AAAGTCCTCG ATTTTTCCGA TTTGGAAGTG CGCGACGCGA TGATTACGCG |
|
| 201 | CAGCCGTATG AACGTTTTAA AAGAAAACGA CAGCATCGAG CGCATCACCG |
|
| 251 | CCTACGTTAT CGATACCGCC CATTCGCGCT TCCCCGTCAT CGGCGAAGAC |
|
| 301 | AAAGACGAAG TTTTGGGCAT TTTGCACGCC AAAGACCTGC TCAAATATAT |
|
| 351 | GTTTAACCCC GAGCAGTTCC ACCTCAAATC CATTCTCCGC CCCGCCGTCT |
|
| 401 | TCGTCCCCGA AGGCAAATCG CTGACCGCCC TTTTAAAAGA GTTCCGCGAA |
|
| 451 | CAGCGCAACC ATATGGCGAT TGTCATCGAC GAATACGGCG GCACATCCGG |
|
| 501 | CTTGGTCACC TTTGAAGACA TCATCGAGCA AATCGTCGGC GAAATCGAAG |
|
| 551 | ACGAGTTTGA CGAAGACGAT AGCGCCGACA ATATCCATGC CGTTTCTTCC |
|
| 601 | GAACGCTGGC GCATCCATGC AGCTACCGAA ATCGAAGACA TCAACACCTT |
|
| 651 | CTTCGGCACG GAATACAGCA GCGAAGAAGC CGACACCATT CGGCCTGGTC |
|
| 701 | ATTCAAGAGT TGGGACATCT GCCCGTGCGC GGCGAAAAAG TCCTTATCGG |
|
| 751 | CGGTTTGCAG TTCACCGTCG CACGCGCCGA CAACCGCCGC CTGCATACGC |
|
| 801 | TGATGGCGAC CCGCGTGAAG TAAGCACCGC CGTTTCTGCA CAGTTTAGGA |
|
| 851 | TGACGGTACG GGCGTTTTCT GTTTCAATCC GCCCCATCCG CCAAACATAA |
[0298]This corresponds to amino acid sequence <SEQ ID 22; ORF5-1>:
[0000]| 1 | MDGAQPKTNF FERLIARLAR EPDSAEDVLN LLRQAHEQEV FDADTLLRLE | |
|
| 51 | KVLDFSDLEV RDAMITRSRM NVLKENDSIE RITAYVIDTA HSRFPVIGED |
|
| 101 | KDEVLGILHA KDLLKYMFNP EQFHLKSILR PAVFVPEGKS LTALLKEFRE |
|
| 151 | QRNHMAIVID EYGGTSGLVT FEDIIEQIVG EIEDEFDEDD SADNIHAVSS |
|
| 201 | ERWRIHAATE IEDINTFFGT EYSSEEADTI RPGHSRVGTS ARARRKSPYR |
|
| 251 | RFAVHRRTRR QPPPAYADGD PREVSTAVSA QFRMTVRAFS VSIRPIRQT* |
[0299]Further work identified the corresponding gene in strain A of N. meningitidis <SEQ ID 23>:
[0000]| 1 | ATGGACGGCG CACAACCGAA AACAAATTTT TTNNAACGCC TGATTGCCCG | |
|
| 51 | ACTCGCCCGC GAACCCGATT CCGCCGAAGA CGTATTGACC CTGTTGCGCC |
|
| 101 | AAGCGCACGA ACAGGAAGTA TTTGATGCGG ATACGCTTTT AAGATTGGAA |
|
| 151 | AAAGTCCTCG ATTTTTCTGA TTTGGAAGTG CGCGACGCGA TGATTACGCG |
|
| 201 | CAGCCGTATG AACGTTTTAA AAGAAAACGA CAGCATCGAA CGCATCACCG |
|
| 251 | CCTACGTTAT CGATACCGCC CATTCGCGCT TCCCCGTCAT CGGTGAAGAC |
|
| 301 | AAAGACGAAG TTTTGGGTAT TTTGCACGCC AAAGACCTGC TCAAATATAT |
|
| 351 | GTTCAACCCC GAGCAGTTCC ACCTCAAATC GATATTGCGC CCTGCCGTCT |
|
| 401 | TCGTCCCCGA AGGCAAATCG CTGACCGCCC TTTTAAAAGA GTTCCGCGAA |
|
| 451 | CAGCGCAACC ATATGGCAAT CGTCATCGAC GAATACGGCG GCACGTCGGG |
|
| 501 | TTTGGTAACT TTTGAAGACA TCATCGAGCA AATCGTCGGC GACATCGAAG |
|
| 551 | ATGAGTTTGA CGAAGACGAA AGCGCGGACA ACATCCACGC CGTTTCCGCC |
|
| 601 | GAACGCTGGC GCATCCACGC GGCTACCGAA ATCGAAGACA TCAACGCCTT |
|
| 651 | TTTCGGCACG GAATACAGCA GCGAAGAAGC CGACACCATC GGCGGCCNTG |
|
| 701 | GTCATTCAGG AATTGGNACA CCTGCCCGTG CGCGGCGAAA AAGTCNTTAT |
|
| 751 | CGGCGNNTTG CANTTCACNG TCGCCNGCGC NGACAACCGC CGCCTGCATA |
|
| 801 | CGCTGATGGC GACCCGCGTG AAGTAAGCTC CGCCGTTTCT GTACAGTTTA |
|
| 851 | GGATGACGGT ACGGGCGTTT TCTGTTTCAA TCCGCCCCAT CCGCCANACA |
|
| 901 | TAA |
[0300]This encodes a protein having amino acid sequence <SEQ ID 24; ORF5a>:
[0000]| 1 | MDGAQPKTNF XXRLIARLAR EPDSAEDVLT LLRQAHEQEV FDADTLLRLE | |
|
| 51 | KVLDFSDLEV RDAMITRSRM NVLKENDSIE RITAYVIDTA HSRFPVIGED |
|
| 101 | KDEVLGILHA KDLLKYMFNP EQFHLKSILR PAVFVPEGKS LTALLKEFRE |
|
| 151 | QRNHMAIVID EYGGTSGLVT FEDIIEQIVG DIEDEFDEDE SADNIHAVSA |
|
| 201 | ERWRIHAATE IEDINAFFGT EYSSEEADTI GGXGHSGIGT PARARRKSXY |
|
| 251 | RRXAXHXRXR XQPPPAYADG DPREVSSAVS VQFRMTVRAF SVSIRPIRXT |
|
| 301 | * |
[0301]The originally-identified partial strain B sequence (ORF5) shows 54.7% identity over a 124aa overlap with ORF5a:
[0000]
[0302]The complete strain B sequence (ORF5-1) and ORF5a show 92.7% identity in 300 aa overlap:
[0000]
[0303]Further work identified the a partial DNA sequence in N. gonorrhoeae <SEQ ID 25> which encodes a protein having amino acid sequence <SEQ ID 26; ORF5ng>:
[0000]| 1 | MDGAQPKTNF FERLIARLAR EPDSAEDVLN LLRQAHEQEV FDADTLTRLE | |
|
| 51 | KVLDFAELEV RDAMITRSRM NVLKENDSIE RITAYVIDTA HSRFPVIGED |
|
| 101 | KDEVLGILHA KDLLKYMFNP EQFHLKSVLR PAVFVPEGKS LTALLKEFRE |
|
| 151 | QRNHMAIVID EYGGTSGLVT FEDIIEQIVG DIEDEFDEDE SADDIHSVSA |
|
| 201 | ERWRIHAATE IEDINAFFGT EYGSEEADTI RRLGHSGIGT PARARRKSPY |
|
| 251 | RRFAVHRRPR RQPPPAHADG DPREVSRACP HRRFCTV* |
[0304]Further analysis revealed the complete gonococcal nucleotide sequence <SEQ ID 27> to be:
[0000]| 1 | ATGGACGGCG CACAACCGAA AACAAATTTT TTTGAACGCC TGATTGCCCG | |
|
| 51 | ACTCGCCCGC GAACCCGATT CCGCCGAAGA CGTATTAAAC CTGCTTCGGC |
|
| 101 | AGGCGCACGA ACAGGAAGTT TTTGATGCCG ACACACTGAC CCGGCTGGAA |
|
| 151 | AAAGTATTGG ACTTTGCCGA GCTGGAAGTG CGCGATGCGA TGATTACGCG |
|
| 201 | CAGCCGCATG AACGTATTGA AAGAAAACGA CAGCATCGAA CGCATCACCG |
|
| 251 | CCTACGTCAT CGATACCGCC CATTCGCGCT TCCCCGTCAT CGGCGAAGAC |
|
| 301 | AAAGACGAAG TTTTGGGCAT TTTGCACGCC AAAGACCTGC TCAAATATAT |
|
| 351 | GTTCAACCCC GAGCAGTTCC ACCTGAAATC CGTCTTGCGC CCTGCCGTTT |
|
| 401 | TCGTGCCCGA AGGCAAATCT TTGACCGCCC TTTTAAAAGA GTTCCGCGAA |
|
| 451 | CAGCGCAACC ATATGGCAAT CGTCATCGAC GAATACGGCG GCACGTCGGG |
|
| 501 | TTTGGTCACC TTTGAAGACA TCATCGAGCA AATCGTCGGT GACATCGAAG |
|
| 551 | ACGAGTTTGA CGAAGACGAA AGCGccgacg acatCCACTC cgTTTccgCC |
|
| 601 | GAACGCTGGC GCATCCacgc ggctaCCGAA ATCGAAGaca TCAACGCCTT |
|
| 651 | TTTCGGTACG GAatacggca gcgaagaagc cgacaccatc cggcggctTG |
|
| 701 | GTCATTCAGG AATTGGGACA CCTGCCCGTG CGCGGCGAAA AAGTCCTTAt |
|
| 751 | cggcgGTTTG Cagttcaccg tCGCCCGCGC CGACAACCGC CGCCTGCACA |
|
| 801 | CGCTGATGGC GACCCGCGTG AAGTAAGCAG AGCCTGCCcg AccgccgttT |
|
| 851 | CTGCacAGTT TAGGatgACG gtaCGGTCGT TTTCTGTTTC AATCCGCCCC |
|
| 901 | ATCCGCCAAA CATAA |
[0305]This encodes a protein having amino acid sequence <SEQ ID 28; ORF5ng-1>:
[0000]| 1 | MDGAQPKTNF FERLIARLAR EPDSAEDVLN LLRQAHEQEV FDADTLTRLE | |
|
| 51 | KVLDFAELEV RDAMITRSRM NVLKENDSIE RITAYVIDTA HSRFPVIGED |
|
| 101 | KDEVLGILHA KDLLKYMFNP EQFHLKSVLR PAVFVPEGKS LTALLKEFRE |
|
| 151 | QRNHMAIVID EYGGTSGLVT FEDIIEQIVG DIEDEFDEDE SADDIHSVSA |
|
| 201 | ERWRIHAATE IEDINAFFGT EYGSEEADTI RRLGHSGIGT PARARRKSPY |
|
| 251 | RRFAVHRRPR RQPPPAHADG DPREVSRACP TAVSAQFRMT VRSFSVSIRP |
|
| 301 | IRQT* |
[0306]The originally-identified partial strain B sequence (ORF5) shows 83.1% identity over a 135aa overlap with the partial gonococcal sequence (ORF5ng):
[0000]
[0307]The complete strain B and gonococcal sequences (ORF5-1 & ORF5ng-1) show 92.4% identity in 304 aa overlap:
[0000]
[0308]Computer analysis of these amino acid sequences indicates a putative leader sequence, and identified the following homologies:
[0000]Homology with Hemolysin Homolog TlyC (Accession U32716) of H. influenzae
[0309]ORF5 and TlyC proteins show 58% aa identity in 77 aa overlap (BLASTp).
[0000]| ORF5 | 2 | HMAIVIDEYGGTSGLVTFEDIIEQIVGEIEDEFDEDDSADNIHAVSSDTWRIHAATEIED | 61 | |
| | HMAIV+DE+G SGLVT EDI+EQIVG+IEDEFDE++ AD I +S T+ + A T+I+D | |
| TlyC | 166 | HMAIVVDEFGAVSGLVTIEDILEQIVGDIEDEFDEEEIAD-IRQLSRHTYAVRALTDIDD | 224 |
|
| ORF5 | 62 | INTFFGTEYSIEEADTI | 78 |
| | N F T++ EE DTI | |
| TlyC | 225 | FNAQFNTDFDDEEVDTI | 241 |
[0310]ORF5ng-1 also shows significant homology with TlyC:
[0000]
[0000]Homology with a Hypothetical Secreted Protein from E. coli:
[0311]ORF5a shows homology to a hypothetical secreted protein from E. coli:
[0000]| sp|P77392|YBEX_ECOLI HYPOTHETICAL 33.3 KD PROTEIN IN CUTE-ASNB INTERGENIC REGION | |
| >gi|1778577 (U82598) similar to H. influenzae [Escherichia coli] >gi|1786879 |
| (AE000170) f292; This 292 aa ORF is 23% identical (9 gaps) to 272 residues of an |
| approx. 440 aa protein YTFL_HAEIN SW: P44717 [Escherichia coli] Length = 292 |
| Score = 212 bits (533), Expect = 3e−54 |
| Identities = 112/230 (48%), Positives = 149/230 (64%), Gaps = 3/230 (1%) |
|
| Query: | 2 | DGAQPKTNFXXRLIARLAR-EPDSAEDVLTLLRQAHEQEVFDADTLLRLEKVLDFSDLEV | 60 | |
| | D K F L+++L EP + +++L L+R + + ++ D DT LE V+D +D V | |
| Sbjct: | 10 | DTISNKKGFFSLLLSQLFHGEPKNRDELLALIRDSGQNDLIDEDTRDMLEGVMDIADQRV | 69 |
|
| Query: | 61 | RDAMITRSRMNVLKENDSIERITAYVIDTAHSRFPVIGEDKDEVLGILHAKDLLKYM-FN | 119 |
| | RD MI RS+M LK N +++ +I++AHSRFPVI EDKD + GIL AKDLL +M + | |
| Sbjct: | 70 | RDIMIPRSQMITLKRNQTLDECLDVIIESAHSRFPVISEDKDHIEGILMAKDLLPFMRSD | 129 |
|
| Query: | 120 | PEQFHLKSILRPAVFVPEGKSLTALLKEFREQRNHMAIVIDEYGGTSGLVTFEDIIEQIV | 179 |
| | E F + +LR AV VPE K + +LKEFR QR HMAIVIDE+GG SGLVT EDI+E IV | |
| Sbjct: | 130 | AEAFSMDKVLRQAVVVPESKRVDRMLKEFRSQRYHMAIVIDEFGGVSGLVTIEDILELIV | 189 |
|
| Query: | 180 | GDIEDEFDEDESADNIHAVSAERWRIHAATEIEDINAFFGTEYSSEEADT | 229 |
| | G+IEDE+DE++ D +S W + A IED N FGT +S EE DT | |
| Sbjct: | 190 | GEIEDEYDEEDDID-FRQLSRHTWTVRALASIEDFNEAFGTHFSDEEVDT | 238 |
[0312]Based on this analysis, including the amino acid homology to the TlyC hemolysin-homologue from H. influenzae (hemolysins are secreted proteins), it was predicted that the proteins from N. meningitidis and N. gonorrhoeae are secreted and could thus be useful antigens for vaccines or diagnostics.
[0313]ORF5-1 (30.7 kDa) was cloned in the pGex vector and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 2A shows the results of affinity purification of the GST-fusion protein. Purified GST-fusion protein was used to immunise mice, whose sera were used for Western blot analysis (FIG. 1B). These experiments confirm that ORF5-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 5
[0314]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 29>:
[0000]| 1 | ATGCGCGGCG GCAGGCCGGA TTCCGTTACC GTGCAGATTA TCGAAGGTTC | |
|
| 51 | GCGTTTTTCG CATATGAGGA AAGTCATCGA CGCAACGCCC GACATCGGAC |
|
| 101 | ACGACACCAA AGGCTGGAGC AATGAAAAAC TGATGGCGGA AGTTGCGCCC |
|
| 151 | GATGCCTTCA GCGGCAATCC TGAAgGGCAG TTTTTCCCCG ACAGCTACGA |
|
| 201 | AATCGATGCG GGCGGCAGTG ATTTGCAGAT TTACCAAACC GCCTACAAgG |
|
| 251 | GCGATGCAAC GCCGCCTGAA TGAgGGCATG GGAAAGCAGG CAGGACGGGC |
|
| 301 | TGCCTTATAA AAACCCTTAT GAAATGCTGA TTATGGCGAr CCTGGTCGAA |
|
| 351 | AAGGAAACAG GGCATGAAGC CGAsCsCGAC CATGTcGCTT CCGTCTTCGT |
|
| 401 | CAACCGCCTG AAAATCGGTA TGCGCCTGCA AACCgAssCG TCCGTGATTT |
|
| 451 | ACGGCATGGG TGCGGCATAC AAGGGCAAAA TCCGTAAAGC CGACCTGCGC |
|
| 501 | CGCGACACGC CGTACAACAC CTACACGCGC GGCGGTCTGC CGCCAACCCC |
|
| 551 | GATTGCGCTG CCC.. |
[0315]This corresponds to the amino acid sequence <SEQ ID 30; ORF7>:
[0000]| 1 | MRGGRPDSVT VQIIEGSRFS HMRKVIDATP DIGHDTKGWS NEKLMAEVAP | |
|
| 51 | DAFSGNPEGQ FFPDSYEIDA GGSDLQIYQT AYKAMQRRLN EAWESRQDGL |
|
| 101 | PYKNPYEMLI MAXLVEKETG HEAXXDHVAS VFVNRLKIGM RLQTXXSVIY |
|
| 151 | GMGAAYKGKI RKADLRRDTP YNTYTRGGLP PTPIALP.. |
[0316]Further sequence analysis revealed the complete DNA sequence <SEQ ID 31>:
[0000]| 1 | ATGTTGAGAA AATTGTTGAA ATGGTCTGCC GTTTTTTTGA CCGTGTCGGC | |
|
| 51 | AGCCGTTTTC GCCGCGCTGC TTTTTGTTCC TAAGGATAAC GGCAGGGCAT |
|
| 101 | ACCGAATCAA AATTGCCAAA AACCAGGGTA TTTCGTCGGT CGGCAGGAAA |
|
| 151 | CTTGCCGAAG ACCGCATCGT GTTCAGCAGG CATGTTTTGA CGGCGGCGGC |
|
| 201 | CTACGTTTTG GGTGTGCACA ACAGGCTGCA TACGGGGACG TACAGATTGC |
|
| 251 | CTTCGGAAGT GTCTGCTTGG GATATCTTGC AGAAAATGCG CGGCGGCAGG |
|
| 301 | CCGGATTCCG TTACCGTGCA GATTATCGAA GGTTCGCGTT TTTCGCATAT |
|
| 351 | GAGGAAAGTC ATCGACGCAA CGCCCGACAT CGGACACGAC ACCAAAGGCT |
|
| 401 | GGAGCAATGA AAAACTGATG GCGGAAGTTG CGCCCGATGC CTTCAGCGGC |
|
| 451 | AATCCTGAAG GGCAGTTTTT CCCCGACAGC TACGAAATCG ATGCGGGCGG |
|
| 501 | CAGTGATTTG CAGATTTACC AAACCGCCTA CAAGGCGATG CAACGCCGCC |
|
| 551 | TGAATGAGGC ATGGGAAAGC AGGCAGGACG GGCTGCCTTA TAAAAACCCT |
|
| 601 | TATGAAATGC TGATTATGGC GAGCCTGGTC GAAAAGGAAA CAGGGCATGA |
|
| 651 | AGCCGACCGC GACCATGTCG CTTCCGTCTT CGTCAACCGC CTGAAAATCG |
|
| 701 | GTATGCGCCT GCAAACCGAC CCGTCCGTGA TTTACGGCAT GGGTGCGGCA |
|
| 751 | TACAAGGGCA AAATCCGTAA AGCCGACCTG CGCCGCGACA CGCCGTACAA |
|
| 801 | CACCTACACG CGCGGCGGTC TGCCGCCAAC CCCGATTGCG CTGCCCGGCA |
|
| 851 | AGGCGGCACT CGATGCCGCC GCCCATCCGT CCGGCGAAAA ATACCTGTAT |
|
| 901 | TTCGTGTCCA AAATGGACGG CACGGGCTTG AGCCAGTTCA GCCATGATTT |
|
| 951 | GACCGAACAC AATGCCGCCG TCCGCAAATA TATTTTGAAA AAATAA |
[0317]This corresponds to the amino acid sequence <SEQ ID 32; ORF7-1>:
[0000]| 1 | MLRKLLKWSA VFLTVSAAVF AALLFVPKDN GRAYRIKIAK NQGISSVGRK | |
|
| 51 | LAEDRIVFSR HVLTAAAYVL GVHNRLHTGT YRLPSEVSAW DILQKMRGGR |
|
| 101 | PDSVTVQIIE GSRFSHMRKV IDATPDIGHD TKGWSNEKLM AEVAPDAFSG |
|
| 151 | NPEGQFFPDS YEIDAGGSDL QIYQTAYKAM QRRLNEAWES RQDGLPYKNP |
|
| 201 | YEMLIMASLV EKETGHEADR DHVASVFVNR LKIGMRLQTD PSVIYGMGAA |
|
| 251 | YKGKIRKADL RRDTPYNTYT RGGLPPTPIA LPGKAALDAA AHPSGEKYLY |
|
| 301 | FVSKMDGTGL SQFSHDLTEH NAAVRKYILK K* |
[0318]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical Protein Encoded by yceg Gene (Accession P44270) of H. influenzae
[0319]ORF7 and yceg proteins show 44% aa identity in 192 aa overlap:
[0000]| ORF7 | 1 | MRGGRPDSVTVQIIEGSRFSHMRKVIDATPDIGHDTKGWSNEKLMA-----EVAPDAFSG | 55 | |
| | + G+ V+ IEG F RK ++ P + K SNE++ A ++ + |
| yceg | 102 | LNSGKEVQFNVKWIEGKTFKDWRKDLENAPHLVQTLKDKSNEEIFALLDLPDIGQNLELK | 161 |
|
| ORF7 | 56 | NPEGQFFPDSYEIDAGGSDLQIYQTAYKAMQRRLNEAWESRQDGLPYKNPYEMLIMAXLV | 115 |
| | N EG +PD+Y +DL++ + + + M++ LN+AW R + LP NPYEMLI+A +V |
| yceg | 162 | NVEGWLYPDTYNYTPKSTDLELLKRSAERMKKALNKAWNERDEDLPLANPYEMLILASIV | 221 |
|
| ORF7 | 116 | EKETGHEAXXDHVASVFVNRLKIGMRLQTXXSVIYGMGAAYKGKIRKADLRRDTPYNTYT | 175 |
| | EKETG VASVF+NRLK M+LQT +VIYGMG Y G IRK DL TPYNTY |
| yceg | 222 | EKETGIANERAKVASVFINRLKAKMKLQTDPTVIYGMGENYNGNIRKKDLETKTPYNTYV | 281 |
|
| ORF7 | 176 | RGGLPPTPIALP | 187 |
| | GLPPTPIA+P |
| yceg | 282 | IDGLPPTPIAMP | 293 |
[0320]The complete length YCEG protein has sequence:
[0000]| 1 | MKKFLIAILL LILILAGVAS FSYYKMTEFV KTPVNVQADE LLTIERGTTS | |
|
| 51 | SKLATLFEQE KLIADGKLLP YLLKLKPELN KIKAGTYSLE NVKTVQDLLD |
|
| 101 | LLNSGKEVQF NVKWIEGKTF KDWRKDLENA PHLVQTLKDK SNEEIFALLD |
|
| 151 | LPDIGQNLEL KNVEGWLYPD TYNYTPKSTD LELLKRSAER MKKALNKAWN |
|
| 201 | ERDEDLPLAN PYEMLILASI VEKETGIANE RAKVASVFIN RLKAKMKLQT |
|
| 251 | DPTVIYGMGE NYNGNIRKKD LETKTPYNTY VIDGLPPTPI AMPSESSLQA |
|
| 301 | VANPEKTDFY YFVADGSGGH KFTRNLNEHN KAVQEYLRWY RSQKNAK |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0321]ORF7 shows 95.2% identity over a 187aa overlap with an ORF (ORF7a) from strain A of N. meningitidis:
[0000]
[0322]The complete length ORF7a nucleotide sequence <SEQ ID 33> is:
[0000]| 1 | ATGTTGAGAA AATTGTTGAA ATGGTCTGCC GTTTTTTTGA CCGTATCGGC | |
|
| 51 | AGCCGTTTTC GCCGCGCTGC TTTTCGTCCC TAAAGACAAC GGCAGGGCAT |
|
| 101 | ACAGGATTAA AATTGCCAAA AACCAGGGTA TTTCGTCGGT CGGCAGGAAA |
|
| 151 | CTTGCCGAAG ACCGCATCGT GTTCAGCAGG CATGTTTTGA CGGCGGCGGC |
|
| 201 | CTACGTTTTG GGTGTGCACA ACAGGCTGCA TACGGGGACG TACAGACTGC |
|
| 251 | CTTCGGAAGT GTCTGCTTGG GATATCTTGC AGAAAATGCG CGGCGGCAGG |
|
| 301 | CCGGATTCCG TTACCGTGCA GATTATCGAA GGTTCGCGTT TTTCGCATAT |
|
| 351 | GAGGAAAGTC ATCGACGCAA CGCCCGACAT CGAACACGAC ACCAAAGGCT |
|
| 401 | GGAGCAATGA AAAACTGATG GCGGAAGTTG CCCCTGATGC CTTCAGCGGC |
|
| 451 | AATCCTGAAG GGCAGTTTTT CCCCGACAGC TACGAAATCG ATGCGGGCGG |
|
| 501 | CAGCGATTTA CGGATTTACC AAATCGCCTA CAAGGCGATG CAACGCCGAC |
|
| 551 | TGAATGAGGC ATGGGAAAGC AGGCAGGACG GGCTGCCTTA TAAAAACCCT |
|
| 601 | TATGAAATGC TGATTATGGC GAGCCTGATC GAAAAGGAAA CAGGGCATGA |
|
| 651 | AGCCGACCGC GACCATGTCG CTTCCGTCTT CGTCAACCGC CTGAAAATCG |
|
| 701 | GTATGCGCCT GCAAACCGAC CCGTCCGTGA TTTACGGCAT GGGTGCGGCA |
|
| 751 | TACAAGGGCA AAATCCGTAA AGCCGACCTG CGCCGCGACA CGCCGTACAA |
|
| 801 | CACCTACACG CGCGGCGGTC TGCCGCCAAC CCCGATCGCG CTGCCCGGCA |
|
| 851 | AGGCGGCACT CGATGCCGCC GCCCATCCGT CCGGTGAAAA ATACCTGTAT |
|
| 901 | TTCGTGTCCA AAATGGACGG TACGGGCTTG AGCCAGTTCA GCCATGATTT |
|
| 951 | GACCGAACAC AACGCCGCCG TTCGCAAATA TATTTTGAAA AAATAA |
[0323]This is predicted to encode a protein having amino acid sequence <SEQ ID 34>:
[0000]| 1 | MLRKLLKWSA VFLTVSAAVF AALLFVPKDN GRAYRIKIAK NQGISSVGRK | |
|
| 51 | LAEDRIVFSR HVLTAAAYVL GVHNRLHTGT YRLPSEVSAW DILQKMRGGR |
|
| 101 | PDSVTVQIIE GSRFSHMRKV IDATPDIEHD TKGWSNEKLM AEVAPDAFSG |
|
| 151 | NPEGQFFPDS YEIDAGGSDL RIYQIAYKAM QRRLNEAWES RQDGLPYKNP |
|
| 201 | YEMLIMASLI EKETGHEADR DHVASVFVNR LKIGMRLQTD PSVIYGMGAA |
|
| 251 | YKGKIRKADL RRDTPYNTYT RGGLPPTPIA LPGKAALDAA AHPSGEKYLY |
|
| 301 | FVSKMDGTGL SQFSHDLTEH NAAVRKYILK K* |
[0324]A leader peptide is underlined.
[0325]ORF7a and ORF7-1 show 98.8% identity in 331 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0326]ORF7 shows 94.7% identity over a 187aa overlap with a predicted ORF (ORF7.ng) from N. gonorrhoeae:
[0000]
[0327]An ORF7ng nucleotide sequence <SEQ ID 35> is predicted to encode a protein having amino acid sequence <SEQ ID 36>:
[0000]| 1 | MRGGRPDSVT VQIIEGSRFS HMRKVIDATP DIGHDTKGWS NEKLMAEVAP | |
|
| 51 | DAFSGNPEGQ FFPDSYEIDA GGSDLQIYQT AYKAMQRRLN EAWAGRQDGL |
|
| 101 | PYKNPYEMLI MASLIEKETG HEADRDHVAS VFVNRLKIGM RLQTDPSVIY |
|
| 151 | GMGAAYKGKI RKADLRRDTP YNTYTGGGLP PTRIALPGKA AMDAAAHPSG |
|
| 201 | EKYLYFVSKM DGTGLSQFSH DLTEHNAAVR KYILKK* |
[0328]Further sequence analysis revealed a partial DNA sequence of ORF7ng <SEQ ID 37>:
[0000]| 1 | ..taccgaatca AGATTGCCAA AAATCAGGGT ATTTCGTCGG TCGGCAGGAA | |
|
| 51 | ACTTGCcgaA GACCGCATCG TGTTCAGCAG GCATGTTTTG ACAGCGGCGG |
|
| 101 | CCTACGTTTT GGGTGTGCAC AACAGGCTGC ATACGGGGAC gTACAGATTG |
|
| 151 | CCTTCGGAAG TGTCTGCTTG GGATATCTTG CAGAAAATGC GCGGCGGCAG |
|
| 201 | GCCGGATTCC GTTACCGTGC AGATTATCGA AGGTTCGCGT TTTTCGCATA |
|
| 251 | TGAGGAAAGT CATCGACGCA ACGCCCGACA TCGGACACGA CACCAAAGGC |
|
| 301 | TGGAGCAATG AAAAACTGAT GGCGGAAGTT GCGCCCGATG CCTTCAGCGG |
|
| 351 | CAATCCTGAA GGGCAGTTTT TTCCCGACAG CTACGAAATC GATGCGGGCG |
|
| 401 | GCAGCGATTT GCAGATTTAC CAAACCGCCT ACAAGGCGAT GCAACGCCGC |
|
| 451 | CTGAACGAGG CATGGGCAGG CAGGCAGGAC GGGCTGCCTT ATAAAAACCC |
|
| 501 | TTATGAAATG CTGATTATGG CGAGCCTGAT CGAAAAGGAA ACGGGGCATG |
|
| 551 | AGGCCGACCG CGACCATGTC GCTTCCGTCT TCGTCAACCG CCTGAAAATC |
|
| 601 | GGTATGCGCC TGCAAACCGA CCCGTCCGTG ATTTACGGCA TGGGTGCGGC |
|
| 651 | ATACAAGGGC AAAATCCGTA AAGCCGACCT GCGCCGCGAC ACGCCGTACA |
|
| 701 | aCAccTAtac gggcgggggc ttgccgccaa cccggattgc gctgcccggC |
|
| 751 | Aaggcggcaa tggatgccgc cgcccacccg tccggcgaAa aatacctgTa |
|
| 801 | tttcgtgtcC AAAATGGACG GCACGGGCTT GAGCCAGTTC AGCCATGATT |
|
| 851 | TGACCGAACA CAACGCCGCc gTcCGCAAAT ATATTTTGAA AAAATAA |
[0329]This corresponds to the amino acid sequence <SEQ ID 38; ORF7ng-1>:
[0000]| 1 | ..YRIKIAKNQG ISSVGRKLAE DRIVFSRHVL TAAAYVLGVH NRLHTGTYRL | |
|
| 51 | PSEVSAWDIL QKMRGGRPDS VTVQIIEGSR FSHMRKVIDA TPDIGHDTKG |
|
| 101 | WSNEKLMAEV APDAFSGNPE GQFFPDSYEI DAGGSDLQIY QTAYKAMQRR |
|
| 151 | LNEAWAGRQD GLPYKNPYEM LIMASLIEKE TGHEADRDHV ASVFVNALKI |
|
| 201 | GMRLQTDPSV IYGMGAAYKG KIRKADLRRD TPYNTYTGGG LPPTRIALPG |
|
| 251 | KAAMDAAAHP SGEKYLYFVS KMDGTGLSQF SHDLTEHNAA VRKYILKK* |
[0330]ORF7ng-1 and ORF7-1 show 98.0% identity in 298 aa overlap:
[0000]
[0331]In addition, ORF7ng-1 shows significant homology with a hypothetical E. coli protein:
[0000]| sp|P28306|YCEG_ECOLI HYPOTHETICAL 38.2 KD PROTEIN IN PABC-HOLB | |
| INTERGENIC REGION |
| gi|1787339 (AE000210) o340; 100% identical to fragment YCEG_ECOLI SW: |
| P28306 but has 97 additional C-terminal residues [Escherichia coli] |
| Length = 340 |
| Score = 79 (36.2 bits), Expect = 5.0e−57, Sum P(2) = 5.0e−57 |
| Identities = 20/87 (22%), Positives = 40/87 (45%) |
|
| Query: | 10 | GISSVGRKLAEDRIVFSRHVLTAAAYVLGVHNRLHTGTYRLPSEVSAWDILQKMRGGRPD | 69 | |
| | G ++G +L D+I+ V + + GTYR +++ ++L+ + G+ |
| Sbjct: | 49 | GRLALGEQLYADKIINRPRVFQWLLRIEPDLSHFKAGTYRFTPQMTVREMLKLLESGKEA | 108 |
|
| Query: | 70 | SVTVQIIEGSRFSHMRKVIDATPDIGH | 96 |
| | ++++EG R S K + P I H |
| Sbjct: | 109 | QFPLRLVEGMRLSDYLKQLREAPYIKH | 135 |
|
| Score = 438 (200.7 bits), Expect = 5.0e−57, Sum P(2) = 5.0e−57 | |
| Identities = 84/155 (54%), Positives = 111/155 (71%) |
|
| Query: | 120 | EGQFFPDSYEIDAGGSDLQIYQTAYKAMQRRLNEAWAGRQDGLPYKNPYEMLIMASLIEK | 179 | |
| | EG F+PD++ A +D+ + + A+K M + ++ AW GR DGLPYK+ +++ MAS+IEK |
| Sbjct: | 158 | EGWFWPDTWMYTANTTDVALLKRAHKKMVKAVDSAWEGRADGLPYKDKNQLVTMASIIEK | 217 |
|
| Query: | 180 | ETGHEADRDHVASVFVNRLKIGMRLQTDPSVIYGMGAAYKGKIRKADLRRDTPYNTYTGG | 239 |
| | ET ++RD VASVF+NRL+IGMRLQTDP+VIYGMG Y GK+ +ADL T YNTYT |
| Sbjct: | 218 | ETAVASERDKVASVFINRLRIGMRLQTDPTVIYGMGERYNGKLSRADLETPTAYNTYTIT | 277 |
|
| Query: | 240 | GLPPTRIALPGKAAMDAAAHPSGEKYLYFVSKMDG | 274 |
| | GLPP IA PG ++ AAAHP+ YLYFV+ G |
| Sbjct: | 278 | GLPPGAIATPGADSLKAAAHPAKTPYLYFVADGKG | 312 |
[0332]Based on this analysis, including the fact that the H. influenzae YCEG protein possesses a possible leader sequence, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 6
[0333]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 39>:
[0000]| 1 | CGTTTCAAAA TGTTAACTGT GTTGACGGCA ACCTTGATTG CCGGACAGGT | |
|
| 51 | ATCTGCCGCC GGAGGCGGTG CGGGGGATAT GAAACAGCCG AAGGAAGTCG |
|
| 101 | GAAAGGTTTT CAGAAAGCAG CAGCGTTACA GCGAGGAAGA AATCAAAAAC |
|
| 151 | GAACGCGCAC GGCTTGCGGC AGTGGGCGAG CGGGTTAATC AGATATTTAC |
|
| 201 | GTTGCTGGGA GGGGAAACCG CCTTGCAAAA GGGGCAGGCG GGAACGGCTC |
|
| 251 | TGGCAACCTA TATGCTGATG TTGGAACGCA CAAAATCCCC CGAAGTCGCC |
|
| 301 | GAACGCGCCT TGGAAATGGC CGTGTCGCTG AACGCGTTTG AACAGGCGGA |
|
| 351 | AATGATTTAT CAGAAATGGC GGCAGATTGA GCCTATACCG GGTAAGGCGC |
|
| 401 | AAAAACGGGC GGGGTGGCTG CGGAACGTGC TGAGGGAAAG AGGAAATCAG |
|
| 451 | CATCTGGACG GACGGGAAGA AGTGCTGGCT CAGGCGGACG AAGGACAG |
[0334]This corresponds to the amino acid sequence <SEQ ID 40; ORF9>:
[0000]| 1 | ..RFKMLTVLTA TLIAGQVSAA GGGAGDMKQP KEVGKVFRKQ QRYSEEEIKN | |
|
| 51 | ERARLAAVGE RVNQIFTLLG GETALQKGQA GTALATYMLM LERTKSPEVA |
|
| 101 | ERALEMAVSL NAFEQAEMIY QKWRQIEPIP GKAQKRAGWL RNVLRERGNQ |
|
| 151 | HLDGREEVLA QADEGQ |
[0335]Further sequence analysis revealed the complete DNA sequence <SEQ ID 41>:
[0000]| 1 | ATGTTACCTA ACCGTTTCAA AATGTTAACT GTGTTGACGG CAACCTTGAT | |
|
| 51 | TGCCGGACAG GTATCTGCCG CCGGAGGCGG TGCGGGGGAT ATGAAACAGC |
|
| 101 | CGAAGGAAGT CGGAAAGGTT TTCAGAAAGC AGCAGCGTTA CAGCGAGGAA |
|
| 151 | GAAATCAAAA ACGAACGCGC ACGGCTTGCG GCAGTGGGCG AGCGGGTTAA |
|
| 201 | TCAGATATTT ACGTTGCTGG GAGGGGAAAC CGCCTTGCAA AAGGGGCAGG |
|
| 251 | CGGGAACGGC TCTGGCAACC TATATGCTGA TGTTGGAACG CACAAAATCC |
|
| 301 | CCCGAAGTCG CCGAACGCGC CTTGGAAATG GCCGTGTCGC TGAACGCGTT |
|
| 351 | TGAACAGGCG GAAATGATTT ATCAGAAATG GCGGCAGATT GAGCCTATAC |
|
| 101 | CGGGTAAGGC GCAAAAACGG GCGGGGTGGC TGCGGAACGT GCTGAGGGAA |
|
| 451 | AGAGGAAATC AGCATCTGGA CGGACTGGAA GAAGTGCTGG CTCAGGCGGA |
|
| 501 | CGAAGGACAG AACCGCAGGG TGTTTTTATT GTTGGCACAA GCCGCCGTGC |
|
| 551 | AACAGGACGG GTTGGCGCAA AAAGCATCGA AAGCGGTTCG CCGCGCGGCG |
|
| 601 | TTGAAATATG AACATCTGCC CGAAGCGGCG GTTGCCGATG TGGTGTTCAG |
|
| 651 | CGTACAGGGA CGCGAAAAGG AAAAGGCAAT CGGAGCTTTG CAGCGTTTGG |
|
| 701 | CGAAGCTCGA TACGGAAATA TTGCCCCCCA CTTTAATGAC GTTGCGTCTG |
|
| 751 | ACTGCACGCA AATATCCCGA AATACTCGAC GGCTTTTTCG AGCAGACAGA |
|
| 801 | CACCCAAAAC CTTTCGGCCG TCTGGCAGGA AATGGAAATT ATGAATCTGG |
|
| 851 | TTTCCCTGCA CAGGCTGGAT GATGCCTATG CGCGTTTGAA CGTGCTGTTG |
|
| 901 | GAACGCAATC CGAATGCAGA CCTGTATATT CAGGCAGCGA TATTGGCGGC |
|
| 951 | AAACCGAAAA GAAGGTGCTT CCGTTATCGA CGGCTACGCC GAAAAGGCAT |
|
| 1001 | ACGGCAGGGG GACGGAGGAA CAGCGGAGCA GGGCGGCGCT AACGGCGGCG |
|
| 1051 | ATGATGTATG CCGACCGCAG GGATTACGCC AAAGTCAGGC AGTGGCTGAA |
|
| 1101 | AAAAGTATCC GCGCCGGAAT ACCTGTTCGA CAAAGGTGTG CTGGCGGCTG |
|
| 1151 | CGGCGGCTGT CGAGTTGGAC GGCGGCAGGG CGGCTTTGCG GCAGATCGGC |
|
| 1201 | AGGGTGCGGA AACTTCCCGA ACAGCAGGGG CGGTATTTTA CGGCAGACAA |
|
| 1251 | TTTGTCCAAA ATACAGATGC TCGCCCTGTC GAAGCTGCCC GATAAACGGG |
|
| 1301 | AGGCTTTGAG GGGGTTGGAC AAGATTATCG AAAAACCGCC TGCCGGCAGT |
|
| 1351 | AATACAGAGT TACAGGCAGA GGCATTGGTA CAGCGGTCAG TTGTTTACGA |
|
| 1401 | TCGGCTTGGC AAGCGGAAAA AAATGATTTC AGATCTTGAA AGGGCGTTCA |
|
| 1451 | GGCTTGCACC CGATAACGCT CAGATTATGA ATAATCTGGG CTACAGCCTG |
|
| 1501 | CTGACCGATT CCAAACGTTT GGACGAAGGT TTCGCCCTGC TTCAGACGGC |
|
| 1551 | ATACCAAATC AACCCGGACG ATACCGCTGT CAACGACAGC ATAGGCTGGG |
|
| 1601 | CGTATTACCT GAAAGGCGAC GCGGAAAGCG CGCTGCCGTA TCTGCGGTAT |
|
| 1651 | TCGTTTGAAA ACGACCCCGA GCCCGAAGTT GCCGCCCATT TGGGCGAAGT |
|
| 1701 | GTTGTGGGCA TTGGGCGAAC GCGATCAGGC GGTTGACGTA TGGACGCAGG |
|
| 1751 | CGGCACACCT TACGGGAGAC AAGAAAATAT GGCGGGAAAC GCTCAAACGT |
|
| 1801 | CACGGCATCG CATTGCCCCA ACCTTCCCGA AAACCTCGGA AATAA |
[0336]This corresponds to the amino acid sequence <SEQ ID 42; ORF9-1>:
[0000]| 1 | MLPNRFKMLT VLTATLIAGQ VSAAGGGAGD MKQPKEVGKV FRKQQRYSEE | |
|
| 51 | EIKNERARLA AVGERVNQIF TLLGGETALQ KGQAGTALAT YMLMLERTKS |
|
| 101 | PEVAERALEM AVSLNAFEQA EMIYQKWRQI EPIPGKAQKR AGWLRNVLRE |
|
| 151 | RGNQHLDGLE EVLAQADEGQ NRRVFLLLAQ AAVQQDGLAQ KASKAVRRAA |
|
| 201 | LKYEHLPEAA VADVVFSVQG REKEKAIGAL QRLAKLDTEI LPPTLMTLRL |
|
| 251 | TARKYPEILD GFFEQTDTQN LSAVWQEMEI MNLVSLHRLD DAYARLNVLL |
|
| 301 | ERNPNADLYI QAAILAANRK EGASVIDGYA EKAYGRGTEE QRSRAALTAA |
|
| 351 | MMYADRRDYA KVRQWLKKVS APEYLFDKGV LAAAAAVELD GGRAALRQIG |
|
| 401 | RVRKLPEQQG RYFTADNLSK IQMLALSKLP DKREALRGLD KIIEKPPAGS |
|
| 451 | NTELQAEALV QRSVVYDRLG KRKKMISDLE RAFRLAPDNA QIMNNLGYSL |
|
| 501 | LTDSKRLDEG FALLQTAYQI NPDDTAVNDS IGWAYYLKGD AESALPYLRY |
|
| 551 | SFENDPEPEV AAHLGEVLWA LGERDQAVDV WTQAAHLTGD KKIWRETLKR |
|
| 601 | HGIALPQPSR KPRK* |
[0337]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0338]ORF9 shows 89.8% identity over a 166aa overlap with an ORF (ORF9a) from strain A of N. meningitidis.
[0000]
[0339]The complete length ORF9a nucleotide sequence <SEQ ID 43> is:
[0000]| 1 | ATGTTACCCG CCCGTTTCAC CATTTTATCT GTGCTCGCGG CAGCCCTGCT | |
|
| 51 | TGCCGGGCAG GCGTATGCCG CCGGCGCGGC GGATGCGAAG CCGCCGAAGG |
|
| 101 | AAGTCGGAAA GGTTTTCAGA AAGCAGCAGC GTTACAGCGA GGAAGAAATC |
|
| 151 | AAAAACGAAC GCGCACGGCT TGCGGCAGTG GGCGAGCGGG TTAATCAGAT |
|
| 201 | ATTTACGTTG CTGGGANGGG AAACCGCCTT GCAAAAGGGG CAGGCGGGAA |
|
| 251 | CGGCTCTGGC AACCTATATG CTGATGTTGG AACGCACAAA ATCCCCCGAA |
|
| 301 | GTCGCCGAAC GCGCCTTGGA AATGGCCGTG TCNCTGAACG CGTTTGAACA |
|
| 351 | GGCGGAAATG ATTTATCAGA AATGGCGGCA GATTGAGCCT ATACCGGGTA |
|
| 401 | AGGCGCAAAA ACGGGCGGGG TGGCTGCGGA ACGTGCTGAG GGAAAGAGGA |
|
| 451 | AATCAGCATC TAGACGGACT GGAAGAANTG CTGGCTCAGG CGGACGAANG |
|
| 501 | ACAGAACCGC AGGGTGTTTT TATTGTTGGC ACAAGCCGCC GTGCAACAGG |
|
| 551 | ACGGGTTGGC GCAAAAAGCA TCGAAAGCGG TTCGCCGCGC GGCGTTGAGA |
|
| 601 | TATGAACATC TGCCCGAAGC GGCGGTTGCC GATGTGGTGT TCAGCGTACA |
|
| 651 | GGNACGCGAA AAGGAAAAGG CAATCGGAGC TTTGCAGCGT TTGGCGAAGC |
|
| 701 | TCGATACGGA AATATTGCCC CCCACTTTAA TGACGTTGCG TCTGACTGCA |
|
| 751 | CGCAAATATC CCGAAATACT CGACGGCTTT TTCGAGCAGA CAGACACCCA |
|
| 801 | AAACCTTTCG GCCGTCTGGC AGGAAATGGA AATTATGAAT CTGGTTTCCC |
|
| 851 | TGCACAGGCT GGATGATGCC TATGCGCGTT TGAACGTGCT GTTGGAACGC |
|
| 901 | AATCCGAATG CAGACCTGTA TATTCAGGCA GCGATATTGG CGGCAAACCG |
|
| 951 | AAAAGAANGT GCTTCCGTTA TCGACGGCTA CGCCGAAAAG GCATACGGCA |
|
| 1001 | GGGGGACGGG GGAACAGCGG GGCAGGGCGG CAATGACGGC GGCGATGATA |
|
| 1051 | TATGCCGACC GAAGGGATTA CACCAAAGTC AGGCAGTGGT TGAAAAAAGT |
|
| 1101 | GTCCGCGCCG GAATACCTGT TCGACAAAGG TGTGCTGGCG GCTGCGGCGG |
|
| 1151 | CTGTCGAGTT GGACNGCGGC AGGGCGGCTT TGCGGCAGAT CGGCAGGGTG |
|
| 1201 | CGGAAACTTC CCGAACAGCA GGGGCGGTAT TTTACGGCAG ACAATTTGTC |
|
| 1251 | CAAAATACAG ATGTTCGCCC TGTCGAAGCT GCCCGACAAA CGGGAGGCTT |
|
| 1301 | TGAGGGGGTT GGACAAGATT ATCGAAAAAC CGCCTGCCGG CAGTAATACA |
|
| 1351 | GAGTTACAGG CAGAGGCATT GGTACAGCGG TCAGTTGTTT ACGATCGGCT |
|
| 1401 | TGGCAAGCGG AAAAAAATGA TTTCAGATCT TGAAAGGGCG TTCAGGCTTG |
|
| 1451 | CACCCGATAA CGCTCAGATT ATGAATAATC TGGGCTACAG CCTGCTTTCC |
|
| 1501 | GATTCCAAAC GTTTGGACGA AGGCTTCGCC CTGCTTCAGA CGGCATACCA |
|
| 1551 | AATCAACCCG GACGATACCG CTGTCAACGA CAGCATAGGC TGGGCGTATT |
|
| 1601 | ACCTGAAANG CGACGCGGAA AGCGCGCTGC CGTATCTGCG GTATTCGTTT |
|
| 1651 | GAAAACGACC CCGAGCCCGA AGTTGCCGCC CATTTGGGCG AAGTGTTGTG |
|
| 1701 | GGCATTGGGC GAACGCGATC AGGCGGTTGA CGTATGGACG CAGGCGGCAC |
|
| 1751 | ACCTTACGGG AGACAAGAAA ATATGGCGGG AAACGCTCAA ACGTCACGGC |
|
| 1801 | ATCGCATTGC CCCAACCTTC CCGAAAACCT CGGAAATAA |
[0340]This encodes a protein having amino acid sequence <SEQ ID 44>:
[0000]| 1 | MLPARFTILS VLAAALLAGQ AYAAGAADAK PPKEVGKVFR KQQRYSEEEI | |
|
| 51 | KNERARLAAV GERVNQIFTL LGXETALQKG QAGTALATYM LMLERTKSPE |
|
| 101 | VAERALEMAV SLNAFEQAEM IYQKWRQIEP IPGKAQKRAG WLRNVLRERG |
|
| 151 | NQHLDGLEEX LAQADEXQNR RVFLLLAQAA VQQDGLAQKA SKAVRRAALR |
|
| 201 | YEHLPEAAVA DVVFSVQXRE KEKAIGALQR LAKLDTEILP PTLMTLRLTA |
|
| 251 | RKYPEILDGF FEQTDTQNLS AVWQEMEIMN LVSLHRLDDA YARLNVLLER |
|
| 301 | NPNADLYIQA AILAANRKEX ASVIDGYAEK AYGRGTGEQR GRAAMTAAMI |
|
| 351 | YADRRDYTKV RQWLKKVSAP EYLFDKGVLA AAAAVELDXG RAALRQIGRV |
|
| 401 | RKLPEQQGRY FTADNLSKIQ MFALSKLPDK REALRGLDKI IEKPPAGSNT |
|
| 451 | ELQAEALVQR SVVYDRLGKR KKMISDLERA FRLAPDNAQI MNNLGYSLLS |
|
| 501 | DSKRLDEGFA LLQTAYQINP DDTAVNDSIG WAYYLKXDAE SALPYLRYSF |
|
| 551 | ENDPEPEVAA HLGEVLWALG ERDQAVDVWT QAAHLTGDKK IWRETLKRHG |
|
| 601 | IALPQPSRKP RK* |
[0341]ORF9a and ORF9-1 show 95.3% identity in 614 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0342]ORF9 shows 82.8% identity over a 163aa overlap with a predicted ORF (ORF9.ng) from N. gonorrhoeae.
[0000]
[0343]The ORF9ng nucleotide sequence <SEQ ID 45> was predicted to encode a protein having including acid sequence <SEQ ID 46>:
[0000]| 1 | MIMLPARFTI LSVLAAALLA GQAYAAGAAD VELPKEVGKV LRKHRRYSEE | |
|
| 51 | EIKNERARLA AVGERVNRVF TLLGGETALQ KGQAGTALAT YMLMLERTKS |
|
| 101 | PEVAERALEM AVSLNAFEQA EMIYQKWRQI EPIPGEAQKP AGWLRNVLKE |
|
| 151 | GGNPHLDRLE EVPAQSDYVH QPMIFLLLVQ AAVQHGGVAQ KPSKAVRPAA |
|
| 201 | YNYEVLPETA GADAVFCVQG PQYEKAIQSF PPCGRNPQTE NIAPPFNELF |
|
| 251 | RPTARPISPK LLQRFFRTEP NLAKPFRPPG PEMETYQTGF PRPLTRNNPT |
[0344]Amino acids 1-28 are a putative leader sequence, and 173-189 are predicted to be a transmembrane domain.
[0345]Further sequence analysis revealed the complete length ORF9ng DNA sequence <SEQ ID 47>:
[0000]| 1 | ATGTTACCCG CCCGTTTCAC TATTTTATCT GTCCTCGCAG CAGCCCTGCT | |
|
| 51 | TGCCGGACAG GCGTATGCTG CCGGCGCGGC GGATGTGGAG CTGCCGAAGG |
|
| 101 | AAGTCGGAAA GGTTTTAAGG AAACATCGGC GTTACAGCGA GGAAGAAATC |
|
| 151 | AAAAACGAAC GCGCACGGCT TGCGGCAGTG GGCGAACGGG TCAACAGGGT |
|
| 201 | GTTTACGCTG TTGGGCGGTG AAACGGCTTT GCAGAAAGGG CAGGCGGGAA |
|
| 251 | CGGCTCTGGC AACCTATATG CTGATGTTGG AACGCACAAA ATCCCCCGAA |
|
| 301 | GTCGCCGAAC GCGCCTTGGA AATGGCCGTG TCGCTGAACG CGTTTGAACA |
|
| 351 | GGCGGAAATG ATTTATCAGA AATGgcggca gatcgagcct ataCcgggtg |
|
| 401 | aggcgcaaaa accgGcgggG tggctgcgga acgtattgaa ggaagggGGa |
|
| 451 | aaTCAGCATC TGGAcgggtt gaaagaggTG CtggcgcaAT cggacgatGT |
|
| 501 | GCAAAAAcgc aggaTATTTT TGCTGCTGGT GCAAGCCGCC GTGCagcagg |
|
| 551 | gTGGGGTGGC TCAAAAAGCA TCGAAAGCGG TTCGCcgtgc GGcgttgaAG |
|
| 601 | TATGAACATC TGCCcgaagc ggcggTTGCC GATGcggTGT TCGGCGTACA |
|
| 651 | GGGACGCGAA AAGGAAAagg caaTCGAAGC TTTGCAGCGT TTGGCGAAGC |
|
| 701 | TCGATACGGA AATATTGCCC CCCACTTTAA TGACGTTGCG TCTGACTGCA |
|
| 751 | CGCAAATATC CCGAAATACT CGACGGCTTT TTCGAGCAGA CAGACACCCA |
|
| 801 | AAACCTTTCG GCCGTCTGGC AGGAAATGGA AATTATGAAT CTGGTTTCCC |
|
| 851 | TGCGTAAGCC GGATGATGCC TATGCGCGTT TGAACGTGCT GTTGGAACAC |
|
| 901 | AACCCGAATG CAAACCTGTA TATTCAGGCG GCGATATTGG CGGCAAACCG |
|
| 951 | AAAAGAAGGT GCGTCCGTTA TCGACGGCTA CGCCGAAAAG GCATACGGCA |
|
| 1001 | GGGGGACGGG GGAACAGCGG GGCagggcgg cAATgacggc GGCGATGATA |
|
| 1051 | TATGCCGACC GCAGGGATTA CGCCAAAGTC AGGCAGTGGT TGAAAAAAGT |
|
| 1101 | GTCCGCGCCG GAATACCTGT TCGACAAAGG CGTGCTGGCG GCTGCGGCGG |
|
| 1151 | CTGCCGAATT GGACGGAGGC CGGGCGGCTT TGCGGCAGAT CGGCAGGGTG |
|
| 1201 | CGGAAACTTC CCGAACAGCA GGGGCGGTAT TTTACGGCAG ACAATTTGTC |
|
| 1251 | CAAAATACAG ATGCTCGCCC TGTCGAAGCT GCCCGACAAA CGGGAAGCCC |
|
| 1301 | TGATCGGGCT GAACAACATC ATCGCCAAAC TTTCGGCGGC GGGAAGCACG |
|
| 1351 | GAACCTTTGG CGGAAGCATT GGCACAGCGT TCCATTATTT ACGaacAGTT |
|
| 1401 | cggCAAACGG GGAAAAATGA TTGCCGACCT tgaAACcgcg CTCAAACTTA |
|
| 1451 | CGCCCGATAA TGCACAAATT ATGAATAATC TGGGCTACAG CCTGCTTTCC |
|
| 1501 | GATTCCAAAC GTTTGGACGA GGGTTTCGCC CTGCTTCAGA CGGCATACCA |
|
| 1551 | AATCAACCCG GACGATACCG CCGTTAACGA CAGCATAGGC TGGGCGTATT |
|
| 1601 | ACCTGAAAGG CGACgcggaA AGCGCGCTGC CGTATCTGcg gtattcgttt |
|
| 1651 | gAAAACGACC CCGAGCCCGA AGTTGCCGCC CATTTGGGCG AAGTGTTGTG |
|
| 1701 | GGCATTGGGC GAACGCGATC AGGCGGTTGA CGTATGGACG CAGGCGGCAC |
|
| 1751 | ACCTTAGGGG AGACAAGAAA ATATGGCGGG AGACGCTCAA ACGCTACGGA |
|
| 1801 | ATCGCCTTGC CCGAGCCTTC CCGAAAACCC CGGAAATAA |
[0346]This encodes a protein having amino acid sequence <SEQ ID 48>:
[0000]| 1 | MLPARFTILS VLAAALLAGQ AYAAGAADVE LPKEVGKVLR KHRRYSEEEI | |
|
| 51 | KNERARLAAV GERVNRVFTL LGGETALQKG QAGTALATYM LMLERTKSPE |
|
| 101 | VAERALEMAV SLNAFEQAEM IYQKWRQIEP IPGEAQKPAG WLRNVLKEGG |
|
| 151 | NQHLDGLKEV LAQSDDVQKR RIFLLLVQAA VQQGGVAQKA SKAVRRAALK |
|
| 201 | YEHLPEAAVA DAVFGVQGRE KEKAIEALQR LAKLDTEILP PTLMTLRLTA |
|
| 251 | RKYPEILDGF FEQTDTQNLS AVWQEMEIMN LVSLRKPDDA YARLNVLLEH |
|
| 301 | NPNANLYIQA AILAANRKEG ASVIDGYAEK AYGRGTGEQR GRAAMTAAMI |
|
| 351 | YADRRDYAKV RQWLKKVSAP EYLFDKGVLA AAAAAELDGG RAALRQIGRV |
|
| 401 | RKLPEQQGRY FTADNLSKIQ MLALSKLPDK REALIGLNNI IAKLSAAGST |
|
| 451 | EPLAEALAQR SIIYEQFGKR GKMIADLETA LKLTPDNAQI MNNLGYSLLS |
|
| 501 | DSKRLDEGFA LLQTAYQINP DDTAVNDSIG WAYYLKGDAE SALPYLRYSF |
|
| 551 | ENDPEPEVAA HLGEVLWALG ERDQAVDVWT QAAHLRGDKK IWRETLKRYG |
|
| 601 | IALPEPSRKP RK* |
[0347]ORF9ng and ORF9-1 show 88.1% identity in 614 aa overlap:
[0000]
[0348]In addition, ORF9ng shows significant homology with a hypothetical protein from P. aeruginosa:
[0000]| sp|P42810|YHE3_PSEAE HYPOTHETICAL 64.8 KD PROTEIN IN HEMM-HEMA | |
| INTERGENIC REGION (ORF3) |
| >gi|1072999|pir||S49376 hypothetical protein 3 - Pseudomonas aeruginosa >gi|557259 |
| (X82071) orf3 [Pseudomonas aeruginosa] Length = 576 |
| Score = 128 bits (318), Expect = 1e−28 |
| Identities = 138/587 (23%), Positives = 228/587 (38%), Gaps = 125/587 (21%) |
|
| Query: | 67 | VFTLLGGETALQKGQAGTALATYMLMLERTKSPEVAERALEMAVSLNAFEQAEMIYQKWR | 126 | |
| | +++LL E A Q+ + AL+ Y++ ++T+ P V+ERA +A L A ++A W |
| Sbjct: | 53 | LYSLLVAELAGQRNRFDIALSNYVVQAQKTRDPGVSERAFRIAEYLGADQEALDTSLLWA | 112 |
|
| Query: | 127 | QIEPIPGEAQKPAG--------------WLRNVLKEGGNQHLDGLKEVLAQSDDVQKRRI | 172 |
| | + P +AQ+ A ++ VL G+ H D L A++D + + |
| Sbjct: | 113 | RSAPDNLDAQRAAAIQLARAGRYEESMVYMEKVLNGQGDTHFDFLALSAAETDPDTRAGL | 172 |
|
| Query: | 173 | FXXXXXXXXXXXXXXXKASKAVRRAALKYEHLPEAAVADAVFGVQGREKEKAIEALQRLA | 232 |
| | ++ KY + + A+ Q ++A+ L+ + |
| Sbjct: | 173 | L------------------QSFDHLLKKYPNNGQLLFGKALLLQQDGRPDEALTLLEDNS | 214 |
|
| Query: | 233 | KLDTEILPPTLMTLRLTARK-----YPEILDGFFEQTDTQNLSAVWQEMEIMNLVSLRKP | 287 |
| | E+ P L + L + K P + G E D + + + + LV + |
| Sbjct: | 215 | ASRHEVAPLLLRSRLLQSMKRSDEALPLLKAGIKEHPDDKRVRLAYARL----LVEQNRL | 270 |
|
| Query: | 288 | DDAYARLNVLLEHNPN---------------------ANLYIQAAI-------------- | 312 |
| | DDA A L++ P+ A +Y++ + |
| Sbjct: | 271 | DDAKAEFAGLVQQFPDDDDDLRFSLALVCLEAQAWDEARIYLEELVERDSHVDAAHFNLG | 330 |
|
| Query: | 313 | -LAANRKEGASVIDGYAEKAYGRGTGEQRGRAAMTAAMIYADRRDYAKVRQWLKKVSAPE | 371 |
| | LA +K+ A +D YA+ G G + T ++ A R D A R + P+ |
| Sbjct: | 331 | RLAEEQKDTARALDEYAQ--VGPGNDFLPAQLRQTDVLLKAGRVDEAAQRLDKARSEQPD | 388 |
|
| Query: | 372 | YLFDKXXXXXXXXXXXXXXXXXXRQIGRVRKLPEQQGRYFTADNLSKIQMLALSKLPDKR | 431 |
| | Y A L I+ ALS + |
| Sbjct: | 389 | Y----------------------------------------AIQLYLIEAEALSNNDQQE | 408 |
|
| Query: | 432 | EALIGLNNIIAKLSAAGSTEPLAEALAQRSIIYEQFGKRGKMIADLETALKLTPDNAQIM | 491 |
| | +A + + + E L L RS++ E+ +M DL + PDNA + |
| Sbjct: | 409 | KAWQAIQEGLKQYP-----EDL-NLLYTRSMLAEKRNDLAQMEKDLRFVIAREPDNAMAL | 462 |
|
| Query: | 492 | NNLGYSLLSDSKRLDEGFALLQTAYQINPDDTAVNDSIGWAYYLKGDAESALPYLRYSFE | 551 |
| | N LGY+L + R E L+ A+++NPDD A+ DS+GW Y +G A YLR + + |
| Sbjct: | 463 | NALGYTLADRTTRYGEARELILKAHKLNPDDPAILDSMGWINYRQGKLADAERYLRQALQ | 522 |
|
| Query: | 552 | NDPEPEVAAHLGEVLWALGERDQAVDVWTQAAHLRGDKKIWRETLKR | 598 |
| | P+ EVAAHLGEVLWA G + A +W + + D + R T+KR |
| Sbjct: | 523 | RYPDHEVAAHLGEVLWAQGRQGDARAIWREYLDKQPDSDVLRRTIKR | 569 |
|
| gi|2983399 (AE000710) hypothetical protein [Aquifex aeolicus] Length = 545 | |
| Score = 81.5 bits (198), Expect = 1e−14 |
| Identities = 61/198 (30%), Positives = 98/198 (48%), Gaps = 19/198 (9%) |
|
| Query: | 408 | GRYFTADNL-SKIQMLALSKLPDKREALIGLNNIIAKLSAAGSTEPLAEALAQ------- | 459 | |
| | G Y A L K ++LA PDK+E L + +K + + L + |
| Sbjct: | 335 | GNYEDAKRLIEKAKVLA----PDKKEILFLEADYYSKTKQYDKALEILKKLEKDYPNDSR | 390 |
|
| Query: | 460 | ----RSIIYEQFGKRGKMIADLETALKLTPDNAQIMNNLGYSLLS--DSKRLDEGFALLQ | 513 |
| | +I+Y+ G L A++L P+N N LGYSLL +R++E L++ |
| Sbjct: | 391 | VYFMEAIVYDNLGDIKNAEKALRKAIELDPENPDYYNYLGYSLLLWYGKERVEEAEELIK | 450 |
|
| Query: | 514 | TAYQINPDDTAVNDSIGWAYYLKGDAESALPYLRYSF-ENDPEPEVAAHLGEVLWALGER | 572 |
| | A + +P++ A DS+GW YYLKGD E A+ YL + E +P V H+G+VL +G + |
| Sbjct: | 451 | KALEKDPENPAYIDSMGWVYYLKGDYERAMQYLLKALREAYDDPVVNEHVGDVLLKMGYK | 510 |
|
| Query: | 573 | DQAVDVWTQAAHLRGDKK | 590 |
| | ++A + + +A L + K |
| Sbjct: | 511 | EEARNYYERALKLLEEGK | 528 |
[0349]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 7
[0350]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 49>:
[0000]| 1 | AACCTCTACG CCGGCCCGCA GACCACATCC GTCATCGCAA ACATCGCCGA | |
|
| 51 | CAACCTGCAA CTGGCCAAAG ACTACGGCAA AGTACACTGG TTCGCCTCCC |
|
| 101 | CGCTCTTCTG GCTCCTGAAC CAACTGCACA ACATCATCGG CAACTGGGGC |
|
| 151 | TGGGCGATTA TCGTTTTAAC CATCATCGTC AAAGCCGTAC TGTATCCATT |
|
| 201 | GACCAACGCC TCTTACCGCT CTATGGCGAA AATGCGTGCC GCCGCACCCA |
|
| 251 | AACTGCAAGC CATCAAAGAG AAATACGGCG ACGACCGTAT GGCGCAACAA |
|
| 301 | CAGGCGATGA TGCAGCTTTA CACAGACGAG AAAATCAACC CGaCTGGGCG |
|
| 351 | GCTGCCTGCC TATGCTGTTG CAAATCCCCG TCTTCATCGG ATTGTATTGG |
|
| 401 | GCATTGTTCG CCTCCGTAGA ATTGCGCCAG GCACCTTGGC TGGGTTGGAT |
|
| 451 | TACCGACCTC AGCCGCGCCG ACCCCTACTA CATCCTGCCC ATCATTATGG |
|
| 501 | CGGCAACGAT GTTCGCCCAA ACTTATCTGA ACCCGCCGCC GAcCGACCCG |
|
| 551 | ATGCagGCGA AAATGATGAA AATCATGCCG TTGGTTTTCT CsGwCrTGTT |
|
| 601 | CTTCTTCTTC CCTGCCGGks TGGTATTGTA CTGGGTAGTC AACAACCTCC |
|
| 651 | TGACCATCGC CCAGCAATGG CACATCAACC GCAGCATCGA AAAACAACGC |
|
| 701 | GCCCAAGGCG AAGTCGTTTC CTAA |
[0351]This corresponds to the amino acid sequence <SEQ ID 50; ORF11>:
[0000]| 1 | ..NLYAGPQTTS VIANIADNLQ LAKDYGKVHW FASPLFWLLN QLHNIIGNWG | |
|
| 51 | WAIIVLTIIV KAVLYPLTNA SYRSMAKMRA AAPKLQAIKE KYGDDRMAQQ |
|
| 101 | QAMMQLYTDE KINPLGGCLP MLLQIPVFIG LYWALFASVE LRQAPWLGWI |
|
| 151 | TDLSRADPYY ILPIIMAATM FAQTYLNPPP TDPMQAKMMK IMPLVFSXXF |
|
| 201 | FFFPAGXVLY WVVNNLLTIA QQWHINRSIE KQRAQGEVVS * |
[0352]Further sequence analysis revealed the complete DNA sequence <SEQ ID 51>:
[0000]| 1 | ATGGATTTTA AAAGACTCAC GGCGTTTTTC GCCATCGCGC TGGTGATTAT | |
|
| 51 | GATCGGCTGG GAAAAGATGT TCCCCACTCC GAAGCCAGTC CCCGCGCCCC |
|
| 101 | AACAGGCAGC ACAACAACAG GCCGTAACCG CTTCCGCCGA AGCCGCGCTC |
|
| 151 | GCGCCCGCAA CGCCGATTAC CGTAACGACC GACACGGTTC AAGCCGTCAT |
|
| 201 | TGATGAAAAA AGCGGCGACC TGCGCCGGCT GACCCTGCTC AAATACAAAG |
|
| 251 | CAACCGGCGA CGAAAATAAA CCGTTCATCC TGTTTGGCGA CGGCAAAGAA |
|
| 301 | TACACCTACG TCGCCCAATC CGAACTTTTG GACGCGCAGG GCAACAACAT |
|
| 351 | TCTAAAAGGC ATCGGCTTTA GCGCACCGAA AAAACAGTAC AGCTTGGAAG |
|
| 401 | GCGACAAAGT TGAAGTCCGC CTGAGCGCGC CTGAAACACG CGGTCTGAAA |
|
| 451 | ATCGACAAAG TTTATACTTT CACCAAAGGC AGCTATCTGG TCAACGTCCG |
|
| 501 | CTTCGACATC GCCAACGGCA GCGGTCAAAC CGCCAACCTG AGCGCGGACT |
|
| 551 | ACCGCATCGT CCGCGACCAC AGCGAACCCG AGGGTCAAGG TTACTTTACC |
|
| 601 | CACTCTTACG TCGGCCCTGT TGTTTATACC CCTGAAGGCA ACTTCCAAAA |
|
| 651 | AGTCAGCTTT TCCGACTTGG ACGACGATGC CAAATCCGGC AAATCCGAGG |
|
| 701 | CCGAATACAT CCGCAAAACC CCGACCGGCT GGCTCGGCAT GATTGAACAC |
|
| 751 | CACTTCATGT CCACCTGGAT TCTCCAACCT AAAGGCAGAC AAAGCGTTTG |
|
| 801 | CGCCGCAGGC GAGTGCAACA TCGACATCAA ACGCCGCAAC GACAAGCTGT |
|
| 851 | ACAGCACCAG CGTCAGCGTG CCTTTAGCCG CCATCCAAAA CGGCGCGAAA |
|
| 901 | GCCGAAGCCT CCATCAACCT CTACGCCGGC CCGCAGACCA CATCCGTCAT |
|
| 951 | CGCAAACATC GCCGACAACC TGCAACTGGC CAAAGACTAC GGCAAAGTAC |
|
| 1001 | ACTGGTTCGC CTCCCCGCTC TTCTGGCTCC TGAACCAACT GCACAACATC |
|
| 1051 | ATCGGCAACT GGGGCTGGGC GATTATCGTT TTAACCATCA TCGTCAAAGC |
|
| 1101 | CGTACTGTAT CCATTGACCA ACGCCTCTTA CCGCTCTATG GCGAAAATGC |
|
| 1151 | GTGCCGCCGC ACCCAAACTG CAAGCCATCA AAGAGAAATA CGGCGACGAC |
|
| 1201 | CGTATGGCGC AACAACAGGC GATGATGCAG CTTTACACAG ACGAGAAAAT |
|
| 1251 | CAACCCGCTG GGCGGCTGCC TGCCTATGCT GTTGCAAATC CCCGTCTTCA |
|
| 1301 | TCGGATTGTA TTGGGCATTG TTCGCCTCCG TAGAATTGCG CCAGGCACCT |
|
| 1351 | TGGCTGGGTT GGATTACCGA CCTCAGCCGC GCCGACCCCT ACTACATCCT |
|
| 1401 | GCCCATCATT ATGGCGGCAA CGATGTTCGC CCAAACTTAT CTGAACCCGC |
|
| 1451 | CGCCGACCGA CCCGATGCAG GCGAAAATGA TGAAAATCAT GCCGTTGGTT |
|
| 1501 | TTCTCCGTCA TGTTCTTCTT CTTCCCTGCC GGTCTGGTAT TGTACTGGGT |
|
| 1551 | AGTCAACAAC CTCCTGACCA TCGCCCAGCA ATGGCACATC AACCGCAGCA |
|
| 1601 | TCGAAAAACA ACGCGCCCAA GGCGAAGTCG TTTCCTAA |
[0353]This corresponds to the amino acid sequence <SEQ ID 52; ORF11-1>:
[0000]| 1 | MDFKRLTAFF AIALVIMIGW EKMFPTPKPV PAPQQAAQQQ AVTASAEAAL | |
|
| 51 | APATPITVTT DTVQAVIDEK SGDLRRLTLL KYKATGDENK PFILFGDGKE |
|
| 101 | YTYVAQSELL DAQGNNILKG IGFSAPKKQY SLEGDKVEVR LSAPETRGLK |
|
| 151 | IDKVYTFTKG SYLVNVRFDI ANGSGQTANL SADYRIVRDH SEPEGQGYFT |
|
| 201 | HSYVGPVVYT PEGNFQKVSF SDLDDDAKSG KSEAEYIRKT PTGWLGMIEH |
|
| 251 | HFMSTWILQP KGRQSVCAAG ECNIDIKRRN DKLYSTSVSV PLAAIQNGAK |
|
| 301 | AEASINLYAG PQTTSVIANI ADNLQLAKDY GKVHWFASPL FWLLNQLHNI |
|
| 351 | IGNWGWAIIV LTIIVKAVLY PLTNASYRSM AKMRAAAPKL QAIKEKYGDD |
|
| 401 | RMAQQQAMMQ LYTDEKINPL GGCLPMLLQI PVFIGLYWAL FASVELRQAP |
|
| 451 | WLGWITDLSR ADPYYILPII MAATMFAQTY LNPPPTDPMQ AKMMKIMPLV |
|
| 501 | FSVMFFFFPA GLVLYWVVNN LLTIAQQWHI NRSIEKQRAQ GEVVS* |
[0354]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a 60 kDa Inner-Membrane Protein (Accession P25754) of Pseudomonas putida
[0355]ORF11 and the 60 kDa protein show 58% aa identity in 229 aa overlap (BLASTp).
[0000]| ORF11 | 2 | LYAGPQTTSVIANIADNLQLAKDYGKVHWFASPLFWLLNQLHNIIGNWGWAIIVLTIIVK | 61 | |
| | LYAGP+ S + ++ L+L DYG + + A P+FWLL +H+++GNWGW+IIVLT+++K |
| 60K | 324 | LYAGPKIQSKLKELSPGLELTVDYGFLWFIAQPIFWLLQHIHSLLGNWGWSIIVLTMLIK | 383 |
|
| ORF11 | 62 | AVLYPLTNASYRSMAKMRAAAPKLQAIKEKYGDDRXXXXXXXXXLYTDEKINPLGGCLPM | 121 |
| | + +PL+ ASYRSMA+MRA APKL A+KE++GDDR LY EKINPLGGCLP+ |
| 60K | 384 | GLFFPLSAASYRSMARMRAVAPKLAALKERFGDDRQKMSQAMMELYKKEKINPLGGCLPI | 443 |
|
| ORF11 | 122 | LLQIPVFIGLYWALFASVELRQAPWLGWITDLSRADPYYILPIIMAATMFAQTYLNPPPT | 181 |
| | L+Q+PVF+ LYW L SVE+RQAPW+ WITDLS DP++ILPIIM ATMF Q LNP P |
| 60K | 444 | LVQMPVFLALYWVLLESVEMRQAPWILWITDLSIKDPFFILPIIMGATMFIQQRLNPTPP | 503 |
|
| ORF11 | 182 | DPMQAKMMKIMPLVXXXXXXXXPAGXVLYWVVNNLLTIAQQWHINRSIE | 230 |
| | DPMQAK+MK+MP++ PAG VLYWVVNN L+I+QQW+I R IE |
| 60K | 504 | DPMQAKVMKMMPIIFTFFFLWFPAGLVLYWVVNNCLSISQQWYITRRIE | 552 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0356]ORF11 shows 97.9% identity over a 240aa overlap with an ORF (ORF11a) from strain A of N. meningitidis.
[0000]
[0357]The complete length ORF11a nucleotide sequence <SEQ ID 53> is:
[0000]| 1 | ANGGATTTTA AAAGACTCAC NGNGTTTTTC GCCATCGCAC TGGTGATTAT | |
|
| 51 | GATCGGATNG NAAANGATGT TCCCCACTCC GAAGCCCGTC CCCGCGCCCC |
|
| 101 | AACAGACGGC ACAACAACAG GCCGTAANCG CTTCCGCCGA AGCCGCGCTC |
|
| 151 | GCGCCCGNAN CGCCGATTAC CGTAACGACC GACACGGTTC AAGCCGTCAT |
|
| 201 | TGATGAAAAA AGCGGCGACC TGCGCCGGCT GACCCTGCTC AAATACAAAG |
|
| 251 | CAACCGGCGA CNAAAATAAA CCGTTCATCC TGTTTGGCGA CGGCAAANAA |
|
| 301 | TACACCTACN TCGCCCANTC CGAACTTTTG GACGCGCAGG GCAACAACAT |
|
| 351 | TCTAAAAGGC ATCGGCTTTA GCGCACCGAA AAAACAGTAC AGCTTGGAAG |
|
| 401 | GCGACAAAGT TGAAGTCCGC CTGAGCGCAC CTGAAACACG CGGTCTGAAA |
|
| 451 | ATCGACAAAG TTTATACTTT CACCAAAGGC AGCTATCTGG TCAACGTCCG |
|
| 501 | CTTCGACATC GCCAACGGCA GCGGTCAAAC CGCCAACCTG AGCGCGGACT |
|
| 551 | ACCGCATCGT CCGCGACCAC AGCGAACCCG AGGGTCAAGG CTACTTTACC |
|
| 601 | CACTCTTACG TCGGCCCTGT TGTTTATACC CCTGAAGGCA ACTTCCAAAA |
|
| 651 | AGTCAGCTTC TCCGACTTGG ACGACGATGC CAANTCCGGN AAATCCGAGG |
|
| 701 | CCGAATACAT CCGCAAAACC CNGACCGGCT GGCTCGGCAT GATTGAACAC |
|
| 751 | CACTTCATGT CCACCTGGAT CCTCCAACCC AAAGGCGGAC AAAGCGTTTG |
|
| 801 | CGCCGCTGGC GACTGCNGTA TNGACATCAA ACGCCGCAAC GACAAGCTGT |
|
| 851 | ACAGCACCAG CGTCAGCGTG CCTTTAGCCG CTATCCAAAA CGGTGCGAAA |
|
| 901 | TCCNAAGCCT CCATCAACCT CTACGCCGGC CCACAGACCA CATCNGTTAT |
|
| 951 | CGCAAACATC GCCGACAACC TGCAACTGGN CAAAGACTAC GGCAAAGTAC |
|
| 1001 | ACTGGTTCGC CTCCCCCCTC TTTTGGCTTT TGAACCAACT GCACAACATC |
|
| 1051 | ATCGGCAACT GGGGCTGGGC GATTATCGTT TTAACCATCA TCGTCAAAGC |
|
| 1101 | CGTACTGTAT CCATTGACCA ACGCCTCTTA CCGTTCGATG GCGAAAATGC |
|
| 1151 | GTGCCGCCGC GCCCAAACTG CAAGCCATCA AAGAGAAATA CGGCGACGAC |
|
| 1201 | CGTATGGCGC AGCAACAAGC CATGATGCAG CTTTACACAG ACGAGAAAAT |
|
| 1251 | CAACCCGCTG GGCGGCTGCC TGCCTATGCT GTTGCAAATC CCCGTCTTCA |
|
| 1301 | TCGGATTGTA TTGGGCATTG TTCGCCTCCG TAGAATTGCG CCAGGCACCT |
|
| 1351 | TGGCTGGGTT GGATTACCGA CCTCAGCCGC GCCGACCCNT ACTACATCCT |
|
| 1401 | GCCCATCATT ATGGCGGCAA CGATGTTCGC CCAAACCTAT CTGAACCCGC |
|
| 1451 | CGCCGACCGA CCCGATGCAG GCGAAAATGA TGAAAATCAT GCCTTTGGTT |
|
| 1501 | NTNTCNNNNA NGTTCTTCNN CTTCCCTGCC GGTCTGGTAT TGTACTGGGT |
|
| 1551 | GATCAACAAC CTCCTGACCA TCGCCCAGCA ATGGCACATC AACCGCAGCA |
|
| 1601 | TCGAAAAACA ACGCGCCCAA GGCGAAGTCG TTTCCTAA |
[0358]This encodes a protein having amino acid sequence <SEQ ID 54>:
[0000]| 1 | XDFKRLTXFF AIALVIMIGX XXMFPTPKPV PAPQQTAQQQ AVXASAEAAL | |
|
| 51 | APXXPITVTT DTVQAVIDEK SGDLRRLTLL KYKATGDXNK PFILFGDGKX |
|
| 101 | YTYXAXSELL DAQGNNILKG IGFSAPKKQY SLEGDKVEVR LSAPETRGLK |
|
| 151 | IDKVYTFTKG SYLVNVRFDI ANGSGQTANL SADYRIVRDH SEPEGQGYFT |
|
| 201 | HSYVGPVVYT PEGNFQKVSF SDLDDDAXSG KSEAEYIRKT XTGWLGMIEH |
|
| 251 | HFMSTWILQP KGGQSVCAAG DCXXDIKRRN DKLYSTSVSV PLAAIQNGAK |
|
| 301 | SXASINLYAG PQTTSVIANI ADNLQLXKDY GKVHWFASPL FWLLNQLHNI |
|
| 351 | IGNWGWAIIV LTIIVKAVLY PLTNASYRSM AKMRAAAPKL QAIKEKYGDD |
|
| 401 | RMAQQQAMMQ LYTDEKINPL GGCLPMLLQI PVFIGLYWAL FASVELRQAP |
|
| 451 | WLGWITDLSR ADPYYILPII MAATMFAQTY LNPPPTDPMQ AKMMKIMPLV |
|
| 501 | XSXXFFXFPA GLVLYWVINN LLTIAQQWHI NRSIEKQRAQ GEVVS* |
[0359]ORF11a and ORF11-1 show 95.2% identity in 544 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0360]ORF11 shows 96.3% identity over a 240aa overlap with a predicted ORF (ORF11.ng) from N. gonorrhoeae:
[0000]
[0361]An ORF11ng nucleotide sequence <SEQ ID 55> was predicted to encode a protein having amino acid sequence <SEQ ID 56>:
[0000]| 1 | MAVNLYAGPQ TTSVIANIAD NLQLAKDYGK VHWFASPLFW LLNQLHNIIG | |
|
| 51 | NWGWAIVVLT IIVKAVLYPL TNASYRSMAK MRAAAPELQT IKEKYGDDRM |
|
| 101 | AQQQAMMQLF EDEEINPLGG CLPMLLQIPV FIGLYWALFA SVELRQAPWL |
|
| 151 | GWITDLSRAD PYYILPIIMA ATMFAQTYLN PPPTDPMQAK MMKIMPLVFS |
|
| 201 | VMFFFFPAGL VLYWVVNNLL TIAQQWHINR SIEKQRAQGE VVS* |
[0362]Further sequence analysis revealed the complete gonococcal DNA sequence <SEQ ID 57> to be:
[0000]| 1 | ATGGATTTTA AAAGACTCAC GGCGTTTTTC GCCATCGCGC TGGTGATTAT | |
|
| 51 | GATCGGCTGG GAAAAAATGT TCCCCACCCC GAAACCCGTC CCCGCGCCCC |
|
| 101 | AACAGGCGGC ACAAAAACAG GCAGCAACCG CTTCCGCCGA AGCCGCGCTC |
|
| 151 | GCGCCCGCAA CGCCGATTAC CGTAACGACC GACACGGTTC AAGCCGTTAT |
|
| 201 | TGATGAAAAA AGTGGCGACC TGCGCCGGCT GACCCTGCTC AAATACAAAG |
|
| 251 | CAACCGGCGA CGAAAACAAA CCGTTCGTCC TGTTTGGCGA CGGCAAAGAA |
|
| 301 | TACACCTACG TCGCCCAATC CGAACTTTTG GACGCGCAGG GCAACAACAT |
|
| 351 | TCTGAAAGGC ATCGGCTTTA GCGCACCGAA AAAACAGTAC ACCCTCAACG |
|
| 401 | GCGACACAGT CGAAGTCCGC CTGAGCGCGC CCGAAACCAA CGGACTGAAA |
|
| 451 | ATCGACAAAG TCTATACCTT TACCAAAGAC AGCTATCTGG TCAACGTCCG |
|
| 501 | CTTCGACATC GCCAACGGCA GCGGTCAAAC CGCCAACCTG AGCGCGGACT |
|
| 551 | ACCGCATCGT CCGCGACCAC AGCGAACCCG AGGGTCAAGG CTACTTTACC |
|
| 601 | CACTCTTACG TCGGCCCTGT TGTTTATACC CCTGAAGGCA ACTTCCAAAA |
|
| 651 | AGTCAGCTTC TCCgacTTgg acgACGATGC gaaaTccggc aaATccgagg |
|
| 701 | ccgaatacaT CCGCAAAACC ccgaccggtt ggctcggcat gattgaacac |
|
| 751 | cacttcatgt ccacctggat cctccAAcct aaaggcggcc aaaacgtttg |
|
| 801 | cgcccaggga gactgccgta tcgacattaa aCgccgcaac gacaagctgt |
|
| 851 | acagcgcaag cgtcagcgtg cctttaaccg ctatcccaac ccgggggcca |
|
| 901 | aaaccgaaaa tggcggTCAA CCTGTATGCC GGTCCGCAAA CCACATCCGT |
|
| 951 | TATCGCAAAC ATCGCcgacA ACCTGCAACT GGCAAAAGAC TACGGTAAAG |
|
| 1001 | TACACTGGTT CGCATCGCCG CTCTTCTGGC TCCTGAACCA ACTGCACAAC |
|
| 1051 | ATTATCGGCA ACTGGGGCTG GGCAATCGTC GTTTTGACCA TCATCGTCAA |
|
| 1101 | AGCCGTACTG TATCCATTGA CCAACGcctc ctACCGTTCG ATGGCGAAAA |
|
| 1151 | TGCGTGccgc cgcacCcaaA CTGCAGACCA TCAAAGAAAA ATAcgGCGAC |
|
| 1201 | GACCGTATGG CGCAACAGCA AGCGATGATG CAGCTTTACA AAgacgAGAA |
|
| 1251 | AATCAACCCG CTGGGCGGCT GTctgcctat gctgttgCAA ATCCCCGTCT |
|
| 1301 | TCATCGGCTT GTACTGGGCA TTGTTCGCCT CCGTAGAATT GCGCCAGGCA |
|
| 1351 | CCTTGGCTGG GCTGGATTAC CGACCTCAGC CGCGCCGACC CCTACTACAT |
|
| 1401 | CCTGCCCATC ATTATGGCGG CAACGATGTT CGCCCAAACC TATCTGAACC |
|
| 1451 | CGCCGCCGAC CGACCCGATG CAGGCGAAAA TGATGAAAAT CATGCCGTTG |
|
| 1501 | GTTTTCTCCG TCATGTTCTT CTTCTTCCCT GCCGGTTTGG TTCTCTACTG |
|
| 1551 | GGTGGTCAAC AACCTCCTGA CCATCGCCCA GCAGTGGCAC ATCAACCGCA |
|
| 1601 | GCATCGAAAA ACAACGCGCC CAAGGCGAAG TCGTTTCCTA A |
[0363]This encodes a protein having amino acid sequence <SEQ ID 58; ORF11ng-1>:
[0000]| 1 | MDFKRLTAFF AIALVIMIGW EKMFPTPKPV PAPQQAAQKQ AATASAEAAL | |
|
| 51 | APATPITVTT DTVQAVIDEK SGDLRRLTLL KYKATGDENK PFVLFGDGKE |
|
| 101 | YTYVAQSELL DAQGNNILKG IGFSAPKKQY TLNGDTVEVR LSAPETNGLK |
|
| 151 | IDKVYTFTKD SYLVNVRFDI ANGSGQTANL SADYRIVRDH SEPEGQGYFT |
|
| 201 | HSYVGPVVYT PEGNFQKVSF SDLDDDAKSG KSEAEYIRKT PTGWLGMIEH |
|
| 251 | HFMSTWILQP KGGQNVCAQG DCRIDIKRRN DKLYSASVSV PLTAIPTRGP |
|
| 301 | KPKMAVNLYA GPQTTSVIAN IADNLQLAKD YGKVHWFASP LFWLLNQLHN |
|
| 351 | IIGNWGWAIV VLTIIVKAVL YPLTNASYRS MAKMRAAAPK LQTIKEKYGD |
|
| 401 | DRMAQQQAMM QLYKDEKINP LGGCLPMLLQ IPVFIGLYWA LFASVELRQA |
|
| 451 | PWLGWITDLS RADPYYILPI IMAATMFAQT YLNPPPTDPM QAKMMKIMPL |
|
| 501 | VFSVMFFFFP AGLVLYWVVN NLLTIAQQWH INRSIEKQRA QGEVVS* |
[0364]ORF11ng-1 and ORF11-1 shown 95.1% identity in 546 aa overlap:
[0000]
[0365]In addition, ORF11ng-1 shows significant homology with an inner-membrane protein from the database (accession number p25754):
[0000]
[0366]Based on this analysis, including the homology to an inner-membrane protein from P. putida and the predicted transmembrane domains (seen in both the meningococcal and gonoccal proteins), it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 8
[0367]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 59>:
[0000]| 1 | ..GCCGTCTTAA TCATCGAATT ATTGACGGGA ACGGTTTATC TTTTGGTTGT | |
|
| 51 | NAGCGCGGCT TTGGCGGGTT CGGGCATTGC TTACGGGCTG ACCGGCAGTA |
|
| 101 | CGCCTGCCGC CGTCTTGACC GNCGCTCTGC TTTCCGCGCT GGGTATTTNG |
|
| 151 | TTCGTACACG CCAAAACCGC CGTTAGAAAA GTTGAAACGG ATTCATATCA |
|
| 201 | GGATTTGGAT GCCGGACAAT ATGTCGAAAT CCTCCGNCAC ACAGGCGGCA |
|
| 251 | ACCGTTACGA AGTT.TTTAT CGCGGTACG. ACTGGCAGGC TCAAAATACG |
|
| 301 | GGGCAAGAAG AGCTTGAACC AGGAACTCGC GCCCTCATTG TCCGCAAGGA |
|
| 351 | AGGCAACCTT CTTATTATCA CACACCCTTA A |
[0368]This corresponds to the amino acid sequence <SEQ ID 60; ORF13>:
[0000]| 1 | ..AVLIIELLTG TVYLLVVSAA LAGSGIAYGL TGSTPAAVLT XALLSALGIX | |
|
| 51 | FVHAKTAVRK VETDSYQDLD AGQYVEILRH TGGNRYEVXY RGTXWQAQNT |
|
| 101 | GQEELEPGTR ALIVRKEGNL LIITHP* |
[0369]Further sequence analysis elaborated the DNA sequence slightly <SEQ ID 61>:
[0000]| 1 | ..GCCGTCTTAA TCATCGAATT ATTGACGGGA ACGGTTTATC TTTTGGTTGT | |
|
| 51 | nAGCGCGGCT TTGGCGGGTT CGGGCATTGC TTACGGGCTG ACCGGCAGTA |
|
| 101 | CGCCTGCCGC CGTCTTGACC GnCGCTCTGC TTTCCGCGCT GGGTATTTnG |
|
| 151 | TTCGTACACG CCAAAACCGC CGTTAGAAAA GTTGAAACGG ATTCATATCA |
|
| 201 | GGATTTGGAT GCCGGACAAT ATGTCGAAAT CCTCCGACAC ACAGGCGGCA |
|
| 251 | ACCGTTACGA AGTTTTtTAT CGCGGTACGc ACTGGCAGGC TCAAAATACG |
|
| 301 | GGGCAAGAAG AGCTTGAACC AGGAACTCGC GCCCTCATTG TCCGCAAGGA |
|
| 351 | AGGCAACCTT CTTATTATCA CACACCCTTA A |
[0370]This corresponds to the amino acid sequence <SEQ ID 62; ORF13-1>:
[0000]| 1 | ..AVLIIELLTG TVYLLVVSAA LAGSGIAYGL TGSTPAAVLT XALLSALGIX | |
|
| 51 | FVHAKTAVRK VETDSYQDLD AGQYVEILRH TGGNRYEVFY RGTHWQAQNT |
|
| 101 | GQEELEPGTR ALIVRKEGNL LIITHP* |
[0371]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0372]ORF13 shows 92.9% identity over a 126aa overlap with an ORF (ORF13a) from strain A of N. meningitidis:
[0000]
[0373]The complete length ORF13a nucleotide sequence <SEQ ID 63> is:
[0000]| 1 | ATGACTGTAT GGTTTGTTGC CGCTGTTGCC GTCTTAATCA TCGAATTATT | |
|
| 51 | GACGGGAACG GTTTATCTTT TGGTTGTCAG CGCGGCTTTG GCGGGTTCGG |
|
| 101 | GCATTGCTTA CGGGCTGACC GGCAGCACGC CTGCCGCCGT CTTGACCGCC |
|
| 151 | GCTCTGCTTT CCGCGCTGGG TATTTGGTTC GTACACGCCA AAACCGCCGT |
|
| 201 | GGGAAAAGTT GAAACGGATT CATATCAGGA TTTGGATGCC GGGCAATATG |
|
| 251 | CCGAAATCCT CCGGCACGCA GGCGGCAACC GTTACGAAGT TTTTTATCGC |
|
| 301 | GGTACGCACT GGCAGGCTCA AAATACGGGG CAAGAAGAGC TTGAACCAGG |
|
| 351 | AACGCGCGCC CTAATCGTCC GCAAGGAAGG CAACCTTCTT ATCATCGCAA |
|
| 401 | AACCTTAA |
[0374]This encodes a protein having amino acid sequence <SEQ ID 64>:
[0000]| 1 | MTVWFVAAVA VLIIELLTGT VYLLVVSAAL AGSGIAYGLT GSTPAAVLTA | |
|
| 51 | ALLSALGIWF VHAKTAVGKV ETDSYQDLDA GQYAEILRHA GGNRYEVFYR |
|
| 101 | GTHWQAQNTG QEELEPGTRA LIVRKEGNLL IIAKP* |
[0375]ORF13a and ORF13-1 show 94.4% identity in 126 aa overlap
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0376]ORF13 shows 89.7% identity over a 126aa overlap with a predicted ORF (ORF13.ng) from N. gonorrhoeae:
[0000]
[0377]The complete length ORF13ng nucleotide sequence <SEQ ID 65> is:
[0000]| 1 | ATGACTGTAT GGTTTGTTGC CGCTGTTGCC GTCTTAATCA TCGAATTATT | |
|
| 51 | GACGGGAACG GTTTATCTTT TGGTTGTCAG CGCGGCTTTG GCGGGTTCGG |
|
| 101 | GCATTGCCTA CGGGCTGACT GGCAGCACGC CTGCCGCCGT CTTGACCGCC |
|
| 151 | GCACTGCTTT CCGCGCTGGG CATTTGGTTC GTACATGCCA AAACCGCCGT |
|
| 201 | GGGAAAAGTT GAAACGGATT CATATCAGGA TTTGGATACC GGAAAATATG |
|
| 251 | CCGAAATCCT CCGATACACA GGCGGCAACC GTTACGAAGT TTTTTATCGC |
|
| 301 | GGTACGCACT GGCAGGCGCA AAATACGGGG CAGGAAGTGT TTGAACCGGG |
|
| 351 | AACGCGCGCC CTCATCGTCC GCAAAGAAGG TAACCTTCTT ATCATCGCAA |
|
| 401 | ACCCTTAA |
[0378]This encodes a protein having amino acid sequence <SEQ ID 66>:
[0000]| 1 | MTVWFVAAVA VLIIELLTGT VYLLVVSAAL AGSGIAYGLT GSTPAAVLTA | |
|
| 51 | ALLSALGIWF VHAKTAVGKV ETDSYQDLDT GKYAEILRYT GGNRYEVFYR |
|
| 101 | GTHWQAQNTG QEVFEPGTRA LIVRKEGNLL IIANP* |
[0379]ORF13ng shows 91.3% identity in 126 aa overlap with ORF13-1:
[0000]
[0380]Based on this analysis, including the extensive leader sequence in this protein, it is predicted that ORF13 and ORF13ng are likely to be outer membrane proteins. It is thus predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 9
[0381]The following DNA sequence was identified in N. meningitidis <SEQ ID 67>:
[0000]| 1 | ATGTwTGATT TCGGTTTrGG CGArCTGGTT TTTGTCGGCA TTATCGCCCT | |
|
| 51 | GATwGtCCTC GGCCCCGAAC GCsTGCCCGA GGCCGCCCGC AyCGCCGGAC |
|
| 101 | GGcTCATCGG CAGGCTGCAA CGCTTTGTCG GcAGCGTCAA ACAGGAATTT |
|
| 151 | GACACTCAAA TCGAACTGGA AGAACTGAGG AAGGCAAAGC AGGAATTTGA |
|
| 201 | AGCTGCCGcC GCTCAGGTTC GAGACAGCCT CAAAGAAACC GGTACGGATA |
|
| 251 | TGGAAGGCAA TCTGCACGAC ATTTCCGACG GTCTGAAGCC TTGGGAAAAA |
|
| 301 | CTGCCCGAAC AGCGGACACC TGCCGATTTC GGTGTCGATG AAAACGGCAA |
|
| 351 | TCCGCT.TCC CGATGCGGCA AACACCCTAT CAGACGGCAT TTCCGACGTT |
|
| 401 | ATGCCGTC.. |
[0382]This corresponds to the amino acid sequence <SEQ ID 68; ORF2>:
[0000]| 1 | MXDFGLGELV FVGIIALIVL GPERXPEAAR XAGRLIGRLQ RFVGSVKQEF | |
|
| 51 | DTQIELEELR KAKQEFEAAA AQVRDSLKET GTDMEGNLHD ISDGLKPWEK |
|
| 101 | LPEQRTPADF GVDENGNPXS RCGKHPIRRH FRRYAV.. |
[0383]Further work revealed the complete nucleotide sequence <SEQ ID 69>:
[0000]| 1 | ATGTTTGATT TCGGTTTGGG CGAGCTGGTT TTTGTCGGCA TTATCGCCCT | |
|
| 51 | GATTGTCCTC GGCCCCGAAC GCCTGCCCGA GGCCGCCCGC ACCGCCGGAC |
|
| 101 | GGCTCATCGG CAGGCTGCAA CGCTTTGTCG GCAGCGTCAA ACAGGAATTT |
|
| 151 | GACACTCAAA TCGAACTGGA AGAACTGAGG AAGGCAAAGC AGGAATTTGA |
|
| 201 | AGCTGCCGCC GCTCAGGTTC GAGACAGCCT CAAAGAAACC GGTACGGATA |
|
| 251 | TGGAAGGCAA TCTGCACGAC ATTTCCGACG GTCTGAAGCC TTGGGAAAAA |
|
| 301 | CTGCCCGAAC AGCGGACACC TGCCGATTTC GGTGTCGATG AAAACGGCAA |
|
| 351 | TCCGCTTCCC GATGCGGCAA ACACCCTATC AGACGGCATT TCCGACGTTA |
|
| 401 | TGCCGTCCGA ACGTTCCTAC GCTTCCGCCG AAACCCTTGG GGACAGCGGG |
|
| 451 | CAAACCGGCA GTACAGCCGA ACCCGCGGAA ACCGACCAAG ACCGCGCATG |
|
| 501 | GCGGGAATAC CTGACTGCTT CTGCCGCCGC ACCCGTCGTA CAGACCGTCG |
|
| 551 | AAGTCAGCTA TATCGATACT GCTGTTGAAA CGCCTGTTCC GCACACCACT |
|
| 601 | TCCCTGCGCA AACAGGCAAT AAGCCGCAAA CGCGATTTTC GTCCGAAACA |
|
| 651 | CCGCGCCAAA CCTAAATTGC GCGTCCGTAA ATCATAA |
[0384]This corresponds to the amino acid sequence <SEQ ID 70; ORF2-1>:
[0000]| 1 | MFDFGLGELV FVGIIALIVL GPERLPEAAR TAGRLIGRLQ RFVGSVKQEF | |
|
| 51 | DTQIELEELR KAKQEFEAAA AQVRDSLKET GTDMEGNLHD ISDGLKPWEK |
|
| 101 | LPEQRTPADF GVDENGNPLP DAANTLSDGI SDVMPSERSY ASAETLGDSG |
|
| 151 | QTGSTAEPAE TDQDRAWREY LTASAAAPVV QTVEVSYIDT AVETPVPHTT |
|
| 201 | SLRKQAISRK RDFRPKHRAK PKLRVRKS* |
[0385]Further work identified the corresponding gene in strain A of N. meningitidis <SEQ ID 71>:
[0000]| 1 | ATGTTTGATT TCGGTTTGGG CGAGCTGGTT TTTGTCGGCA TTATCGCCCT | |
|
| 51 | GATTGTCCTC GGCCCCGAAC GCCTGCCCGA GGCCGCCCGC ACCGCCGGAC |
|
| 101 | GGCTCATCGG CAGGCTGCAA CGCTTTGTCG GCAGCGTCAA ACAGGAATTT |
|
| 151 | GACACGCAAA TCGAACTGGA AGAACTAAGG AAGGCAAAGC AGGAATTTGA |
|
| 201 | AGCTGCCGCT GCTCAGGTTC GAGACAGCCT CAAAGAAACC GGTACGGATA |
|
| 251 | TGGAGGGTAA TCTGCACGAC ATTTCCGACG GTCTGAAGCC TTGGGAAAAA |
|
| 301 | CTGCCCGAAC AGCGCACGCC TGCTGATTTC GGTGTCGATG AAAACGGCAA |
|
| 351 | TCCCTTTCCC GATGCGGCAA ACACCCTATT AGACGGCATT TCCGACGTTA |
|
| 401 | TGCCGTCCGA ACGTTCCTAC GCTTCCGCCG AAACCCTTGG GGACAGCGGG |
|
| 451 | CAAACCGGCA GTACAGCCGA ACCCGCGGAA ACCGACCAAG ACCGTGCATG |
|
| 501 | GCGGGAATAC CTGACTGCTT CTGCCGCCGC ACCCGTCGTA CAGACCGTCG |
|
| 551 | AAGTCAGCTA TATCGATACC GCTGTTGAAA CCCCTGTTCC GCATACCACT |
|
| 601 | TCGCTGCGTA AACAGGCAAT AAGCCGCAAA CGCGATTTGC GTCCTAAATC |
|
| 651 | CCGCGCCAAA CCTAAATTGC GCGTCCGTAA ATCATAA |
[0386]This encodes a protein having amino acid sequence <SEQ ID 72; ORF2a>:
[0000]| 1 | MFDFGLGELV FVGIIALIVL GPERLPEAAR TAGRLIGRLQ RFVGSVKQEF | |
|
| 51 | DTQIELEELR KAKQEFEAAA AQVRDSLKET GTDMEGNLHD ISDGLKPWEK |
|
| 101 | LPEQRTPADF GVDENGNPFP DAANTLLDGI SDVMPSERSY ASAETLGDSG |
|
| 151 | QTGSTAEPAE TDQDRAWREY LTASAAAPVV QTVEVSYIDT AVETPVPHTT |
|
| 201 | SLRKQAISRK RDLRPKSRAK PKLRVRKS* |
[0387]The originally-identified partial strain B sequence (ORF2) shows 97.5% identity over a 118aa overlap with ORF2a:
[0000]
[0388]The complete strain B sequence (ORF2-1) and ORF2a show 98.2% identity in 228 aa overlap:
[0000]
[0389]Further work identified a partial DNA sequence <SEQ ID 73> in N. gonorrhoeae encoding the following amino acid sequence <SEQ ID 74; ORF2ng>:
[0000]| 1 | MFDFGLGELI FVGIIALIVL GPERLPEAAR TAGRLIGRLQ RFVGSVKQEL | |
|
| 51 | DTQIELEELR KVKQAFEAAA AQVRDSLKET DTDMQNSLHD ISDGLKPWEK |
|
| 101 | LPEQRTPADF GVDEKGNSLS RYGKHRIRRH FRRYAV* |
[0390]Further work identified the complete gonococcal gene sequence <SEQ ID 75>:
[0000]| 1 | ATGTTTGATT TCGGTTTGGG CGAGCTGATT TTTGTCGGCA TTATCGCCCT | |
|
| 51 | GATTGTCCTT GGTCCAGAAC GCCTGCCCGA AGCCGCCCGC ACTGCCGGAC |
|
| 101 | GGCTTATCGG CAGGCTGCAA CGCTTTGTAG GAAGCGTCAA ACAAGAACTT |
|
| 151 | GACACTCAAA TCGAACTGGA AGAGCTGAGG AAGGTCAAGC AGGCATTCGA |
|
| 201 | AGCTGCCGCC GCTCAGGTTC GAGACAGCCT CAAAGAAACC GATACGGATA |
|
| 251 | TGCAGAACAG TCTGCACGAC ATTTCCGACG GTCTGAAGCC TTGGGAAAAA |
|
| 301 | CTGCCCGAAC AGCGCACGCc tgccgatttc gGTGTCGATg AAAacggcaa |
|
| 351 | tccccttccc gATACGGCAA ACACCGTATC AGACGGCATT TCCGACGTTA |
|
| 401 | TGCCGTCTGA ACGTTCCGAT ACTtccgcCG AAACCCTTGG GGACGACAGG |
|
| 451 | CAAACCGGCA GTACAGCCGA ACCTGCGGAA ACCGACAAAG ACCGCGCATG |
|
| 501 | GCGGGAATAC CTGactgctt ctgccgccgc acctgtcgta Cagagggccg |
|
| 551 | tcgaagtcag ctaTATCGAT ACTGCTGTTG AAacgcctgT tccgcaCacc |
|
| 601 | acttccctgc gcaAACAGGC AATAAACCGC AAACGCGATT TttgtccgaA |
|
| 651 | ACACCGCGCc aAACCGAAat tgcgcgtcCG TAAATCATAA |
[0391]This encodes a protein having the amino acid sequence <SEQ ID 76; ORF2ng-1>:
[0000]| 1 | MFDFGLGELI FVGIIALIVL GPERLPEAAR TAGRLIGRLQ RFVGSVKQEL | |
|
| 51 | DTQIELEELR KVKQAFEAAA AQVRDSLKET DTDMQNSLHD ISDGLKPWEK |
|
| 101 | LPEQRTPADF GVDENGNPLP DTANTVSDGI SDVMPSERSD TSAETLGDDR |
|
| 151 | QTGSTAEPAE TDKDRAWREY LTASAAAPVV QRAVEVSYID TAVETPVPHT |
|
| 201 | TSLRKQAINR KRDFCPKHRA KPKLRVRKS* |
[0392]The originally-identified partial strain B sequence (ORF2) shows 87.5% identity over a 136aa overlap with ORF2ng:
[0000]
[0393]The complete strain B and gonococcal sequences (ORF2-1 & ORF2ng-1) show 91.7% identity in 229 aa overlap:
[0000]
[0394]Computer analysis of these amino acid sequences indicates a transmembrane region (underlined), and also revealed homology (59% identity) between the gonococcal sequence and the TatB protein of E. coli:
[0000]| gnl|PID|e1292181 (AJ005830) TatB protein [Escherichia coli] Length = 171 | |
| Score = 56.6 bits (134), Expect = 1e−07 |
| Identities = 30/88 (34%), Positives = 52/88 (59%), Gaps = 1/88 (1%) |
|
| Query: | 1 | MFDFGLGELIFVGIIALIVLGPERLPEAARTAGRLIGRLQRFVGSVKQELDTQIELEELR | 60 |
| | MFD G EL+ V II L+VLGP+RLP A +T I L+ +V+ EL +++L+E + |
| Sbjct: | 1 | MFDIGFSELLLVFIIGLVVLGPQRLPVAVKTVAGWIRALRSLATTVQNELTQELKLQEFQ | 60 |
|
| Query: | 61 | -KVKQAFEAAAAQVRDSLKETDTDMQNS | 87 |
| | +K+ +A+ + LK + +++ + |
| Sbjct: | 61 | DSLKKVEKASLTNLTPELKASMDELRQA | 88 |
[0395]Based on this analysis, it was predicted that ORF2, ORF2a and ORF2ng are likely to be membrane proteins and so the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0396]ORF2-1 (16 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 3A shows the results of affinity purification of the GST-fusion protein, and FIG. 3B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for Western blots (FIG. 3C), ELISA (positive result), and FACS analysis (FIG. 3D). These experiments confirm that ORF37-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 10
[0397]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 77>:
[0000]| 1 | ATGCAAGCAC GGCTGCTGAT ACCTATTCTT TTTTCAGTTT TTATTTTATC | |
|
| 51 | CGC.TGCGGG ACACTGACAG GTATTCCATC GCATGGCGgA GkTAAACgCT |
|
| 101 | TTgCGGTCGA ACAAGAACTT GTGGCCGCTT CTGCCAGAGC TGCCGTTAAA |
|
| 151 | GACATGGATT TACAGGCATT ACACGGACGA AAAGTTGCAT TGTACATTGC |
|
| 201 | CACTATGGGC GACCAAGGTT CAGGcAGTTT GACAGGGGGG TCGCTACTCC |
|
| 251 | ATTGATGCAC kGrTwCsTGG CGAATACATA AACAGCCCTG CCGTCCGTAC |
|
| 301 | CGATTACACC TATCCACGTT ACGAAACCAC CGCTGAAACA ACATCAGGCG |
|
| 351 | GTTTGACAGG TTTAACCACT TCTTTATCTA CACTTAATGC CCCTGCACTC |
|
| 401 | TCTCGCACCC AATCAGACGG TAGCGGAAGT AAAAGCAGTC TGGGCTTAAA |
|
| 451 | TATTGGCGGG ATGGGGGATT ATCGAAATGA AACCTTGACG ACTAACCCGC |
|
| 501 | GCGACACTGC CTTTCTTTCC CACTTGGTAC AGACCGTATT TTTCCTGCGC |
|
| 551 | GGCATAGACG TTGTTTCTCC TGCCAATGCC GATACAGATG TGTTTATTAA |
|
| 601 | CATCGACGTA TTCGGAACGA TACGCAACAG AACCGAAATG.. |
[0398]This corresponds to the amino acid sequence <SEQ ID 78; ORF15>:
[0000]| 1 | MQARLLIPIL FSVFILSACG TLTGIPSHGG XKRFAVEQEL VAASARAAVK | |
|
| 51 | DMDLQALHGR KVALYIATMG DQGSGSLTGG RYSIDAXXXG EYINSPAVRT |
|
| 101 | DYTYPRYETT AETTSGGLTG LTTSLSTLNA PALSRTQSDG SGSKSSLGLN |
|
| 151 | IGGMGDYRNE TLTTNPRDTA FLSHLVQTVF FLRGIDVVSP ANADTDVFIN |
|
| 201 | IDVFGTIRNR TEM.. |
[0399]Further work revealed the complete nucleotide sequence <SEQ ID 79>:
[0000]| 1 | ATGCAAGCAC GGCTGCTGAT ACCTATTCTT TTTTCAGTTT TTATTTTATC | |
|
| 51 | CGCCTGCGGG ACACTGACAG GTATTCCATC GCATGGCGGA GGTAAACGCT |
|
| 101 | TTGCGGTCGA ACAAGAACTT GTGGCCGCTT CTGCCAGAGC TGCCGTTAAA |
|
| 151 | GACATGGATT TACAGGCATT ACACGGACGA AAAGTTGCAT TGTACATTGC |
|
| 201 | CACTATGGGC GACCAAGGTT CAGGCAGTTT GACAGGGGGT CGCTACTCCA |
|
| 251 | TTGATGCACT GATTCGTGGC GAATACATAA ACAGCCCTGC CGTCCGTACC |
|
| 301 | GATTACACCT ATCCACGTTA CGAAACCACC GCTGAAACAA CATCAGGCGG |
|
| 351 | TTTGACAGGT TTAACCACTT CTTTATCTAC ACTTAATGCC CCTGCACTCT |
|
| 401 | CTCGCACCCA ATCAGACGGT AGCGGAAGTA AAAGCAGTCT GGGCTTAAAT |
|
| 451 | ATTGGCGGGA TGGGGGATTA TCGAAATGAA ACCTTGACGA CTAACCCGCG |
|
| 501 | CGACACTGCC TTTCTTTCCC ACTTGGTACA GACCGTATTT TTCCTGCGCG |
|
| 551 | GCATAGACGT TGTTTCTCCT GCCAATGCCG ATACAGATGT GTTTATTAAC |
|
| 601 | ATCGACGTAT TCGGAACGAT ACGCAACAGA ACCGAAATGC ACCTATACAA |
|
| 651 | TGCCGAAACA CTGAAAGCCC AAACAAAACT GGAATATTTC GCAGTAGACA |
|
| 701 | GAACCAATAA AAAATTGCTC ATCAAACCAA AAACCAATGC GTTTGAAGCT |
|
| 751 | GCCTATAAAG AAAATTACGC ATTGTGGATG GGGCCGTATA AAGTAAGCAA |
|
| 801 | AGGAATTAAA CCGACGGAAG GATTAATGGT CGATTTCTCC GATATCCGAC |
|
| 851 | CATACGGCAA TCATACGGGT AACTCCGCCC CATCCGTAGA GGCTGATAAC |
|
| 901 | AGTCATGAGG GGTATGGATA CAGCGATGAA GTAGTGCGAC AACATAGACA |
|
| 951 | AGGACAACCT TGA |
[0400]This corresponds to the amino acid sequence <SEQ ID 80; ORF15-1>:
[0000]| 1 | MQARLLIPIL FSVFILSACG TLTGIPSHGG GKRFAVEQEL VAASARAAVK | |
|
| 51 | DMDLQALHGR KVALYIATMG DQGSGSLTGG RYSIDALIRG EYINSPAVRT |
|
| 101 | DYTYPRYETT AETTSGGLTG LTTSLSTLNA PALSRTQSDG SGSKSSLGLN |
|
| 151 | IGGMGDYRNE TLTTNPRDTA FLSHLVQTVF FLRGIDVVSP ANADTDVFIN |
|
| 201 | IDVFGTIRNR TEMHLYNAET LKAQTKLEYF AVDRTNKKLL IKPKTNAFEA |
|
| 251 | AYKENYALWM GPYKVSKGIK PTEGLMVDFS DIRPYGNHTG NSAPSVEADN |
|
| 301 | SHEGYGYSDE VVRQHRQGQP * |
[0401]Further work identified the corresponding gene in strain A of N. meningitidis <SEQ ID 81>:
[0000]| 1 | ATGCAAGCAC GGCTGCTGAT ACCTATTCTT TTTTCAGTTT TTATTTTATC | |
|
| 51 | CGCCTGCGGG ACACTGACAG GTATTCCATC GCATGGCGGA GGTAAACGCT |
|
| 101 | TTGCGGTCGA ACAAGAACTT GTGGCCGCTT CTGCCAGAGC TGCCGTTAAA |
|
| 151 | GACATGGATT TACAGGCATT ACACGGACGA AAAGTTGCAT TGTACATTGC |
|
| 201 | AACTATGGGC GACCAAGGTT CAGGCAGTTT GACAGGGGGT CGCTACTCCA |
|
| 251 | TTGATGCACT GATTCGTGGC GAATACATAA ACAGCCCTGC CGTCCGTACC |
|
| 301 | GATTACACCT ATCCACGTTA CGAAACCACC GCTGAAACAA CATCAGGCGG |
|
| 351 | TTTGACAGGT TTAACCACTT CTTTATCTAC ACTTAATGCC CCTGCACTCT |
|
| 401 | CGCGCACCCA ATCAGACGGT AGCGGAAGTA AAAGCAGTCT GGGCTTAAAT |
|
| 451 | ATTGGCGGGA TGGGGGATTA TCGAAATGAA ACCTTGACGA CTAACCCGCG |
|
| 501 | CGACACTGCC TTTCTTTCCC ACTTGGTACA GACCGTATTT TTCCTGCGCG |
|
| 551 | GCATAGACGT TGTTTCTCCT GCCAATGCCG ATACGGATGT GTTTATTAAC |
|
| 601 | ATCGACGTAT TCGGAACGAT ACGCAACAGA ACCGAAATGC ACCTATACAA |
|
| 651 | TGCCGAAACA CTGAAAGCCC AAACAAAACT GGAATATTTC GCAGTAGACA |
|
| 701 | GAACCAATAA AAAATTGCTC ATCAAACCAA AAACCAATGC GTTTGAAGCT |
|
| 751 | GCCTATAAAG AAAATTACGC ATTGTGGATG GGACCGTATA AAGTAAGCAA |
|
| 801 | AGGAATTAAA CCGACAGAAG GATTAATGGT CGATTTCTCC GATATCCAAC |
|
| 851 | CATACGGCAA TCATATGGGT AACTCTGCCC CATCCGTAGA GGCTGATAAC |
|
| 901 | AGTCATGAGG GGTATGGATA CAGCGATGAA GCAGTGCGAC GACATAGACA |
|
| 951 | AGGGCAACCT TGA |
[0402]This encodes a protein having amino acid sequence <SEQ ID 82; ORF15a>:
[0000]| 1 | MQARLLIPIL FSVFILSACG TLTGIPSHGG GKRFAVEQEL VAASARAAVK | |
|
| 51 | DMDLQALHGR KVALYIATMG DQGSGSLTGG RYSIDALIRG EYINSPAVRT |
|
| 101 | DYTYPRYETT AETTSGGLTG LTTSLSTLNA PALSRTQSDG SGSKSSLGLN |
|
| 151 | IGGMGDYRNE TLTTNPRDTA FLSHLVQTVF FLRGIDVVSP ANADTDVFIN |
|
| 201 | IDVFGTIRNR TEMHLYNAET LKAQTKLEYF AVDRTNKKLL IKPKTNAFEA |
|
| 251 | AYKENYALWM GPYKVSKGIK PTEGLMVDFS DIQPYGNHMG NSAPSVEADN |
|
| 301 | SHEGYGYSDE AVRRHRQGQP * |
[0403]The originally-identified partial strain B sequence (ORF15) shows 98.1% identity over a 213aa overlap with ORF15a:
[0000]
[0404]The complete strain B sequence (ORF15-1) and ORF15a show a 98.8% identity in 320 aa overlap:
[0000]
[0405]Further work identified the corresponding gene in N. gonorrhoeae <SEQ ID 83>:
[0000]| 1 | ATGCGGGCAC GGCTGCTGAT ACCTATTCTT TTTTCAGTTT TTATTTTATC | |
|
| 51 | CGCCTGCGGG ACACTGACAG GTATTCCATC GCATGGCGGA GGCAAACGCT |
|
| 101 | TCGCGGTCGA ACAAGAACTT GTGGCCGCTT CTGCCAGAGC TGCCGTTAAA |
|
| 151 | GACATGGATT TACAGGCATT ACACGGACGA AAAGTTGCAT TGTACATTGC |
|
| 201 | AACTATGGGC GACCAAGGTT CAGGCAGTTT GACAGGGGGT CGCTACTCCA |
|
| 251 | TTGATGCACT GATTCGCGGC GAATACATAA ACAGCCCTGC CGTCCGCACC |
|
| 301 | GATTACACCT ATCCGCGTTA CGAAACCACC GCTGAAACAA CATCAGGCGG |
|
| 351 | TTTGACGGGT TTAACCACTT CTTTATCTAC ACTTAATGCC CCTGCACTCT |
|
| 401 | CGCGCACCCA ATCAGACGGT AGCGGAAGTA GGAGCAGTCT GGGCTTAAAT |
|
| 451 | ATTGGCGGGA TGGGGGATTA TCGAAATGAA ACCTTGACGA CCAACCCGCG |
|
| 501 | CGACACTGCC TTTCTTTCCC ACTTGGTGCA GACCGTATTT TTCCTGCGCG |
|
| 551 | GCATAGACGT TGTTTCTCCT GCCAATGCCG ATACAGATGT GTTTATTAAC |
|
| 601 | ATCGACGTAT TCGGAACGAT ACGCAACAGA ACCGAAATGC ACCTATACAA |
|
| 651 | TGCCGAAACA CTGAAAGCCC AAACAAAACT GGAATATTTC GCAGTAGACA |
|
| 701 | GAACCAATAA AAAATTGCTC ATCAAACCCA AAACCAATGC GTTTGAAGCT |
|
| 751 | GCCTATAAAG AAAATTACGC ATTGTGGATG GGGCCGTATA AAGTAAGCAA |
|
| 801 | AGGAATCAAA CCGACGGAAG GATTGATGGT CGATTTCTCC GATATCCAAC |
|
| 851 | CATACGGCAA TCATACGGGT AACTCCGCCC CATCCGTAGA GGCTGATAAC |
|
| 901 | AGTCATGAGG GGTATGGATA CAGCGATGAA GCAGTGCGAC AACATAGACA |
|
| 951 | AGGGCAACCT TGA |
[0406]This encodes a protein having amino acid sequence <SEQ ID 84; ORF15ng>:
[0000]| 1 | MRARLLIPIL FSVFILSACG TLTGIPSHGG GKRFAVEQEL VAASARAAVK | |
|
| 51 | DMDLQALHGR KVALYIATMG DQGSGSLTGG RYSIDALIRG EYINSPAVRT |
|
| 101 | DYTYPRYETT AETTSGGLTG LTTSLSTLNA PALSRTQSDG SGSRSSLGLN |
|
| 151 | IGGMGDYRNE TLTTNPRDTA FLSHLVQTVF FLRGIDVVSP ANADTDVFIN |
|
| 201 | IDVFGTIRNR TEMHLYNAET LKAQTKLEYF AVDRTNKKLL IKPKTNAFEA |
|
| 251 | AYKENYALWM GPYKVSKGIK PTEGLMVDFS DIQPYGNHTG NSAPSVEADN |
|
| 301 | SHEGYGYSDE AVRQHRQGQP * |
[0407]The originally-identified partial strain B sequence (ORF 15) shows 97.2% identity over a 213aa overlap with ORF15ng:
[0000]
[0408]The complete strain B sequence (ORF15-1) and ORF15ng show 98.8% identity in 320 aa overlap:
[0000]
[0409]Computer analysis of these amino acid sequences reveals an ILSAC motif (putative membrane lipoprotein lipid attachment site, as predicted by the MOTIFS program).
[0000]indicates a putative leader sequence, and it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0410]ORF15-1 (31.7 kDa) was cloned in pET and pgex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 4A shows the results of affinity purification of the GST-fusion protein, and FIG. 4B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for Western blot (FIG. 4C) and ELISA (positive result). These experiments confirm that ORFX-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 11
[0411]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 85>:
[0000]| 1 | ..GG.CAGCACA AAAAACAGGC GGTTGAACGG AAAAACCGTA TTTACGATGA | |
|
| 51 | TGCCGGGTAT GATATTCGGC GTATTCACGG GCGCATTCTC CGCAAAATAT |
|
| 101 | ATCCCCGCGT TCGGGCTTCA AATTTTCTTC ATCCTGTTTT TAACCGCCGT |
|
| 151 | CGCATTCAAA ACACTGCATA CCGACCCTCA GACGGCATCC CGCCCGCTGC |
|
| 201 | CCGGACTGCC CrGACTGACT GCGGTTTCCA CACTGTTCGG CACAATGTCG |
|
| 251 | AGCTGGGTCG GCATAGGCGG CGGTTCACTT TCCGTCCCCT TCTTAATCCA |
|
| 301 | CTGCGGCTTC CCCGCCCATA AAGCCATCGG CACATCATCC GGCCTTGCCT |
|
| 351 | GGCCGATTGC ACTCTCCGGC GCAATATCGT ATCTGCTCAA CGGCCTGAAT |
|
| 401 | ATTGCAGGAT TGCCCGAAGG GTCACTGGGC TTCCTTTACC TGCCCGCCGT |
|
| 451 | CGCCGTCCTC AGCGCGGCAA CCATTGCCTT TGCCCCGCTC GGTGTCAAAA |
|
| 501 | CCGCCCACAA ACTTTCTTCT GCCAAACTCA AAAAATC.TT CGGCATTATG |
|
| 551 | TTGCTTTTGA TTGCCGGAAA AATGCTGTAC AACCTGCTTT AA |
[0412]This corresponds to the amino acid sequence <SEQ ID 86; ORF17>:
[0000]| 1 | ..GQHKKQAVNG KTVFTMMPGM IFGVFTGAFS AKYIPAFGLQ IFFILFLTAV | |
|
| 51 | AFKTLHTDPQ TASRPLPGLP XLTAVSTLFG TMSSWVGIGG GSLSVPFLIH |
|
| 101 | CGFPAHKAIG TSSGLAWPIA LSGAISYLLN GLNIAGLPEG SLGFLYLPAV |
|
| 151 | AVLSAATIAF APLGVKTAHK LSSAKLKKSF GIMLLLIAGK MLYNLL* |
[0413]Further work revealed the complete nucleotide sequence <SEQ ID 87>:
[0000]| 1 | ATGTGGCATT GGGACATTAT CTTAATCCTG CTTGCCGTAG GCAGTGCGGC | |
|
| 51 | AGGTTTTATT GCCGGCCTGT TCGGCGTAGG CGGCGGCACG CTGATTGTCC |
|
| 101 | CTGTCGTTTT ATGGGTGCTT GATTTGCAGG GTTTGGCACA ACATCCTTAC |
|
| 151 | GCGCAACACC TCGCCGTCGG CACATCCTTC GCCGTCATGG TCTTCACCGC |
|
| 201 | CTTTTCCAGT ATGCTGGGGC AGCACAAAAA ACAGGCGGTC GACTGGAAAA |
|
| 251 | CCGTATTTAC GATGATGCCG GGTATGATAT TCGGCGTATT CACGGGCGCA |
|
| 301 | CTCTCCGCAA AATATATCCC CGCGTTCGGG CTTCAAATTT TCTTCATCCT |
|
| 351 | GTTTTTAACC GCCGTCGCAT TCAAAACACT GCATACCGAC CCTCAGACGG |
|
| 401 | CATCCCGCCC GCTGCCCGGA CTGCCCGGAC TGACTGCGGT TTCCACACTG |
|
| 451 | TTCGGCACAA TGTCGAGCTG GGTCGGCATA GGCGGCGGTT CACTTTCCGT |
|
| 501 | CCCCTTCTTA ATCCACTGCG GCTTCCCCGC CCATAAAGCC ATCGGCACAT |
|
| 551 | CATCCGGCCT TGCCTGGCCG ATTGCACTCT CCGGCGCAAT ATCGTATCTG |
|
| 601 | CTCAACGGCC TGAATATTGC AGGATTGCCC GAAGGGTCAC TGGGCTTCCT |
|
| 651 | TTACCTGCCC GCCGTCGCCG TCCTCAGCGC GGCAACCATT GCCTTTGCCC |
|
| 701 | CGCTCGGTGT CAAAACCGCC CACAAACTTT CTTCTGCCAA ACTCAAAAAA |
|
| 751 | Tc.TTCGGCA TTATGTTGCT TTTGATTGCC GGAAAAATGC TGTACAACCT |
|
| 801 | GCTTTAA |
[0414]This corresponds to the amino acid sequence <SEQ ID 88; ORF17-1>:
[0000]| 1 | MWHWDIILIL LAVGSAAGFI AGLFGVGGGT LIVPVVLWVL DLQGLAQHPY | |
|
| 51 | AQHLAVGTSF AVMVFTAFSS MLGQHKKQAV DWKTVFTMMP GMIFGVFTGA |
|
| 101 | LSAKYIPAFG LQIFFILFLT AVAFKTLHTD PQTASRPLPG LPGLTAVSTL |
|
| 151 | FGTMSSWVGI GGGSLSVPFL IHCGFPAHKA IGTSSGLAWP IALSGAISYL |
|
| 201 | LNGLNIAGLP EGSLGFLYLP AVAVLSAATI AFAPLGVKTA HKLSSAKLKK |
|
| 251 | XFGIMLLLIA GKMLYNLL* |
[0415]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical H. influenzae Transmembrane Protein HI0902 (Accession Number P44070)
[0416]ORF17 and HI0902 proteins show 28% aa identity in 192 aa overlap:
[0000]| ORF17 | 3 | HKKQAVNGKTVFTMMPGMIFGVFT-GAFSAKYIPAFGLQIF--FILFLTAVAFKTLHTDP | 59 | |
| | HK + + V + P ++ VF G F + +IF +++L ++ D |
| HI0902 | 72 | HKLGNIVWQAVRILAPVIMLSVFICGLFIGRLDREISAKIFACLVVYLATKMVLSIKKD- | 130 |
|
| ORF17 | 60 | QTASRPLPGLPXLTAVSTLFGTMSSWVGIGGGSLSVPFLIHCGFPAHKAIGTSSGLAWPI | 119 |
| | Q ++ L L + L G SS GIGGG VPFL G +AIG+S+ + |
| HI0902 | 131 | QVTTKSLTPLSSVIG-GILIGMASSAAGIGGGGFIVPFLTARGINIKQAIGSSAFCGMLL | 189 |
|
| ORF17 | 120 | ALSGAISYLLNGLNIAGLPEGSLGFLYLPAVAVLSAATIAFAPLGVXXXXXXXXXXXXXX | 179 |
| | +SG S++++G +PE SLG++YLPAV ++A + + LG |
| HI0902 | 190 | GISGMFSFIVSGWGNPLMPEYSLGYIYLPAVLGITATSFFTSKLGASATAKLPVSTLKKG | 249 |
|
| ORF17 | 180 | FGIMLLLIAGKM | 191 |
| | F + L+++A M |
| HI0902 | 250 | FALFLIVVAINM | 261 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0417]ORF17 shows 96.9% identity over a 196aa overlap with an ORF (ORF17a) from strain A of N. meningitidis:
[0000]
[0418]The complete length ORF17a nucleotide sequence <SEQ ID 89> is:
[0000]| 1 | ATGTGGCATT GGGACATTAT CTTAATCCTG CTTGCCGTAG GCAGTGCGGC | |
|
| 51 | AGGTTTTATT GCCGGCCTGT TCGGCGTAGG CGGCGGCACG CTGATTGTCC |
|
| 101 | CTGTCGTTTT ATGGGTGCTT GATTTGCAGG GTTTGGCACA ACATCCTTAC |
|
| 151 | GCGCAACACC TCGCCGTCGG CACATCCTTC GCCGTCATGG TCTTCACCGC |
|
| 201 | CTTTTCCAGT ATGCTGGGGC AGCACAAAAA ACAGGCGGTC GACTGGAAAA |
|
| 251 | CCGTATTTAC GATGATGCCG GGTATGGTAT TCGGCGTATT CGCTGGCGCA |
|
| 301 | CTCTCCGCAA AATATATCCC AGCGTTCGGG CTTCAAATTT TCTTCATCCT |
|
| 351 | GTTTTTAACC GCCGTCGCAT TCAAAACACT GCATACCGAC CCTCAGACGG |
|
| 401 | CATCCCGCCC GCTGCCCGGA CTGCCCGGAC TGACTGCGGT TTCCACACTG |
|
| 451 | TTCGGCACAA TGTCGAGCTG GGTCGGCATA GGCGGCGGTT CACTTTCCGT |
|
| 501 | CCCCTTCTTA ATCCACTGCG GCTTCCCCGC CCATAAAGCC ATCGGCACAT |
|
| 551 | CATCCGGCCT TGCCTGGCCG ATTGCACTCT CCGGCGCAAT ATCGTATCTG |
|
| 601 | CTCAACGGCC TGAATATTGC AGGATTGCCC GAAGGGTCAC TGGGCTTCCT |
|
| 651 | TTACCTGCCC GCCGTCGCCG TCCTCAGCGC GGCAACCATT GCCTTTGCCC |
|
| 701 | CGCTCGGTGT CAAAACCGCC CACAAACTTT CTTCTGCCAA ACTCAAAAAA |
|
| 751 | TCCTTCGGCA TTATGTTGCT TTTGATTGCC GGAAAAATGC TGTACAACCT |
|
| 801 | GCTTTAA |
[0419]This encodes a protein having amino acid sequence <SEQ ID 90>:
[0000]| 1 | MWHWDIILIL LAVGSAAGFI AGLFGVGGGT LIVPVVLWVL DLQGLAQHPY | |
|
| 51 | AQHLAVGTSF AVMVFTAFSS MLGQHKKQAV DWKTVFTMMP GMVFGVFAGA |
|
| 101 | LSAKYIPAFG LQIFFILFLT AVAFKTLHTD PQTASRPLPG LPGLTAVSTL |
|
| 151 | FGTMSSWVGI GGGSLSVPFL IHCGFPAHKA IGTSSGLAWP IALSGAISYL |
|
| 201 | LNGLNIAGLP EGSLGFLYLP AVAVLSAATI AFAPLGVKTA HKLSSAKLKK |
|
| 251 | SFGIMLLLIA GKMLYNLL* |
[0420]ORF17a and ORF17-1 show 98.9% identity in 268 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0421]ORF17 shows 93.9% identity over a 196aa overlap with a predicted ORF (ORF17.ng) from N. gonorrhoeae:
[0000]
[0422]An ORF17ng nucleotide sequence <SEQ ID 91> is predicted to encode a protein having amino acid sequence <SEQ ID 92>:
[0000]| 1 | MWHWDIILIL LAVGSAAGFI AGLFGVGGGT LIVPVVLWVL DLQGLAQHPY | |
|
| 51 | AQHLAVGTSF AVMVFTAFSS MLGQHKKQAV DWKTIFAMMP GMIFGVFAGA |
|
| 101 | LSAKYIPAFG LQIFFILFLT AVAFKTLHTG RQTASRPLPG LPGLTAVSTL |
|
| 151 | FGAMSSWVGI GGGSLSVPFL IHCGFPAHKA IGTSSGLAWP IALSGAISYL |
|
| 201 | VNGLNIAGLP EGSLGFLYLP AVAVLSAATI AFAPLGVKTA HKLSSAKLKE |
|
| 251 | SFGIMLLLIA GKMLYNLL* |
[0423]Further work revealed the complete gonococcal DNA sequence <SEQ ID 93>:
[0000]| 1 | ATGTGGCATT GGGACATTAT CTTAATCCTG CTTGCcgtag gcAGTGCGGC | |
|
| 51 | AGGTTTTATT GCCGGCCTGT Tcggtgtagg cggcgGTACG CTGATTGTCC |
|
| 101 | CTGTCGTTTT ATGGGTGCTT GATTTGCAGG GTTTGGCACA ACATCCTTAC |
|
| 151 | GCGCAACACC TCGCCGTCGG CAcaTccttc gcCGTCATGG TCTTCACCGC |
|
| 201 | CTTTTCCAGT ATGTTGGGGC AGCACAAAAA ACAGGCGGTC GACTGGAAAA |
|
| 251 | CCATATTTGC GATGATGCCG GGTATGATAT TCGGCGTATT CGCTGGCGCA |
|
| 301 | CTCTCCGCAA AATATATCCC CGCGTTCGGG CTTCAAATTT TCTTCATCCT |
|
| 351 | GTTTTTAACC GCCGTCGCAT TCAAAACACT GCATACCGGT CGTCAGACGG |
|
| 401 | CATCCCGCCC GCTGCCCGGG CTGCCCGGAC TGACTGCGGT TTCCACACTG |
|
| 451 | TTCGGCGCAA TGTCGAGCTG GGTCGGCATA GGCGGCGGTT CACTTTCCGT |
|
| 501 | CCCCTTCTTA ATCCACTGCG GCTTCCCCGC CCATAAAGCC ATCGGCACAT |
|
| 551 | CATCCGGCCT TGCCTGGCCG ATTGCACTCT CCGGCGCAAT ATCGTATCTG |
|
| 601 | GTCAACGGTC TGAATATTGC AGGATTGCCC GAAGGGTCGC TGGGCTTCCT |
|
| 651 | TTACCTGCCC GCCGTCGCCG TCCTCAGCGC GGCAACCATT GCCTTTGCCC |
|
| 701 | CGCTCGGTGT CAAAACCGCC CACAAACTTT CTTCTGCCAA ACTCAAAGAA |
|
| 751 | TCCTTCGGCA TTATGTTGCT TTTGATTGCC GGAAAAATGC TGTACAACCT |
|
| 801 | GCTTTAA |
[0424]This corresponds to the amino acid sequence <SEQ ID 94; ORF17ng-1>:
[0000]| 1 | MWHWDIILIL LAVGSAAGFI AGLFGVGGGT LIVPVVLWVL DLQGLAQHPY | |
|
| 51 | AQHLAVGTSF AVMVFTAFSS MLGQHKKQAV DWKTIFAMMP GMIFGVFAGA |
|
| 101 | LSAKYIPAFG LQIFFILFLT AVAFKTLHTG RQTASRPLPG LPGLTAVSTL |
|
| 151 | FGAMSSWVGI GGGSLSVPFL IHCGFPAHKA IGTSSGLAWP IALSGAISYL |
|
| 201 | VNGLNIAGLP EGSLGFLYLP AVAVLSAATI AFAPLGVKTA HKLSSAKLKE |
|
| 251 | SFGIMLLLIA GKMLYNLL* |
[0425]ORF17ng-1 and ORF17-1 show 96.6% identity in 268 aa overlap:
[0000]
[0426]In addition, ORF17ng-1 shows significant homology with a hypothetical H. influenzae protein:
[0000]| sp|P44070|Y902_HAEIN HYPOTHETICAL PROTEIN HI0902 pir||G64015 | |
| hypothetical protein HI0902 - Haemophilus influenzae (strain Rd KW20) |
| gi|1573922 (U32772) H. influenzae predicted coding region HI0902 |
| [Haemophilus influenzae] Length = 264 |
| Score = 74 (34.9 bits), Expect = 1.6e−23, Sum P(2) = 1.6e−23 |
| Identities = 15/43 (34%), Positives = 23/43 (53%) |
|
| Query: | 55 | AVGTSFAVMVFTAFSSMLGQHKKQAVDWKTIFAMMPGMIFGVF | 97 | |
| | A+GTSFA +V T S HK + W+ + + P ++ VF |
| Sbjct: | 52 | ALGTSFATIVITGIGSAQRHHKLGNIVWQAVRILAPVIMLSVF | 94 |
|
| Score = 195 (91.9 bits), Expect = 1.6e−23, Sum P(2) = 1.6e−23 | |
| Identities = 44/114 (38%), Positives = 65/114 (57%) |
|
| Query: | 150 | LFGAMSSWVGIGGGSLSVPFLIHCGFPAHKAIGTSSGLAWPIALSGAISYLVNGLNIAGL | 209 | |
| | L G SS GIGGG VPFL G +AIG+S+ + +SG S++V+G + |
| Sbjct: | 148 | LIGMASSAAGIGGGGFIVPFLTARGINIKQAIGSSAFCGMLLGISGMFSFIVSGWGNPLM | 207 |
|
| Query: | 210 | PEGSLGFLYLPAVAVLSAATIAFAPLGVKTAFIKLSSAKLKESFGIMLLLIAGKM | 263 |
| | PE SLG++YLPAV ++A + + LG KL + LK+ F + L+++A M |
| Sbjct: | 208 | PEYSLGYIYLPAVLGITATSFFTSKLGASATAKLPVSTLKKGFALFLIVVAINM | 261 |
[0427]This analysis, including the homology with the hypothetical H. influenzae transmembrane protein, suggests that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 12
[0428]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 95>:
[0000]| 1 | ..GGAAACGGAT GGCAGGCAGA CCCCGAACAT CCGCTGCTCG GGCTTTTTGC | |
|
| 51 | CGTCAGTAAT GTATCGATGA CGCTTGCTTT TGTCGGAATA TGTGCGTTGG |
|
| 101 | TGCATTATTG CTTTTCGGGA ACGGTTCAAG TGTTTGTGTT TGCGGCACTG |
|
| 151 | CTCAAACTTT ATGCGCTGAA GCCGGTTTAT TGGTTCGTGT TGCAGTTTGT |
|
| 201 | GCTGATGGCG GTTGCCTATG TCCACCGCTG CGGTATAGAC CGGCAGCCGC |
|
| 251 | CGTCAACGTT CGGCGGCTCG CAGCTGCGAC TCGGCGGGTT GACGGCAGCG |
|
| 301 | TTGATGCAGG TCTCGGTACT GGTGCTGCTG CTTTCAGAAA TTGGAAGATA |
|
| 351 | A |
[0429]This corresponds to the amino acid sequence <SEQ ID 96; ORF18>:
[0000]| 1 | ..GNGWQADPEH PLLGLFAVSN VSMTLAFVGI CALVHYCFSG TVQVFVFAAL | |
|
| 51 | LKLYALKPVY WFVLQFVLMA VAYVHRCGID RQPPSTFGGS QLRLGGLTAA |
|
| 101 | LMQVSVLVLL LSEIGR* |
[0430]Further work revealed the complete nucleotide sequence <SEQ ID 97>:
[0000]| 1 | ATGATTTTGC TGCATTTGGA TTTTTTGTCT GCCTTACTGT ATGCGGCGGT | |
|
| 51 | TTTTCTGTTT CTGATATTCC GCGCAGGAAT GTTGCAATGG TTTTGGGCGA |
|
| 101 | GTATTATGCT GTGGCTGGGC ATATCGGTTT TGGGGGCAAA GCTGATGCCC |
|
| 151 | GGCATATGGG GAATGACCCG CGCCGCGCCC TTGTTCATCC CCCATTTTTA |
|
| 201 | CCTGACTTTG GGCAGCATAT TTTTTTTCAT CGGGCATTGG AACCGGAAAA |
|
| 251 | CAGATGGAAA CGGATGGCAG GCAGACCCCG AACATCCGCT GCTCGGGCTT |
|
| 301 | TTTGCCGTCA GTAATGTATC GATGACGCTT GCTTTTGTCG GAATATGTGC |
|
| 351 | GTTGGTGCAT TATTGCTTTT CGGGAACGGT TCAAGTGTTT GTGTTTGCGG |
|
| 401 | CACTGCTCAA ACTTTATGCG CTGAAGCCGG TTTATTGGTT CGTGTTGCAG |
|
| 451 | TTTGTGCTGA TGGCGGTTGC CTATGTCCAC CGCTGCGGTA TAGACCGGCA |
|
| 501 | GCCGCCGTCA ACGTTCGGCG GCTCGCAGCT GCGACTCGGC GGGTTGACGG |
|
| 551 | CAGCGTTGAT GCAGGTCTCG GTACTGGTGC TGCTGCTTTC AGAAATTGGA |
|
| 601 | AGATAA |
[0431]This corresponds to the amino acid sequence <SEQ ID 98; ORF18-1>:
[0000]| 1 | MILLHLDFLS ALLYAAVFLF LIFRAGMLQW FWASIMLWLG ISVLGAKLMP | |
|
| 51 | GIWGMTRAAP LFIPHFYLTL GSIFFFIGHW NRKTDGNGWQ ADPEHPLLGL |
|
| 101 | FAVSNVSMTL AFVGICALVH YCFSGTVQVF VFAALLKLYA LKPVYWFVLQ |
|
| 151 | FVLMAVAYVH RCGIDRQPPS TFGGSQLRLG GLTAALMQVS VLVLLLSEIG |
|
| 201 | R* |
[0432]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0433]ORF18 shows 98.3% identity over a 116aa overlap with an ORF (ORF18a) from strain A of N. meningitidis:
[0000]
[0434]The complete length ORF18a nucleotide sequence <SEQ ID 99> is:
[0000]| 1 | ATGATTTTGC TGCATTTGGA TTTTTTGTCT GCCTTACTGT ATGCGGCGGT | |
|
| 51 | TTTTCTGTTT CTGATATTCC GCGCAGGAAT GTTGCAATGG TTTTGGGCGA |
|
| 101 | GTATTATGCT GTGGCTGGGC ATATCGGTTT TGGGGGCAAA GCTGATGCCC |
|
| 151 | GGCATATGGG GAATGACCCG CGCCGCGCCC TTGTTCATCC CCCATTTTTA |
|
| 201 | CCTGACTTTG GGCAGCATAT TTTTTTTCAT CGGGCATTGG AACCGGAAAA |
|
| 251 | CGGATGGAAA CGGATGGCAG GCAGACCCCG AACATCCTCT GCTCGGGCTG |
|
| 301 | TTTGCCGTCA GTAATGTATC GATGACGCTT GCTTTTGTCG GAATATGTGC |
|
| 351 | GTTGGTGCAT TATTGCTTTT CGNGAACGGT TCAAGTGTTT GTGTTTGCGG |
|
| 401 | CACTGCTCAA ACTTTATGCG CTGAAGCCGG TTTATTGGTT CGTGTTGCAG |
|
| 451 | TTTGTGCTGA TGGCGGTTGC CTATGTCCAC CGCTGCGGTA TAGACCGGCA |
|
| 501 | GCCGCCGTCA ACGTTCGGCG GNTCGCAGCT GCGACTCGGC GGGTTGACGG |
|
| 551 | CAGCGTTGAT GCAGNTCTCG GTACTGGTGC TGCTGCTTTC AGAAATTGGA |
|
| 601 | AGATAA |
[0435]This encodes a protein having amino acid sequence <SEQ ID 100>:
[0000]| 1 | MILLHLDFLS ALLYAAVFLF LIFRAGMLQW FWASIMLWLG ISVLGAKLMP | |
|
| 51 | GIWGMTRAAP LFIPHFYLTL GSIFFFIGHW NRKTDGNGWQ ADPEHPLLGL |
|
| 101 | FAVSNVSMTL AFVGICALVH YCFSXTVQVF VFAALLKLYA LKPVYWFVLQ |
|
| 151 | FVLMAVAYVH RCGIDRQPPS TFGGSQLRLG GLTAALMQXS VLVLLLSEIG |
|
| 201 | R* |
[0436]ORF18a and ORF18-1 show 99.0% identity in 201 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0437]ORF18 shows 93.1% identity over a 116aa overlap with a predicted ORF (ORF18.ng) from N. gonorrhoeae.
[0000]
[0438]The complete length ORF18ng nucleotide sequence is <SEQ ID 101>:
[0000]| 1 | ATGATTTTGC TGCATTTGGA TTTTTTGTCT GCCTTACTGt aTGCGGcggt | |
|
| 51 | tttTctgTTT CTGATATTCC GCGCAGGAAT GTTGCAATGG TTTTGGGCGA |
|
| 101 | GTATTGCGTT GTGGCTCGGC ATCTCGGTTT TAGGGGTAAA GCTGATGCCG |
|
| 151 | GGGATGTGGG GAATGACCCG CGCCGCGCCT TTGTTCATCC CCCATTTTTA |
|
| 201 | CCTGACTTTG GGCAGCATAT TTTTTTTCAT CGGGTATTGG AACCGGAAAA |
|
| 251 | CAGATGGAAA CGGATGGCAG GCAGACCCCG AACATCCGCT GCTCGGGCTT |
|
| 301 | TTTGCCGTCA GTAATGTATC GATGACGCTT GCTTTTGTCG GAATATGTGC |
|
| 351 | GTTGGTGCAT TATTGCTTTT CGGGAACGGT TCAAGTGTTT GTGTTTGCGG |
|
| 401 | CATTGCTCAA ACTTTATGCG CTGAAGCCGG TTTATTGGTT CGTGTTGCAG |
|
| 451 | TTTGTATTGA TGGCGGttgC CTATGTCCAC CGCTGCGGTA TAGACCGGCA |
|
| 501 | GCCGCCGTCA ACGTTCGGCG GTTCGCAGCT GCGACTCGGC GTGTTGGCGG |
|
| 551 | CGATGTTGAT GCAGGTTGCG GTAACGGCGA TGCTGCTTGC CGAAATCGGC |
|
| 601 | AGATGA |
[0439]This encodes a protein having amino acid sequence <SEQ ID 102>:
[0000]| 1 | MILLHLDFLS ALLYAAVFLF LIFRAGMLQW FWASIALWLG ISVLGVKLMP | |
|
| 51 | GMWGMTRAAP LFIPHFYLTL GSIFFFIGYW NRKTDGNGWQ ADPEHPLLGL |
|
| 101 | FAVSNVSMTL AFVGICALVH YCFSGTVQVF VFAALLKLYA LKPV YWFVLQ |
|
| 151 | FVLMAVAYVH RCGIDRQPPS TFGGSQLRLG VLAAMLMQVA VTAMLLAEIG |
|
| 201 | R* |
[0440]This ORF18ng protein sequence shows 94.0% identity in 201 aa overlap with ORF18-1:
[0000]
[0441]Based on this analysis, including the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 13
[0442]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 103>:
[0000]| 1 | ATGAAAACCC CACTCCTCAA GCCTCTGCTN ATTACCTCGC TTCCCGTTTT | |
|
| 51 | CGCCAGTGTT TTTACCGCCG CCTCCATCGT CTGGCAGCTA GGCGAACCCA |
|
| 101 | AGCTCGCCAT GCCCTTCGTA CTCGGCATCA TCGCCGGCGG CCTTGTCGAT |
|
| 151 | TTGGACAACC NCNTGACCGG ACGGCTNAAA AACATCATCA CCACCGTCGC |
|
| 201 | CCTGTTCACC CTCTCCTCGC TCACGGCACA AAGCACCCTC GGCACAGGGC |
|
| 251 | TGCCCTTCAT CCTCGCCATG ACCCTGATGA CTT.CG.CTT CACCATTTTA |
|
| 301 | GGCGCGGNCG ... |
[0443]This corresponds to the amino acid sequence <SEQ ID 104; ORF19>:
[0000]| 1 | MKTPLLKPLL ITSLPVFASV FTAASIVWQL GEPKLAMPFV LGIIAGGLVD | |
|
| 51 | LDNXXTGRLK NIITTVALFT LSSLTAQSTL GTGLPFILAM TLMTXXFTIL |
|
| 101 | GAX... |
[0444]Further work revealed the complete nucleotide sequence <SEQ ID 105>:
[0000]| 1 | ATGAAAACCC CACTCCTCAA GCCTCTGCTC ATTACCTCGC TTCCCGTTTT | |
|
| 51 | CGCCAGTGTT TTTACCGCCG CCTCCATCGT CTGGCAGCTA GGCGAACCCA |
|
| 101 | AGCTCGCCAT GCCCTTCGTA CTCGGCATCA TCGCCGGCGG CCTTGTCGAT |
|
| 151 | TTGGACAACC GCCTGACCGG ACGGCTGAAA AACATCATCA CCACCGTCGC |
|
| 201 | CCTGTTCACC CTCTCCTCGC TCACGGCACA AAGCACCCTC GGCACAGGGC |
|
| 251 | TGCCCTTCAT CCTCGCCATG ACCCTGATGA CCTTCGGCTT CACCATTTTA |
|
| 301 | GGCGCGGTCG GGCTCAAATA CCGCACCTTC GCCTTCGGTG CACTCGCCGT |
|
| 351 | CGCCACCTAC ACCACACTTA CCTACACCCC CGAAACCTAC TGGCTGACCA |
|
| 401 | ACCCCTTCAT GATTTTATGC GGCACCGTAC TGTACAGCAC CGCCATCCTC |
|
| 451 | CTGTTCCAAA TCGTCCTGCC CCACCGCCCC GTCCAAGAAA GCGTCGCCAA |
|
| 501 | CGCCTACGAC GCACTCGGCG GCTACCTCGA AGCCAAAGCC GACTTCTTCG |
|
| 551 | ACCCCGATGA GGCAGCCTGG ATAGGCAACC GCCACATCGA CCTCGCCATG |
|
| 601 | AGCAACACCG GCGTCATCAC CGCCTTCAAC CAATGCCGTT CCGCCCTGTT |
|
| 651 | TTACCGCCTT CGCGGCAAAC ACCGCCACCC GCGCACCGCC AAAATGCTGC |
|
| 701 | GTTACTACTT TGCCGCCCAA GACATACACG AACGCATCAG CTCCGCCCAC |
|
| 751 | GTCGATTATC AGGAAATGTC CGAAAAATTC AAAAACACCG ACATCATCTT |
|
| 801 | CCGCATCCAC CGCCTGCTCG AAATGCAGGG ACAAGCCTGC CGCAACACCG |
|
| 851 | CCCAAGCCCT GCGCGCAAGC AAAGACTACG TTTACAGCAA ACGCCTCGGC |
|
| 901 | CGCGCCATCG AAGGCTGCCG CCAATCGCTG CGCCTCCTTT CAGACAGCAA |
|
| 951 | CGACAGTCCC GACATCCGCC ACCTGCGCCG CCTTCTCGAC AACCTCGGCA |
|
| 1001 | GCGTCGACCA GCAGTTCCGC CAACTCCAGC ACAACGGCCT GCAGGCAGAA |
|
| 1051 | AACGACCGCA TGGGCGACAC CCGCATCGCC GCCCTCGAAA CCAGCAGCCT |
|
| 1101 | CAAAAACACC TGGCAGGCAA TCCGTCCGCA GCTAAACCTC GAATCAGGCG |
|
| 1151 | TATTCCGCCA TGCCGTCCGC CTGTCCCTCG TCGTTGCCGC CGCCTGCACC |
|
| 1201 | ATCGTCGAAG CCCTCAACCT CAACCTCGGC TACTGGATAC TACTGACCGC |
|
| 1251 | CCTTTTCGTC TGCCAACCCA ACTACACCGC CACCAAAAGC CGCGTCCGCC |
|
| 1301 | AGCGCATCGC CGGCACCGTA CTCGGCGTAA TCGTCGGCTC GCTCGTCCCC |
|
| 1351 | TACTTCACCC CGTCTGTCGA AACCAAACTC TGGATTGTCA TCGCCAGTAC |
|
| 1401 | CACCCTCTTT TTCATGACCC GCACCTACAA ATACAGTTTC TCCACCTTCT |
|
| 1451 | TCATTACCAT TCAAGCCCTG ACCAGCCTCT CCCTCGCAGG TTTGGACGTA |
|
| 1501 | TACGCCGCCA TGCCCGTACG CATCATCGAC ACCATTATCG GCGCATCCCT |
|
| 1551 | TGCCTGGGCG GCAGTCAGCT ACCTGTGGCC AGACTGGAAA TACCTCACGC |
|
| 1601 | TCGAACGCAC CGCCGCCCTT GCCGTATGCA GCAACGGTGC CTATCTCGAA |
|
| 1651 | AAAATCACCG AACGCCTCAA AAGCGGCGAA ACCGGCGACG ACGTCGAATA |
|
| 1701 | CCGCGCCACC CGCCGCCGCG CCCACGAACA CACCGCCGCC CTCAGCAGCA |
|
| 1751 | CCCTTTCCGA CATGAGCAGC GAACCCGCAA AATTCGCCGA CAGCCTGCAA |
|
| 1801 | CCCGGCTTTA CCCTGCTCAA AACCGGCTAC GCCCTGACCG GCTACATCTC |
|
| 1851 | CGCCCTCGGC GCATACCGCA GCGAAATGCA CGAAGAATGC AGCCCCGACT |
|
| 1901 | TTACCGCACA GTTCCACCTC GCCGCCGAAC ACACCGCCCA CATCTTCCAA |
|
| 1951 | CACCTGCCCG AAACCGAACC CGACGACTTT CAGACAGCAC TGGATACACT |
|
| 2001 | GCGCGGCGAA CTCGACACCC TCCGCACCCA CAGCAGCGGA ACACAAAGCC |
|
| 2051 | ACATCCTCCT CCAACAGCTC CAACTCATCG CCCGACAGCT CGAACCCTAC |
|
| 2101 | TACCGCGCCT ACCGCCAAAT TCCGCACAGG CAGCCCCAAA ATGCAGCCTG |
|
| 2151 | A |
[0445]This corresponds to the amino acid sequence <SEQ ID 106; ORF19-1>:
[0000]| 1 | MKTPLLKPLL ITSLPVFASV FTAASIVWQL GEPKLAMPFV LGIIAGGLVD | |
|
| 51 | LDNRLTGRLK NIITTVALFT LSSLTAQSTL GTGLPFILAM TLMTFGFTIL |
|
| 101 | GAVGLKYRTF AFGALAVATY TTLTYTPETY WLTNPFMILC GTVLYSTAIL |
|
| 151 | LFQIVLPHRP VQESVANAYD ALGGYLEAKA DFFDPDEAAW IGNRHIDLAM |
|
| 201 | SNTGVITAFN QCRSALFYRL RGKHRHPRTA KMLRYYFAAQ DIHERISSAH |
|
| 251 | VDYQEMSEKF KNTDIIFRIH RLLEMQGQAC RNTAQALRAS KDYVYSKRLG |
|
| 301 | RAIEGCRQSL RLLSDSNDSP DIRHLRRLLD NLGSVDQQFR QLQHNGLQAE |
|
| 351 | NDRMGDTRIA ALETSSLKNT WQAIRPQLNL ESGVFRHAVR LSLVVAAACT |
|
| 401 | IVEALNLNLG YWILLTALFV CQPNYTATKS RVRQRIAGTV LGVIVGSLVP |
|
| 451 | YFTPSVETKL WIVIASTTLF FMTRTYKYSF STFFITIQAL TSLSLAGLDV |
|
| 501 | YAAMPVRIID TIIGASLAWA AVSYLWPDWK YLTLERTAAL AVCSNGAYLE |
|
| 551 | KITERLKSGE TGDDVEYRAT RRRAHEHTAA LSSTLSDMSS EPAKFADSLQ |
|
| 601 | PGFTLLKTGY ALTGYISALG AYRSEMHEEC SPDFTAQFHL AAEHTAHIFQ |
|
| 651 | HLPETEPDDF QTALDTLRGE LDTLRTHSSG TQSHILLQQL QLIARQLEPY |
|
| 701 | YRAYRQIPHR QPQNAA* |
[0446]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Predicted Transmembrane Protein YHFK of H. influenzae (Accession Number P44289)
[0447]ORF19 and YHFK proteins show 45% aa identity in 97 aa overlap:
[0000]| orf19 | 6 | LKPLLITSLPVFASVFTAASIVWQLGEPKLAMPFVLGIIAGGLVDLDNXXTGRLKNIITT | 65 |
| | L +I+++PVF +V AA +W +MP +LGIIAGGLVDLDN TGRLKN+ T |
| YHFK | 5 | LNAKVISTIPVFIAVNIAAVGIWFFDISSQSMPLILGIIAGGLVDLDNRLTGRLKNVFFT | 64 |
|
| orf19 | 66 | VALFTLSSLTAQSTLGTGLPFILAMTLMTXXFTILGA | 102 |
| | + F++SS Q +G + +I+ MT++T FT++GA |
| YHFK | 65 | LIAFSISSFIVQLHIGKPIQYIVLMTVLTFIFTMIGA | 101 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0448]ORF19 shows 92.2% identity over a 102aa overlap with an ORF (ORF19a) from strain A of N. meningitidis.
[0000]
[0449]The complete length ORF19a nucleotide sequence <SEQ ID 107> is:
[0000]| 1 | ATGAAAACCC CACCCCTCAA GCCTCTGCTC ATTACCTCGC TTCCCGTTTT | |
|
| 51 | CGCCAGTGTC TTTACCGCCG CCTCCATCGT CTGGCAGCTG GGCGAACCCA |
|
| 101 | AGCTCGCCAT GCCCTTCGTA CTCGGCATCA TCGCTGGCGG CCTGGTCGAT |
|
| 151 | TTGGACAACC GCCTGACCGG ACGGCTGAAA AACATCATCG CCACCGTCGC |
|
| 201 | CCTGTTCACC CTCTCCTCAC TTGTCGCGCA AAGCACCCTC GGCACAGGTT |
|
| 251 | TGCCATTCAT CCTCGCCATG ACCCTGATGA CTTTCGGCTT TACCATCATG |
|
| 301 | GGCGCGGTCG GGCTGAAATA CCGCACCTTC GCCTTCGGCG CACTCGCCGT |
|
| 351 | CGCCACCTAC ACCACACTTA CCTACACCCC CGAAACCTAC TGGCTGACCA |
|
| 401 | ACCCCTTTAT GATTCTGTGC GGAACCGTAC TGTACAGCAC CGCCATCATC |
|
| 451 | CTGTTCCAAA TCATCCTGCC CCACCGCCCC GTTCAAGAAA ACGTCGCCAA |
|
| 501 | CGCCTACGAA GCACTCGGCA GCTACCTCGA AGCCAAAGCC GACTTTTTCG |
|
| 551 | ATCCCGACGA AGCCGAATGG ATAGGCAACC GCCACATCGA CCTCGCCATG |
|
| 601 | AGCAACACCG GCGTCATCAC CGCCTTCAAC CAATGCCGTT CCGCCCTGTT |
|
| 651 | TTACCGCCTT CGCGGCAAAC ACCGCCACCC GCGCACCGCC AAAATGCTGC |
|
| 701 | GCTACTACTT CGCCGCCCAA GACATACACG AACGCATCAG CTCCGCCCAC |
|
| 751 | GTCGACTACC AAGAGATGTC CGAAAAATTC AAAAACACCG ACATCATCTT |
|
| 801 | CCGCATCCAC CGCCTGCTCG AAATGCAGGG ACAAGCCTGC CGCAACACCG |
|
| 851 | CCCAAGCCCT GCGCGCAAGC AAAGACTACG TTTACAGCAA ACGCCTCGGC |
|
| 901 | CGCGCCATCG AAGGCTGCCG CCAATCGCTG CGCCTCCTTT CAGACAGCAA |
|
| 951 | CGACAATCCC GACATCCGCC ACCTGCGCCG CCTTCTCGAC AACCTCGGCA |
|
| 1001 | GCGTCGACCA GCAGTTCCGC CAACTCCAGC ACAACGGCCT GCAGGCAGAA |
|
| 1051 | AACGACCGCA TGGGCGACAC CCGCATCGCC GCCCTCGAAA CCGGCAGCCT |
|
| 1101 | CAAAAACACC TGGCAGGCAA TCCGTCCGCA GCTAAACCTC GAATCAGGCG |
|
| 1151 | TATTCCGCCA TGCCGTCCGC CTGTCCCTTG TCGTTGCCGC CGCCTGCACC |
|
| 1201 | ATCGTCGAAG CCCTCAACCT CAACCTCGGC TACTGGATAC TACTGACCGC |
|
| 1251 | CCTTTTCGTC TGCCAACCCA ACTACACCGC CACCAAAAGC CGCGTCCGCC |
|
| 1301 | AGCGCATCGC CGGCACCGTA CTCGGCGTAA TCGTCGGCTC GCTCGTCCCC |
|
| 1351 | TACTTTACCC CCTCCGTCGA AACCAAACTC TGGATCGTCA TCGCCAGTAC |
|
| 1401 | CACCCTCTTT TTCATGACCC GCACCTACAA ATACAGCTTC TCGACATTTT |
|
| 1451 | TCATCACCAT TCAAGCCCTG ACCAGCCTCT CCCTCGCAGG GTTGGACGTA |
|
| 1501 | TACGCCGCCA TGCCCGTACG CATCATCGAC ACCATTATCG GCGCATCCCT |
|
| 1551 | TGCCTGGGCG GCAGTCAGCT ACCTGTGGCC AGACTGGAAA TACCTCACGC |
|
| 1601 | TCGAACGCAC CGCCGCCCTT GCCGTATGCA GCAACGGCGC CTATCTCGAA |
|
| 1651 | AAAATCACCG AACGCCTCAA AAGCGGCGAA ACCGGCGACG ACGTCGAATA |
|
| 1701 | CCGCGCCACC CGCCGCCGCG CCCACGAACA CACCGCCGCC CTCAGCAGCA |
|
| 1751 | CCCTTTCCGA CATGAGCAGC GAACCCGCAA AATTCGCCGA CAGCCTGCAA |
|
| 1801 | CCCGGCTTTA CCCTGCTCAA AACCGGCTAC GCCCTGACCG GCTACATCTC |
|
| 1851 | CGCCCTCGGC GCATACCGCA GCGAAATGCA CGAAGAATGC AGCCCCGACT |
|
| 1901 | TTACCGCACA GTTCCACCTC GCCGCCGAAC ACACCGCCCA CATCTTCCAA |
|
| 1951 | CACCTGCCCG AAACCGAACC CGACGACTTT CAGACAGCAC TGGATACACT |
|
| 2001 | GCGCGGCGAA CTCGACACCC TCCGCACCCA CAGCAGCGGA ACACAAAGCC |
|
| 2051 | ACATCCTCCT CCAACAGCTC CAACTCATCG CCCGGCAGCT CGAACCCTAC |
|
| 2101 | TACCGCGCCT ACCGACAAAT TCCGCACAGG CAGCCCCAAA ACGCAGCCTG |
|
| 2151 | A |
[0450]This encodes a protein having amino acid sequence <SEQ ID 108>:
[0000]| 1 | MKTPPLKPLL ITSLPVFASV FTAASIVWQL GEPKLAMPFV LGIIAGGLVD | |
|
| 51 | LDNRLTGRLK NIIATVALFT LSSLVAQSTL GTGLPFILAM TLMTFGFTIM |
|
| 101 | GAVGLKYRTF AFGALAVATY TTLTYTPETY WLTNPFMILC GTVLYSTAII |
|
| 151 | LFQIILPHRP VQENVANAYE ALGSYLEAKA DFFDPDEAEW IGNRHIDLAM |
|
| 201 | SNTGVITAFN QCRSALFYRL RGKHRHPRTA KMLRYYFAAQ DIHERISSAH |
|
| 251 | VDYQEMSEKF KNTDIIFRIH RLLEMQGQAC RNTAQALRAS KDYVYSKRLG |
|
| 301 | RAIEGCRQSL RLLSDSNDNP DIRHLRRLLD NLGSVDQQFR QLQHNGLQAE |
|
| 351 | NDRMGDTRIA ALETGSLKNT WQAIRPQLNL ESGVFRHAVR LSLVVAAACT |
|
| 401 | IVEALNLNLG YWILLTALFV CQPNYTATKS RVRQRIAGTV LGVIVGSLVP |
|
| 451 | YFTPSVETKL WIVIASTTLF FMTRTYKYSF STFFITIQAL TSLSLAGLDV |
|
| 501 | YAAMPVRIID TIIGASLAWA AVSYLWPDWK YLTLERTAAL AVCSNGAYLE |
|
| 551 | KITERLKSGE TGDDVEYRAT RRRAHEHTAA LSSTLSDMSS EPAKFADSLQ |
|
| 601 | PGFTLLKTGY ALTGYISALG AYRSEMHEEC SPDFTAQFHL AAEHTAHIFQ |
|
| 651 | HLPETEPDDF QTALDTLRGE LDTLRTHSSG TQSHILLQQL QLIARQLEPY |
|
| 701 | YRAYRQIPHR QPQNAA* |
[0451]ORF19a and ORF19-1 show 98.3% identity in 716 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0452]ORF19 shows 95.1% identity over a 102aa overlap with a predicted ORF (ORF19.ng) from N. gonorrhoeae:
[0000]
[0453]An ORF19ng nucleotide sequence <SEQ ID 109> is predicted to encode a protein having amino acid sequence <SEQ ID 110>:
[0000]| 1 | MKTPLLKPLL ITSLPVFASV FTAASIVWQL GEPKLAMPFV LGIIAGGLVD | |
|
| 51 | LDNRLTGRLK NIIATVALFT LSSLTAQSTL GTGLPFILAM TLMTFGFTIL |
|
| 101 | GAVGLKYRTF AFGALAVATY TTLTYTPETY WLTNPFMILC GTVLYSTAII |
|
| 151 | LFQIILPHRP VQESVANAYE ALGGYLEAKA DFFDPDEAAW IGNRHIDLAM |
|
| 201 | SNTGVITAFN QCRSALFYRL RGKHRHPRTA KMLRYYFAAQ DIHERISSAH |
|
| 251 | VDYQEMSEKF KNTDIIFRIR RLLEMQGQAC RNTAQAIRSG KDYVYSKRLG |
|
| 301 | RAIEGCRQSL RLLSDGNDSP DIRHLSRLLD NLGSVDQQFR QLRHSDSPAE |
|
| 351 | NDRMGDTRIA ALETGSFKNT * |
[0454]Further work revealed the complete nucleotide sequence <SEQ ID 111>:
[0000]| 1 | ATGAAAACCC CACTCCTCAA GCCTCTGCTC ATTACCTCGC TTCCCGTTTT | |
|
| 51 | CGCCAGTGTC TTTACCGCCG CCTCCATCGT CTGGCAGCTA GGCGAACCCA |
|
| 101 | AGCTCGCCAT GCCCTTCGTA CTCGGCATCA TCGCCGGCGG CCTGGTCGAT |
|
| 151 | TTGGACAACC GCCTGACCGG ACGGCTGAAA AACATCATCG CCACCGTCGC |
|
| 201 | CCTGTTTACC CTCTCCTCGC TCACGGCGCA AAGCACCCTC GGCACAGGGC |
|
| 251 | TGCCCTTCAT CCTCGCCATG ACCCTGATGA CCTTCGGCTT TACCATTTTA |
|
| 301 | GGCGCGGTCG GGCTGAAATA CCGCACCTTC GCCTTCGGCG CACTCGCCGT |
|
| 351 | CGCCACCTAC ACCACGCTTA CCTACACCCC CGAAACCTAC TGGCTGACCA |
|
| 401 | ACCCCTTCAT GATTTTATGC GGCACCGTAC TGTACAGCAC CGCCATCATC |
|
| 451 | CTGTTCCAAA TCATCCTGCC CCACCGCCCC GTCCAAGAAA GCGTCGCCAA |
|
| 501 | TGCCTACGAA GCACTCGGCG GCTACCTCGA AGCCAAAGCC GACTTCTTCG |
|
| 551 | ACCCCGATGA GGCAGCCTGG ATAGGCAACC GCCACATCGA CCTCGCCATG |
|
| 601 | AGCAACACCG GCGTCATCAC CGCCTTCAAC CAATGCCGTT CCGCCCTGTT |
|
| 651 | TTACCGTTTG CGCGGCAAAC ACCGCCACCC GCGCACCGCC AAAATGCTGC |
|
| 701 | GCTACTACTT CGCCGCCCAA GACATCCACG AACGCATCAG CTCCGCCCAC |
|
| 751 | GTCGACTACC AAGAGATGTC CGAAAAATTC AAAAACACCG ACATCATCTT |
|
| 801 | CCGCATCCGC CGCCTGCTCG AAATGCAGGG GCAGGCGTGC CGCAACACCG |
|
| 851 | CCCAAGCCAT CCGGTCGGGC AAAGACTAcg tTTACAGCAA ACGCCTCGGA |
|
| 901 | CGCGCCATcg aaggctgCCG CCAGTCGCtg cgcctCCTTt cagacggcaA |
|
| 951 | CGACAGTCCC GACATCCGCC ACCTGAGccg CCTTCTCGAC AACCTCGgca |
|
| 1001 | GCGTcgacca gcagtTCcgc caactCCGAC ACAgcgactC CCCCGCcgaa |
|
| 1051 | Aacgaccgca tgggcgacaC CCGCATCGCC GCCCtcgaaa ccggcagctT |
|
| 1101 | caaaaaCAcc tggcaggCAA TCCGTCCGCa gctgaaCCTC GAATCatgCG |
|
| 1151 | TATTCCGCCA TGCCGTCCGC CTGTCCCTCG TCGTTGCCGC CGCCTGCACC |
|
| 1201 | ATCGTCgaag cCCTCAACCT CAACCTCGGC TACTGGATAC TGCTGACCGC |
|
| 1251 | CCTTTTCGTC TGCCAACCCA ACTACACCGC CACCAAAAGC CGCGTGTACC |
|
| 1301 | AACGCATCGC CGGCACCGTA CTCGGCGTAA TCGTCGGCTC GCTCGTCCCC |
|
| 1351 | TACTTCACCC CCTCCGTCGA AACCAAACTC TGGATTGTCA TCGCCGGTAC |
|
| 1401 | CACCCTGTTC TTCATGACCC GCACCTACAA ATACAGTTTC TCCACCTTCT |
|
| 1451 | TCATCACCAT TCAGGCACTG ACCAGCCTCT CCCTCGCAGG TTTGGACGTA |
|
| 1501 | TACGCCGCCA TGCCCGTGCG CATCATcgaC ACCATTATCG GCGCATCCCT |
|
| 1551 | TGCCTGGGCG GCGGTCAGCT ACCTGTGGCC AGACTGGAAA TACCTCACGC |
|
| 1601 | TCGAACGCAC CGCCGCCCTT GCCGTATGCA GCAGCGGCAC ATACCTCCAA |
|
| 1651 | AAAATTGCCG AACGCCTCAA AACCGGCGAA ACCGGCGACG ACATAGAATA |
|
| 1701 | CCGCATCACC CGCCGCCGCG CCCACGAACA CACCGCCGCC CTCAGCAGCA |
|
| 1751 | CCCTTTCCGA CATGAGCAGC GAACCCGCAA AATTCGCCGA CAGCCTGCAA |
|
| 1801 | CCCGGCTTTA CCCTGCTCAA AACCGGCTAC GCCCTGACCG GCTACATCTC |
|
| 1851 | CGCCCTCGGC GCATACCGCA GCGAAATGCA CGAAGAATGC AGCCCCGACT |
|
| 1901 | TTACCGCACA GTTCCACCTT GCCGCCGAAC ACACCGCCCA CATCTTCCAA |
|
| 1951 | CACCTGCCCG ACATGGGACC CGACGACTTT CAGACGGCAT TGGATACACT |
|
| 2001 | GCGCGGCGAA CTCGGCACCC TCCGCACCCG CAGCAGCGGA ACACAAAGCC |
|
| 2051 | ACATCCTCCT CCAACAGCTC CAACTCATCG CccgGCAACT CGAACCCTAC |
|
| 2101 | TACCGCGCCT ACCGACAAAT TCCGCACAGG CAGCCCCAAA ACGCAGCCTG |
|
| 2151 | A |
[0455]This corresponds to the amino acid sequence <SEQ ID 112; ORF19ng-1>:
[0000]| 1 | MKTPLLKPLL ITSLPVFASV FTAASIVWQL GEPKLAMPFV LGIIAGGLVD | |
|
| 51 | LDNRLTGRLK NIIATVALFT LSSLTAQSTL GTGLPFILAM TLMTFGFTIL |
|
| 101 | GAVGLKYRTF AFGALAVATY TTLTYTPETY WLTNPFMILC GTVLYSTAII |
|
| 151 | LFQIILPHRP VQESVANAYE ALGGYLEAKA DFFDPDEAAW IGNRHIDLAM |
|
| 201 | SNTGVITAFN QCRSALFYRL RGKHRHPRTA KMLRYYFAAQ DIHERISSAH |
|
| 251 | VDYQEMSEKF KNTDIIFRIR RLLEMQGQAC RNTAQAIRSG KDYVYSKRLG |
|
| 301 | RAIEGCRQSL RLLSDGNDSP DIRHLSRLLD NLGSVDQQFR QLRHSDSPAE |
|
| 351 | NDRMGDTRIA ALETGSFKNT WQAIRPQLNL ESCVFRHAVR LSLVVAAACT |
|
| 401 | IVEALNLNLG YWILLTALFV CQPNYTATKS RVYQRIAGTV LGVIVGSLVP |
|
| 451 | YFTPSVETKL WIVIAGTTLF FMTRTYKYSF STFFITIQAL TSLSLAGLDV |
|
| 501 | YAAMPVRIID TIIGASLAWA AVSYLWPDWK YLTLERTAAL AVCSSGTYLQ |
|
| 551 | KIAERLKTGE TGDDIEYRIT RRRAHEHTAA LSSTLSDMSS EPAKFADSLQ |
|
| 601 | PGFTLLKTGY ALTGYISALG AYRSEMHEEC SPDFTAQFHL AAEHTAHIFQ |
|
| 651 | HLPDMGPDDF QTALDTLRGE LGTLRTRSSG TQSHILLQQL QLIARQLEPY |
|
| 701 | YRAYRQIPHR QPQNAA* |
[0456]ORF19ng-1 and ORF19-1 show 95.5% identity in 716 aa overlap:
[0000]
[0457]In addition, ORF19ng-1 shows significant homology to a hypothetical gonococcal protein previously entered in the databases:
[0000]| sp|O33369|YOR2_NEIGO HYPOTHETICAL 45.5 KD PROTEIN (ORF2) gnl|PID|e1154438 |
| (AJ002423) hypothetical protein [Neisseria gonorrh] Length = 417 |
| Score = 1512 (705.6 bits), Expect = 5.3e−203, P = 5.3e−203 |
| Identities = 301/326 (92%), Positives = 306/326 (93%) |
|
| Query: | 307 | RQSLRLLSDGNDSPDIRHLSRLLDNLGSVDQQFRQLRHSDSPAENDRMGDTRIAALETGS | 366 |
| | RQSLRLLSDGNDS DIRHLSRLLDNLGSVDQQFRQLRHSDSPAENDRMGDTRIAALETGS |
| Sbjct: | 1 | RQSLRLLSDGNDSXDIRHLSRLLDNLGSVDQQFRQLRHSDSPAENDRMGDTRIAALETGS | 60 |
|
| Query: | 367 | FKNTWQAIRPQLNLESCVFRHAVRLSLVVAAACTIVEALNLNLGYWILLTALFVCQPNYT | 426 |
| | FKNTWQAIRPQLNLES VFRHAVRLSLVVAAACTIVEALNLNLGYWILLT LFVCQPNYT |
| Sbjct: | 61 | FKNTWQAIRPQLNLESGVFRHAVRLSLVVAAACTIVEALNLNLGYWILLTRLFVCQPNYT | 120 |
|
| Query: | 427 | ATKSRVYQRIAGTVLGVIVGSLVPYFTPSVETKLWIVIAGTTLFFMTRTYKYSFSTFFIT | 486 |
| | ATKSRVYQRIAGTVLGVIVGSLVPYFTPSVETKLWIVIAGTTLFFMTRTYKYSFSTFFIT |
| Sbjct: | 121 | ATKSRVYQRIAGTVLGVIVGSLVPYFTPSVETKLWIVIAGTTLFFMTRTYKYSFSTFFIT | 180 |
|
| Query: | 487 | IQALTSLSLAGLDVYAAMPVRIIDTIIGASLAWAAVSYLWPDWKYLTLERTAALAVCSSG | 546 |
| | IQALTSLSLAGLDVYAAMPVRIIDTIIGASLAWAAVSYLWPDWKYLTLERTAALAVCSSG |
| Sbjct: | 181 | IQALTSLSLAGLDVYAAMPVRIIDTIIGASLAWAAVSYLWPDWKYLTLERTAALAVCSSG | 240 |
|
| Query: | 547 | TYLQKIAERLKTGETGDDIEYRITRRRAHEHTAALSSTLSDMSSEPAKFADSLQPGFTLL | 606 |
| | TYLQKIAERLKTGETGDDIEYRITRRRAHEHTAALSSTLSDMSSEPAKFAD+ P |
| Sbjct: | 241 | TYLQKIAERLKTGETGDDIEYRITRRRAHEHTAALSSTLSDMSSEPAKFADTCNPALPCS | 300 |
|
| Query: | 607 | KTGYALTGYISALGAYRSEMHEECSP | 632 |
| | K ALTGYISALG ++ + +P |
| Sbjct: | 301 | KPATALTGYISALGHTAAKCTKNAAP 326 |
[0458]Based on this analysis, including the presence of several putative transmembrane domains in the gonococcal protein (the first of which is also seen in the meningococcal protein), and on homology with the YHFK protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 14
[0459]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 113>.
[0000]| 1 | ATGAATATGC TGGGAGCTTT GGCAAAAGTC GGCAGCCTGA CGATGGTGTC | |
|
| 51 | GCGCGTTTTG GGATTTGTGC GCGATACGGT CATTGCGCGG GCATTCGGCG |
|
| 101 | CGGGTATGGC GACGGATGCG TTTTTTGTCG CGTTCAAACT GCCCAACCTG |
|
| 151 | CTTCGCCGCG TGTTTGCGGA GGGGGCGTTT GCCCAAGCGT TTGTGCCGAT |
|
| 201 | TTTGGCGGAA TACAAGGAAA CGCGTTCAAA AGAGGCGG.C GAAGCCTTTA |
|
| 251 | TCCGCCATGT GGCGGGGATG CTGTCGTTTG TACTGGTTAT CGTTACCGCG |
|
| 301 | CTGGGCATAC TTGCCGCGCC TTGGGTGATT TATGTTTCCG CACCCGAGTT |
|
| 351 | TTGCCCAAGA TGCCGACAAA TTTCAGCTCT CCATCGATTT GCTGCGGATT |
|
| 401 | ACGTTTCCTT ATATATTATT GATTTCCCTG TCTTCATTTG TCGGCTCGGT |
|
| 451 | ACTCAATTCT TATCATAAGT TCGGCATTCC GGCGTTTACG CCAC.GTTTC |
|
| 501 | TGAACGTGTC GTTTATCGTA TTCGCGCTGT TTTTCGTGCC GTATTTCGAT |
|
| 551 | CCGCCCGTTA CCGCGCyGGC GTGGGCGGTC TTTGTCGGCG GCATTTTGCA |
|
| 601 | ACTCGrmTTC CAACTGCCCT GGCTGGCGAA ACTGGGCTTT TTGAAACTGC |
|
| 651 | CCAAACtGAG TTTCAAAGAT GCGGCGGTCA ACCGCGTGAT GAAACAGATG |
|
| 701 | GCGCCTGCgA TTTTgGGCGT GAgCGTGGCG CAGGTTTCTT TGGTGATCAA |
|
| 751 | CACGATTTTc GCGTCTTATC TGCAATCGGG CAGCGTTTCA TGGATGTATT |
|
| 801 | ACGCCGACCG CATGATGGAG CTGCCCAGCG GCGTGCTGGG GGCGGCACTC |
|
| 851 | GGTACGATTT TGCTGCCGAC TTTGTCCAAA CACTCGGCAA ACCaAGATAC |
|
| 901 | GGaACAGTTT TCCGCCCTGC TCGACTGGGG TTTGCGCCTG TGCATGCtgc |
|
| 951 | TGACGCTGCC GGCGgcGGTC GGACTGGCGG TGTTGTCGTT cCCgCtGGTG |
|
| 1001 | GCGACGCTGT TTATGTACCG CGwATTTACG CTGTTTGACG CGCAGATGAC |
|
| 1051 | GCAACACGCG CTGATTGCCT ATTCTTTCGG TTTAATCGGC TTAATCATGA |
|
| 1101 | TTAAAGTGTT GGCACCCGGC TTCTATGCGC GGCAAAACAT CAAwAmGCCC |
|
| 1151 | GTCAAAATCG CCATCTTCAC GCTCATCTGC mCGCAGTTGA TGAACCTTGs |
|
| 1201 | CTTTAyCGGC CCACTrrAAC rCasTCGGAC TTTCGCTTGC CATCGGTCTG |
|
| 1251 | GGCGCGTGTA TCAATGCCGG ATTGTTGTTT TACCTGTTGC GCAGACACGG |
|
| 1301 | TATTTACCAA CCTGG.CAAG GGTTGGGCAG CGTTCTT.AG CAAAAATGCT |
|
| 1351 | GcTCTCGCTC GCCGTGA |
[0460]This corresponds to the amino acid sequence <SEQ ID 114; ORF20>:
[0000]| 1 | MNMLGALAKV GSLTMVSRVL GFVRDTVIAR AFGAGMATDA FFVAFKLPNL | |
|
| 51 | LRRVFAEGAF AQAFVPILAE YKETRSKEAX EAFIRHVAGM LSFVLVIVTA |
|
| 101 | LGILAAPWVI YVSAPSFAQD ADKFQLSIDL LRITFPYILL ISLSSFVGSV |
|
| 151 | LNSYHKFGIP AFTPXFLNVS FIVFALFFVP YFDPPVTAXA WAVFVGGILQ |
|
| 201 | LXFQLPWLAK LGFLKLPKLS FKDAAVNRVM KQMAPAILGV SVAQVSLVIN |
|
| 251 | TIFASYLQSG SVSWMYYADR MMELPSGVLG AALGTILLPT LSKHSANQDT |
|
| 301 | EQFSALLDWG LRLCMLLTLP AAVGLAVLSF PLVATLFMYR XFTLFDAQMT |
|
| 351 | QHALIAYSFG LIGLIMIKVL APGFYARQNI XXPVKIAIFT LICXQLMNLX |
|
| 401 | FXGPLXXIGL SLAIGLGACI NAGLLFYLLR RHGIYQPXQG LGSVLXQKCC |
|
| 451 | SRSP* |
[0461]These sequences were elaborated, and the complete DNA sequence <SEQ ID 115> is:
[0000]| 1 | ATGAATATGC TGGGAGCTTT GGCAAAAGTC GGCAGCCTGA CGATGGTGTC | |
|
| 51 | GCGCGTTTTG GGATTTGTGC GCGATACGGT CATTGCGCGG GCATTCGGCG |
|
| 101 | CGGGTATGGC GACGGATGCG TTTTTTGTCG CGTTCAAACT GCCCAACCTG |
|
| 151 | CTTCGCCGCG TGTTTGCGGA GGGGGCGTTT GCCCAAGCGT TTGTGCCGAT |
|
| 201 | TTTGGCGGAA TACAAGGAAA CGCGTTCAAA AGAGGCGGCG GAGGCTTTTA |
|
| 251 | TCCGCCATGT GGCGGGGATG CTGTCGTTTG TACTGGTTAT CGTTACCGCG |
|
| 301 | CTGGGCATAC TTGCCGCGCC TTGGGTGATT TATGTTTCCG CACCCGGTTT |
|
| 351 | TGCCCAAGAT GCCGACAAAT TTCAGCTCTC CATCGATTTG CTGCGGATTA |
|
| 401 | CGTTTCCTTA TATATTATTG ATTTCCCTGT CTTCATTTGT CGGCTCGGTA |
|
| 451 | CTCAATTCTT ATCATAAGTT CGGCATTCCG GCGTTTACGC CCACGTTTCT |
|
| 501 | GAACGTGTCG TTTATCGTAT TCGCGCTGTT TTTCGTGCCG TATTTCGATC |
|
| 551 | CGCCCGTTAC CGCGCTGGCG TGGGCGGTCT TTGTCGGCGG CATTTTGCAA |
|
| 601 | CTCGGCTTCC AACTGCCCTG GCTGGCGAAA CTGGGCTTTT TGAAACTGCC |
|
| 651 | CAAACTGAGT TTCAAAGATG CGGCGGTCAA CCGCGTGATG AAACAGATGG |
|
| 701 | CGCCTGCGAT TTTGGGCGTG AGCGTGGCGC AGGTTTCTTT GGTGATCAAC |
|
| 751 | ACGATTTTCG CGTCTTATCT GCAATCGGGC AGCGTTTCAT GGATGTATTA |
|
| 801 | CGCCGACCGC ATGATGGAGC TGCCCAGCGG CGTGCTGGGG GCGGCACTCG |
|
| 851 | GTACGATTTT GCTGCCGACT TTGTCCAAAC ACTCGGCAAA CCAAGATACG |
|
| 901 | GAACAGTTTT CCGCCCTGCT CGACTGGGGT TTGCGCCTGT GCATGCTGCT |
|
| 951 | GACGCTGCCG GCGGCGGTCG GACTGGCGGT GTTGTCGTTC CCGCTGGTGG |
|
| 1001 | CGACGCTGTT TATGTACCGC GAATTTACGC TGTTTGACGC GCAGATGACG |
|
| 1051 | CAACACGCGC TGATTGCCTA TTCTTTCGGT TTAATCGGCT TAATCATGAT |
|
| 1101 | TAAAGTGTTG GCACCCGGCT TCTATGCGCG GCAAAACATC AAAACGCCCG |
|
| 1151 | TCAAAATCGC CATCTTCACG CTCATCTGCA CGCAGTTGAT GAACCTTGCC |
|
| 1201 | TTTATCGGCC CACTGAAACA CGTCGGACTT TCGCTTGCCA TCGGTCTGGG |
|
| 1251 | CGCGTGTATC AATGCCGGAT TGTTGTTTTA CCTGTTGCGC AGACACGGTA |
|
| 1301 | TTTACCAACC TGGCAAGGGT TGGGCAGCGT TCTTAGCAAA AATGCTGCTC |
|
| 1351 | TCGCTCGCCG TGATGTGCGG CGGACTGTGG GCAGCGCAGG CTTACCTGCC |
|
| 1401 | GTTTGAATGG GCGCACGCCG GCGGAATGCG GAAAGCGGGG CAGCTCTGCA |
|
| 1451 | TCCTGATTGC CGTCGGCGGC GGACTGTATT TCGCATCACT GGCGGCTTTG |
|
| 1501 | GGCTTCCGTC CGCGCCATTT CAAACGCGTG GAAAACTGA |
[0462]This corresponds to the amino acid sequence <SEQ ID 116; ORF20-1>:
[0000]| 1 | MNMLGALAKV GSLTMVSRVL GFVRDTVIAR AFGAGMATDA FFVAFKLPNL | |
|
| 51 | LRRVFAEGAF AQAFVPILAE YKETRSKEAA EAFIRHVAGM LSFVLVIVTA |
|
| 101 | LGILAAPWVI YVSAPGFAQD ADKFQLSIDL LRITFPYILL ISLSSFVGSV |
|
| 151 | LNSYHKFGIP AFTPTFLNVS FIVFALFFVP YFDPPVTALA WAVFVGGILQ |
|
| 201 | LGFQLPWLAK LGFLKLPKLS FKDAAVNRVM KQMAPAILGV SVAQVSLVIN |
|
| 251 | TIFASYLQSG SVSWMYYADR MMELPSGVLG AALGTILLPT LSKHSANQDT |
|
| 301 | EQFSALLDWG LRLCMLLTLP AAVGLAVLSF PLVATLFMYR EFTLFDAQMT |
|
| 351 | QHALIAYSFG LIGLIMIKVL APGFYARQNI KTPVKIAIFT LICTQLMNLA |
|
| 401 | FIGPLKHVGL SLAIGLGACI NAGLLFYLLR RHGIYQPGKG WAAFLAKMLL |
|
| 451 | SLAVMCGGLW AAQAYLPFEW AHAGGMRKAG QLCILIAVGG GLYFASLAAL |
|
| 501 | GFRPRHFKRV EN* |
[0463]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the MviN Virulence Factor of S. typhimurium (Accession Number P37169)
[0464]ORF20 and MviN proteins show 63% aa identity in 440aa overlap:
[0000]| Orf20 | 1 | MNMLGALAKVGSLTMVSRVLGFVRDTVIARAFGAGMATDAFFVAFKLPNLLRRVFAEGAF | 60 | |
| | MN+L +LA V S+TM SRVLGF RD ++AR FGAGMATDAFFVAFKLPNLLRR+FAEGAF |
| MviN | 14 | MNLLKSLAAVSSMTMFSRVLGFARDAIVARIFGAGMATDAFFVAFKLPNLLRRIFAEGAF | 73 |
|
| Orf20 | 61 | AQAFVPILAEYKETRSKEAXEAFIRHVAGMLSFVLVIVTALGILAAPWVIYVSAPSFAQD | 120 |
| | +QAFVPILAEYK + +EA F+ +V+G+L+ L +VT G+LAAPWVI V+AP FA |
| MviN | 74 | SQAFVPILAEYKSKQGEEATRIFVAYVSGLLTLALAVVTVAGMLAAPWVIMVTAPGFADT | 133 |
|
| Orf20 | 121 | ADKFQLSIDLLRITFPYILLISLSSFVGSVLNSYHKFGIPAFTPXFLNVSFIVFALFFVP | 180 |
| | ADKF L+ LLRITFPYILLISL+S VG++LN++++F IPAF P FLN+S I FALF P |
| MviN | 134 | ADKFALTTQLLRITFPYILLISLASLVGAILNTWNRFSIPAFAPTFLNISMIGFALFAAP | 193 |
|
| Orf20 | 181 | YFDPPVTAXAWAVFVGGILQLXFQLPWLAKLGFLKLPKLSFKDAAVNRVMKOMAPAILGV | 240 |
| | YF+PPV A AWAV VGG+LQL +QLP+L K+G L LP+++F+D RV+KQM PAILGV |
| MviN | 194 | YFNPPVLALAWAVTVGGVLQLVYQLPYLKKIGMLVLPRINFRDTGAMRVVKQMGPAILGV | 253 |
|
| Orf20 | 241 | SVAQVSLVINTIFASYLQSGSVSWMYYADRMMELPSGVLGAALGTILLPTLSKHSANQDT | 300 |
| | SV+Q+SL+INTIFAS+L SGSVSWMYYADR+ME PSGVLG ALGTILLP+LSK A+ + |
| MviN | 254 | SVSQISLIINTIFASFLASGSVSWMYYADRLMEFPSGVLGVALGTILLPSLSKSFASGNH | 313 |
|
| Orf20 | 301 | EQFSALLDWGLRLCMLLTLPAAVGLAVLSFPLVATLFMYRXFTLFDAQMWHALIMSFG | 360 |
| | +++ L+DWGLRLC LL LP+AV L +L+ PL +LF Y FT FDA MTQ ALIAYS G |
| MviN | 314 | DEYCRLMDWGLRLCFLLALPSAVALGILAKPLTVSLFQYGKFTAFDAAMTQRALIAYSVG | 373 |
|
| Orf20 | 361 | LIGLIMIKVLAPGFYARQNIXXPVKIAIFTLICXQLMNLXFXXXXXXXXXXXXXXXXXCI | 420 |
| | LIGLI++KVLAPGFY+RQ+I PVKIAI TLI QLMNL F C+ |
| MviN | 374 | LIGLIVVKVLAPGFYSRQDIKTPVKIAIVTLIMTQLMNLAFIGPLKHAGLSLSIGLAACL | 433 |
|
| Orf20 | 421 | NAGLLFYLLRRHGIYQPXQG | 440 |
| | NA LL++ LR+ I+ P G |
| MviN | 434 | NASLLYWQLRKQNIFTPQPG | 453 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0465]ORF20 shows 93.5% identity over a 447aa overlap with an ORF (ORF20a) from strain A of N. meningitidis.
[0000]
[0466]The complete length ORF20a nucleotide sequence <SEQ ID 117> is:
[0000]| 1 | ATGAATATGC TGGGAGCTTT GGTAAAAGTC GGCAGCCTGA CGATGGTGTC | |
|
| 51 | GCGCGTTTTG GGATTTGTGC GCGATACGGT CATTGCGCGC GCATTCGGCG |
|
| 101 | CAGGCATGGC GACGGATGCG TTCTTTGTCG CGTTCAAACT GCCCAACCTG |
|
| 151 | CTTCGCCGCG TGTTTGCGGA GGGGGCGTTT GCCCAAGCGT TTGTGCCGAT |
|
| 201 | TTTGGCGGAA TATAAGGAAA CGCGTTCTAA AGAGGCGACG GAGGCTTTTA |
|
| 251 | TCCGCCATGT GGCGGGGATG CTGTCGTTTG TACTGGTCAT CGTTACCGCG |
|
| 301 | CTGGGCATAC TTGCCGCGCC TTGGGTGATT TATGTTTCCG CACCCGGTTT |
|
| 351 | TGCCAAAGAT GCCGACAAAT TTCAGCTCTC TATCGATTTG CTGCGGATTA |
|
| 401 | CGTTTCCTTA TATCTTATTG ATTTCACTTT CCTCTTTTGT CGGCTCGGTA |
|
| 451 | CTCAATTCCT ATCATAAATT CAGCATTCCT GCGTTTACGC CCACGTTCCT |
|
| 501 | GAACGTGTCG TTTATCGTAT TCGCGCTGTT TTTCGTGCCG TATTTCGATC |
|
| 551 | CTCCCGTTAC CGCGCTGGCT TGGGCGGTTT TTGTCGGCGG CATTTTGCAA |
|
| 601 | CTCGGCTTCC AACTGCCCTG GCTGGCGAAA CTGGGTTTTT TGAAACTGCC |
|
| 651 | CAAACTGAGT TTCAAAGATG CGGCGGTCAA CCGCGTGATG AAACAGATGG |
|
| 701 | CGCCTGCGAT TTTGGGCGTG AGCGTGGCGC AGATTTCTTT GGTGATCAAC |
|
| 751 | ACGATTTTCG CGTCTTATCT GCAATCGGGC AGCGTTTCAT GGATGTATTA |
|
| 801 | CGCCGACCGC ATGATGGAAC TGCCCGGCGG CGTGCTGGGG GCGGCACTCG |
|
| 851 | GTACGATTTT GCTGCCGACT TTGTCCAAAC ACTCGGCAAA CCAAGATACG |
|
| 901 | GAACAGTTTT CCGCCCTGCT CGACTGGGGT TTGCGCNTGT GCATGCTGCT |
|
| 951 | GACGCTGCCG GCGGCGGTCG GAATGGCGGT GTTGTCGTTC CCGCTGGTGG |
|
| 1001 | CAACCTTGTT TATGTACCGA GAATTCACGC TGTTTGACGC GCAGATGACG |
|
| 1051 | CAACACGCGC TGATTGCCTA TTCTTTCGGT TTAATCGGTT TAATCATGAT |
|
| 1101 | TAAAGTGTTG GCGCCCGGCT TTTATGCGCG GCAAAACATC AAAACGCCCG |
|
| 1151 | TCAAAATCGC CATCTTCACG CTCATTTGCA CGCAGTTGAT GAACCTTGCC |
|
| 1201 | TTTATCGGCC CACTGAAACA CGTCGGACTT TCGCTTGCCA TCGGTCTGGG |
|
| 1251 | CGCGTGTATC AATGCCGGAT TGTTGTTTTA CCTGTTGCGC AGACACGGTA |
|
| 1301 | TTTACCAACC TGGCAAGGGT TGGGCAGCGT TCTTGGCAAA AATGCTGCTC |
|
| 1351 | TCGCTCGCCG TGATGGGAGG CGGCCTGTAT GCCGCCCAAA TCTGGCTGCC |
|
| 1401 | GTTCGACTGG GCACACGCCG GCGGAATGCA AAAGGCCGCC CGGCTCTTCA |
|
| 1451 | TCCTGATTGC CGTCGGCGGC GGACTGTATT TCGCATCACT GGCGGCTTTG |
|
| 1501 | GGCTTCCGTC CGCGCCATTT CAAACGCGTG GAAAGCTGA |
[0467]This encodes a protein having amino acid sequence <SEQ ID 118>:
[0000]| 1 | MNMLGALVKV GSLTMVSRVL GFVRDTVIAR AFGAGMATDA FFVAFKLPNL | |
|
| 51 | LRRVFAEGAF AQAFVPILAE YKETRSKEAT EAFIRHVAGM LSFVLVIVTA |
|
| 101 | LGILAAPWVI YVSAPGFAKD ADKFQLSIDL LRITFPYILL ISLSSFVGSV |
|
| 151 | LNSYHKFSIP AFTPTFLNVS FIVFALFFVP YFDPPVTALA WAVFVGGILQ |
|
| 201 | LGFQLPWLAK LGFLKLPKLS FKDAAVNRVM KQMAPAILGV SVAQISLVIN |
|
| 251 | TIFASYLQSG SVSWMYYADR MMELPGGVLG AALGTILLPT LSKHSANQDT |
|
| 301 | EQFSALLDWG LRXCMLLTLP AAVGMAVLSF PLVATLFMYR EFTLFDAQMT |
|
| 351 | QHALIAYSFG LIGLIMIKVL APGFYARQNI KTPVKIAIFT LICTQLMNLA |
|
| 401 | FIGPLKHVGL SLAIGLGACI NAGLLFYLLR RHGIYQPGKG WAAFLAKMLL |
|
| 451 | SLAVMGGGLY AAQIWLPFDW AHAGGMQKAA RLFILIAVGG GLYFASLAAL |
|
| 501 | GFRPRHFKRV ES* |
[0468]ORF20a and ORF20-1 show 96.5% identity in 512 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0469]ORF20 shows 92.1% identity over a 454aa overlap with a predicted ORF (ORF20ng) from N. gonorrhoeae.
[0000]
[0470]An ORF20ng nucleotide sequence <SEQ ID 119> was predicted to encode a protein having amino acid sequence <SEQ ID 120>:
[0000]| 1 | MNMLGALAKV GSLTMVSRVL GFVRDTVIAR AFGAGMATDA FFVAFKLPNL | |
|
| 51 | LRRVFAEGAF AQAFVPILAE YKETRSKEAT EAFIRHVAGM LSFVLIVVTA |
|
| 101 | LGILAAPWVI YVSAPGFTKD ADKFQLSISL LRITFPYILL ISLSSFVGSI |
|
| 151 | LNSYHKFGIP AFTPTFLNIS FIVFALFFVP YFDPPVTALA WAVFVGGILQ |
|
| 201 | LGFQLPWLAK LGFLKLPKLN FKDAAVNRVM KQMAPAILGV SVAQISLVIN |
|
| 251 | TIFASYLQSG SVSWMYYADR MMELPGGVLG AALGTILLPT LSKHSANQDT |
|
| 301 | EQFSALLDWG LRLCMLLTLP AAAGLAVLSF PLVATLFMYR EFTLFDAQMT |
|
| 351 | QHALIAYSFG LIGLIMIKVL ASGFYARQNI KTPVKIAIFT LICTQLMNLA |
|
| 401 | FIGPLKHAGL SLAIGLGACI NAGLLFFLFR KHGIYRPGQG LGQPSWRKCC |
|
| 451 | SRSP* |
[0471]Further DNA sequence analysis revealed the following DNA sequence <SEQ ID 121>:
[0000]| 1 | ATGAATATGC TTGGAGCTTT GGCAAAAGTC GGCAGCCTGA CGATGGTGTC | |
|
| 51 | GCGCGTTTTG GGATTTGTGC GCGATACGGT CATTGCGCGG GCATTCGGCG |
|
| 101 | CGGGTATGGC GACGGATGCG TTTTTTGTCG CGTTCAAACT GCCCAACCTG |
|
| 151 | CTTCGCCGCG TGTTTGCGGA GGGGGCGTTT GCCCAAGCGT TTGTGCCGAT |
|
| 201 | TTTGGCGGAA TATAAGGAAA CGCGTTCTAA AGAGGCGAcg gAGGCTTTTA |
|
| 251 | TCCGCCACGt tgcgggAatg CTGTCGTTTG TGCTGATcgt cGttacCGCG |
|
| 301 | CTGGGCATAC TTGCCGCgcc tTGGGTGATT TATGTTtccg CgcccGGCTT |
|
| 351 | TACCAAAGAC GCGGACAAGT TCCAACTTTC CATCAGCCTG CTGCGGATTA |
|
| 401 | CGTTTCCTTA TATATTATTG ATTTCTTTGT CTTCTTTTGT CGGCTCGATA |
|
| 451 | CTCAATTCCT ACCATAAGTT CGGCATTCCC GCGTTTACGC CCACGTTTTT |
|
| 501 | AAACATCTCT TTTATCGTAT TCGCACTGTT TTTCGTGCCG TATTTCGATC |
|
| 551 | CGCCCGTTAC CGCGCTGGCG TGGGCGGTTT TTGTCGGCGG TATTTTGCAG |
|
| 601 | CTCGGTTTCC AACTGCCGTG GCTGGCGAAA CTGGGCTTTT TGAAACTGCC |
|
| 651 | CAAACTGAAT TTCAAAGATG CGGCGGTCAA CCGCGTCATG AAACAGATGG |
|
| 701 | CGCCTGCGAT TTTGGGCGTG agcgTGGCGC AAATTTCTTT GgttATCAAC |
|
| 751 | ACGATTTTCG CGTCTTATCT GCAATCGGGC AGCGTTTCAT GGATGTatta |
|
| 801 | cgCCGACCGC ATGATGGAGc tgcgccGGGG CGTGCTGGGG GCTGCACTCG |
|
| 851 | GTACAATTTT GCTGCCGACT TTGTCCAAAC ACTCGGCAAA CCAAGATACG |
|
| 901 | GAACAGTTTT CCGCCCTGCT CGACTGGGGT TTGCGCCTGT GCATGCTGCT |
|
| 951 | GACGCTGCCG GCGGCGGccg GACTGGCGGT ATTGTCGTTC CCGCTGGTGG |
|
| 1001 | CGACGCTGTT TATGTACCGA GAATTCACGC TGTTTGACGC ACAAATGACG |
|
| 1051 | CAACACGCGC TGATTGCCTA TTCTTTCGGT TTAATCGGTT TAATTATGAT |
|
| 1101 | TAAAGTGTTG GCATCCGGCT TTTATGCGCG GCAAAACATC AAAACGCCCG |
|
| 1151 | TCAAAATCGC CATCTTCACG CTCATCTGCA CGCAGTTGAT GAACCTCGCC |
|
| 1201 | TTTATCGGTC CGTTGAAACA CGCCGGGCTT TCGCTCGCCA TCGGCCTGGG |
|
| 1251 | CGCGTGCATC AACGCCGGAT TGTTGTTCTT CCTGTTGCGC AAACACGGTA |
|
| 1301 | TTTACCGGCC cggcaggggt tgggcggcgt TCTTGGCGAA AATGCTGCTC |
|
| 1351 | GCGCTCGCCG TGATGTGCGG CGGACTGTGG GCGGCGCAGG CTTGCCTGCC |
|
| 1401 | GTTCGAATGG GCGCACGCCG GCGGAATGCG GAAAGCGGGG CAGCTCTGCA |
|
| 1451 | TCCTGATTGC CGTCGGCGGC GGACTGTATT TCGCATCTCT GGCGGCTTTG |
|
| 1501 | GGCTTCCGTC CGCGCCATTT CAAACGCGTG GAAAGCTGA |
[0472]This encodes the following amino acid sequence <SEQ ID 122; ORF20ng-1>:
[0000]| 1 | MNMLGALAKV GSLTMVSRVL GFVRDTVIAR AFGAGMATDA FFVAFKLPNL | |
|
| 51 | LRRVFAEGAF AQAFVPILAE YKETRSKEAT EAFIRHVAGM LSFVLIVVTA |
|
| 101 | LGILAAPWVI YVSAPGFTKD ADKFQLSISL LRITFPYILL ISLSSFVGSI |
|
| 151 | LNSYHKFGIP AFTPTFLNIS FIVFALFFVP YFDPPVTALA WAVFVGGILQ |
|
| 201 | LGFQLPWLAK LGFLKLPKLN FKDAAVNRVM KQMAPAILGV SVAQISLVIN |
|
| 251 | TIFASYLQSG SVSWMYYADR MMELRRGVLG AALGTILLPT LSKHSANQDT |
|
| 301 | EQFSALLDWG LRLCMLLTLP AAAGLAVLSF PLVATLFMYR EFTLFDAQMT |
|
| 351 | QHALIAYSFG LIGLIMIKVL ASGFYARQNI KTPVKIAIFT LICTQLMNLA |
|
| 401 | FIGPLKHAGL SLAIGLGACI NAGLLFFLLR KHGIYRPGRG WAAFLAKMLL |
|
| 451 | ALAVMCGGLW AAQACLPFEW AHAGGMRKAG QLCILIAVGG GLYFASLAAL |
|
| 501 | GFRPRHFKRV ES* |
[0473]ORF20ng-1 and ORF20-1 show 95.7% identity in 512 aa overlap:
[0000]
[0474]In addition, ORF20ng-1 shows significant homology with a virulence factor of S. typhimurium:
[0000]| sp|P37169|MVIN_SALTY VIRULENCE FACTOR MVIN pir||S40271 mviN protein - | |
| Salmonella typhimurium gi|438252 (Z26133) mviB gene product |
| [Salmonella typhimurium] gnl|PID|d1005521 (D25292) ORF2 |
| [Salmonella typhimurium] Length = 524 |
| Score = 1573 (750.1 bits), Expect = 1.1e−220, Sum P(2) = 1.1e−220 |
| Identities = 309/467 (66%), Positives = 368/467 (78%) |
|
| Query: | 1 | MNMLGALAKVGSLTMVSRVLGFVRDTVIARAFGAGMATDAFFVAFKLPNLLRRVFAEGAF | 60 | |
| | MN+L +LA V S+TM SRVLGF RD ++AR FGAGMATDAFFVAFKLPNLLRR+FAEGAF |
| Sbjct: | 14 | MNLLKSLAAVSSMTMFSRVLGFARDAIVARIFGAGMATDAFFVAFKLPNLLRRIFAEGAF | 73 |
|
| Query: | 61 | AQAFVPILAEYKETRSKEATEAFIRHVAGMLSFVLIVVTALGILAAPWVIYVSAPGFTKD | 120 |
| | +QAFVPILAEYK + +EAT F+ +V+G+L+ L VVT G+LAAPWVI V+APGF |
| Sbjct: | 74 | SQAFVPILAEYKSKQGEEATRIFVAYVSGLLTLALAVVTVAGMLAAPWVIMVTAPGFADT | 133 |
|
| Query: | 121 | ADKFQLSISLLRITFPYILLISLSSFVGSILNSYHKFGIPAFTPTFLNISFIVFALFFVP | 180 |
| | ADKF L+ LLRITFPYILLISL+S VG+ILN++++F IPAF PTFLNIS I FALF P |
| Sbjct: | 134 | ADKFALTTQLLRITFPYILLISLASLVGAILNTWNRFSIPAFAPTFLNISMIGFALFAAP | 193 |
|
| Query: | 181 | YFDPPVTALAWAVFVGGILQLGFQLPWLAKLGFLKLPKLNFKDAAVNRVMKQMAPAILGV | 240 |
| | YF+PPV ALAWAV VGG+LQL +QLP+L K+G L LP++NF+D RV+KQM PAILGV |
| Sbjct: | 194 | YFNPPVLALAWAVTVGGVLQLVYQLPYLKKIGMLVLPRINFRDTGAMRVVKQMGPAILGV | 253 |
|
| Query: | 241 | SVAQISLVINTIFASYLQSGSVSWMYYADRMMELRRGVLGAALGTILLPTLSKHSANQDT | 300 |
| | SV+QISL+INTIFAS+L SGSVSWMYYADR+ME GVLG ALGTILLP+LSK A+ + |
| Sbjct: | 254 | SVSQISLIINTIFASFLASGSVSWMYYADRLMEFPSGVLGVALGTILLPSLSKSFASGNH | 313 |
|
| Query: | 301 | EQFSALLDWGLRLCMLLTLPAAAGLAVLSFPLVATLFMYREFTLFDAQMTQHALIAYSFG | 360 |
| | +++ L+DWGLRLC LL LP+A L +L+ PL +LF Y +FT FDA MTQ ALIAYS G |
| Sbjct: | 314 | DEYCRLMDWGLRLCFLLALPSAVALGILAKPLTVSLFQYGKFTAFDAAMTQRALIAYSVG | 373 |
|
| Query: | 361 | LIGLIMIKVLASGFYARQNIKTPVKIAIFTLICTQLMNLAFIGPLKHAGLSLAIGLGACI | 420 |
| | LIGLI++KVLA GFY+RQ+IKTPVKIAI TLI TQLMNLAFIGPLKHAGLSL+IGL AC+ |
| Sbjct: | 374 | LIGLIVVKVLAPGFYSRQDIKTPVKIAIVTLIMTQLMNLAFIGPLKHAGLSLSIGLAACL | 433 |
|
| Query: | 421 | NAGLLFFLLRKHGIYRPGRGWXXXXXXXXXXXXVMCGGLWAAQACLP | 467 |
| | NA LL++ LRK I+ P GW VM L+ +P |
| Sbjct: | 434 | NASLLYWQLRKQNIFTPQPGWMWFLMRLIISVLVMAAVLFGVLHIMP | 480 |
|
| Score = 70 (33.4 bits), Expect = 1.1e−220, Sum P(2) = 1.1e−220 | |
| Identities = 14/41 (34%), Positives = 23/41 (56%) |
|
| Query: | 469 | EWAHAGGMRKAGQLCILIAVGGGLYFASLAALGFRPRHFKR | 509 | |
| | EW+ + + +L ++ G YFA+LA LGF+ + F R |
| Sbjct: | 481 | EWSQGSMLWRLLRLMAVVIAGIAAYFAALAVLGFKVKEFVR | 521 |
[0475]Based on this analysis, including the homology with a virulence factor from S. typhimurium, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 15
[0476]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 123>:
[0000]| 1 | atGATTAAAA TCAAAAAAGG TCTAAACCTG CCCATCGCGG GCAGACCGGA | |
|
| 51 | GCAAGCCGTT tACGACGGCC CGGCCaTTAC CGAAGtCGCG TTGCTTGGCG |
|
| 101 | AAGAATATGC CGGTATGCGC CCCTCGATGA AAGTCAAGGA AGGCGATGCC |
|
| 151 | GTcAAAAAAG GCCAAGTGCT GTTTGAAGAC AAAAAGAATC CGGGCGTGGT |
|
| 201 | GTTTACTGCG CCGGCTTCAG GcAAAATCGC CGCGATTCAC CGTGGCGAAA |
|
| 251 | AGCGCGTACT TCAGTCAGTC GTGATTGCCG TTGAArGCAA CGACGAAATC |
|
| 301 | GAGTTTGAAC GCTACGCACC TGAAGCGCTG GCAAACTTAA GCGGCGAAGA |
|
| 351 | AGTGCGCCGC AACCTGATCC AATCCGGTTT GTGGACTGCG CTGCGCACCC |
|
| 401 | GTCCGTTCAG CAAAATTCCT GCCGTCGATG CCGAGCCGTT CGCCATCTTC |
|
| 451 | GTCAATGCGA tGGACACCAA TCCG.. |
[0477]This corresponds to the amino acid sequence <SEQ ID 124; ORF22>:
[0000]| 1 | MIKIKKGLNL PIAGRPEQAV YDGPAITEVA LLGEEYAGMR PSMKVKEGDA | |
|
| 51 | VKKGQVLFED KKNPGVVFTA PASGKIAAIH RGEKRVLQSV VIAVEXNDEI |
|
| 101 | EFERYAPEAL ANLSGEEVRR NLIQSGLWTA LRTRPFSKIP AVDAEPFAIF |
|
| 151 | VNAMDTNP.. |
[0478]Further work revealed the complete nucleotide sequence <SEQ ID 125>:
[0000]| 1 | ATGATTAAAA TCAAAAAAGG TCTAAACCTG CCCATCGCGG GCAGACCGGA | |
|
| 51 | GCAAGCCGTT TACGACGGCC CGGCCATTAC CGAAGTCGCG TTGCTTGGCG |
|
| 101 | AAGAATATGC CGGTATGCGC CCCTCGATGA AAGTCAAGGA AGGCGATGCC |
|
| 151 | GTCAAAAAAG GCCAAGTGCT GTTTGAAGAC AAAAAGAATC CGGGCGTGGT |
|
| 201 | GTTTACTGCG CCGGCTTCAG GCAAAATCGC CGCGATTCAC CGTGGCGAAA |
|
| 251 | AGCGCGTACT TCAGTCAGTC GTGATTGCCG TTGAAGGCAA CGACGAAATC |
|
| 301 | GAGTTTGAAC GCTACGCACC TGAAGCGCTG GCAAACTTAA GCGGCGAAGA |
|
| 351 | AGTGCGCCGC AACCTGATCC AATCCGGTTT GTGGACTGCG CTGCGCACCC |
|
| 401 | GTCCGTTCAG CAAAATTCCT GCCGTCGATG CCGAGCCGTT CGCCATCTTC |
|
| 451 | GTCAATGCGA TGGACACCAA TCCGCTGGCT GCCGACCCTA CGGTCATTAT |
|
| 501 | CAAAGAAGCC GCCGAGGATT TCAAACGCGG CCTGTTGGTA TTGAGCCGTT |
|
| 551 | TGACCGAACG CAAAATCCAT GTTTGTAAGG CAGCTGGCGC AGACGTGCCG |
|
| 601 | TCTGAAAATG CTGCCAACAT CGAAACACAT GAATTCGGCG GCCCGCATCC |
|
| 651 | TGCCGGTTTG AGTGGCACGC ACATTCATTT CATCGAGCCG GTCGGCGCGA |
|
| 701 | ATAAAACCGT GTGGACCATC AATTATCAAG ATGTAATTAC CATTGGCCGT |
|
| 751 | TTGTTTGCAA CAGGCCGTCT GAACACCGAG CGCGTGATTG CCCTAGGTGG |
|
| 801 | TTCTCAAGTC AACAAACCGC GCCTCTTGCG TACCGTTTTG GGTGCGAAAG |
|
| 851 | TATCGCAAAT TACTGCGGGC GAATTGGTTG ACACAGACAA CCGCGTGATT |
|
| 901 | TCCGGTTCGG TATTGAACGG CGCGATTACA CAAGGCGCGC ACGATTATTT |
|
| 951 | GGGACGCTAC CACAATCAGA TTTCCGTTAT CGAAGAAGGC CGCAGCAAAG |
|
| 1001 | AGCTGTTCGG CTGGGTTGCG CCGCAGCCGG ACAAATACTC CATCACGCGT |
|
| 1051 | ACAACCCTCG GCCATTTCCT GAAAAACAAA CTCTTCAAGT TCAACACAGC |
|
| 1101 | CGTCAACGGC GGCGACCGCG CCATGGTGCC GATTGGTACT TACGAGCGCG |
|
| 1151 | TGATGCCCTT GGATATCCTG CCCACCCTGC TTTTGCGCGA TTTAATCGTC |
|
| 1201 | GGCGATACCG ACAGCGCGCA GGCATTGGGT TGCTTGGAAT TGGACGAAGA |
|
| 1251 | AGACCTCGCT TTGTGCAGCT TCGTCTGCCC GGGCAAATAC GAATACGGCC |
|
| 1301 | CGCTGTTGCG CAAAGTGCTG GAAACCATTG AGAAGGAAGG CTGA |
[0479]This corresponds to the amino acid sequence <SEQ ID 126; ORF22-1>:
[0000]| 1 | MIKIKKGLNL PIAGRPEQAV YDGPAITEVA LLGEEYAGMR PSMKVKEGDA | |
|
| 51 | VKKGQVLFED KKNPGVVFTA PASGKIAAIH RGEKRVLQSV VIAVEGNDEI |
|
| 101 | EFERYAPEAL ANLSGEEVRR NLIQSGLWTA LRTRPFSKIP AVDAEPFAIF |
|
| 151 | VNAMDTNPLA ADPTVIIKEA AEDFKRGLLV LSRLTERKIH VCKAAGADVP |
|
| 201 | SENAANIETH EFGGPHPAGL SGTHIHFIEP VGANKTVWTI NYQDVITIGR |
|
| 251 | LFATGRLNTE RVIALGGSQV NKPRLLRTVL GAKVSQITAG ELVDTDNRVI |
|
| 301 | SGSVLNGAIT QGAHDYLGRY HNQISVIEEG RSKELFGWVA PQPDKYSITR |
|
| 351 | TTLGHFLKNK LFKFNTAVNG GDRAMVPIGT YERVMPLDIL PTLLLRDLIV |
|
| 401 | GDTDSAQALG CLELDEEDLA LCSFVCPGKY EYGPLLRKVL ETIEKEG* |
[0480]Further work identified the corresponding gene in strain A of N. meningitidis <SEQ ID 127>:
[0000]| 1 | ATGATTAAAA TCAAAAAAGG TCTAAACCTG CCCATCGCGG GCAGACCGGA | |
|
| 51 | GCAAGTCATT TATGACGGGC CCGTCATTAC CGAAGTCGCG TTGCTTGGCG |
|
| 101 | AAGAATATGC CGGTATGCGC CCCTNGATGA AAGTCAAGGA AGGCGATGCC |
|
| 151 | GTCAAAAAAG GCCAAGTGCT GTTTGAAGAC AAAAAGNATC CGGGCGTGGT |
|
| 201 | GTTTACCGCG CCNGTTTCAG GCAAAATCGC CGCCATCCAT CGCGGCGAAA |
|
| 251 | AGCGCGTACT TCAGTCGGTC GTGATTGCCG TTGAAGGCAA CGACGAAATC |
|
| 301 | GAGTTCGAAC GCTACGCGCC CGAAGCGTTG GCAAACTTAA GCGGCGANGA |
|
| 351 | ANTNNGNNGC AATCTGATCC AATCCGGTTT GTGGACTGCG CTGCGTANCC |
|
| 401 | GTCCGTTCAG CAAAATCCCT GCCGTCGATG CCGAGCCGTT CGCCATCTTC |
|
| 451 | GTCAATGCGA TGGACACCAA TCCGCTNGCG GCAGACCCTG TGGTTGTGAT |
|
| 501 | CAAAGAAGCC GNCGANGATT TCAGACGANG TNTGCTGGTA TTGAGCCGTT |
|
| 551 | TGACCGAGCG TAAAATCCAT GTGTGTAAGG CAGCTGGCGC AGACGTGCCG |
|
| 601 | TCTGAAAATG CTGCCAACAT CGAAACACAT GAATTCGGCG GCCCGCATCC |
|
| 651 | GGCCGGTTTG AGTGGCACGC ACATTCATTT CATTGAGCCG GTCGGTGCAA |
|
| 701 | ACAAAACCGT TTGGACCATC AATTATCAAG ATGTAATTGC CATCGGACGT |
|
| 751 | TTGTTTGCAA CAGGCCGTCT GAACACCGAG CGCGTGATTG CTTTGGGTGG |
|
| 801 | TTCTCAAGTC AACAAACCAC GCCTCTTGCG TACCGTTTTG GGTGCGAAAG |
|
| 851 | TATCGCAAAT TACTGCGGGC GAATTGGTTG ACGCAGACAA CCGCGTGATT |
|
| 901 | TCCGGTTCGG TATTGAACGG CGCGATTACA CAAGGCGCGC ACGATTATTT |
|
| 951 | GGGACGCTAC CACAATCAGA TTTCCGTTAT CGAAGAAGGC CGCAGCAAAG |
|
| 1001 | AGCTGTTCGG CTGGGTTGCG CCGCAGCCGG ACAAATACTC CATCACGCGT |
|
| 1051 | ACGACCCTCG GCCATTTCCT GAAAAACAAA CTCTTCAAGT TCACGACAGC |
|
| 1101 | CGTCAACGGT GGCGACCGCG CCATGGTGCC GATTGGTACT TACGAGCGCG |
|
| 1151 | TAATGCCGCT AGACATCCTG CCTACCCTGC TTTTGCGCGA TTTAATCGTC |
|
| 1201 | GGCGATACCG ACAGCGCGCA AGCATTGGGT TGCTTGGAAT TGGACGAAGA |
|
| 1251 | AGACCTCGCT TTGTGCAGCT TCGTCTGCCC GGGCAAATAC GAATANGGCC |
|
| 1301 | CGCTGTTGCG TAAGGTGCTG GAAACCNTTG AGAAGGAAGG CTGA |
[0481]This encodes a protein having amino acid sequence <SEQ ID 128; ORF22a>:
[0000]| 1 | MIKIKKGLNL PIAGRPEQVI YDGPVITEVA LLGEEYAGMR PXMKVKEGDA | |
|
| 51 | VKKGQVLFED KKXPGVVFTA PVSGKIAAIH RGEKRVLQSV VIAVEGNDEI |
|
| 101 | EFERYAPEAL ANLSGXEXXX NLIQSGLWTA LRXRPFSKIP AVDAEPFAIF |
|
| 151 | VNAMDTNPLA ADPVVVIKEA XXDFRRXXLV LSRLTERKIH VCKAAGADVP |
|
| 201 | SENAANIETH EFGGPHPAGL SGTHIHFIEP VGANKTVWTI NYQDVIAIGR |
|
| 251 | LFATGRLNTE RVIALGGSQV NKPRLLRTVL GAKVSQITAG ELVDADNRVI |
|
| 301 | SGSVLNGAIT QGAHDYLGRY HNQISVIEEG RSKELFGWVA PQPDKYSITR |
|
| 351 | TTLGHFLKNK LFKFTTAVNG GDRAMVPIGT YERVMPLDIL PTLLLRDLIV |
|
| 401 | GDTDSAQALG CLELDEEDLA LCSFVCPGKY EXGPLLRKVL ETXEKEG* |
[0482]The originally-identified partial strain B sequence (ORF22) shows 94.2% identity over a 158aa overlap with ORF22a:
[0000]
[0483]The complete strain B sequence (ORF22-1) and ORF22a show 94.9% identity in 447 aa overlap:
[0000]
[0484]Further work identified a partial gene sequence <SEQ ID 129> from N. gonorrhoeae, which encodes the following amino acid sequence <SEQ ID 130; ORF22ng>:
[0000]| 1 | MIKIKKGLNL PIAGRPEQVI YDGPAITEVA LLGEEYVGMR PSMKIKEGEA | |
|
| 51 | VKKGQVLFED KKNPGVVFTA PASGKIAAIH RGEKRVLQSV VIAVEGNDEI |
|
| 101 | EFERYVPEAL AKLSSEKVRR NLIQSGLWTA LRTRPFSKIP AVDAEPFAIF |
|
| 151 | VNAMDTNPLA ADPTVIIKEA AEDFKRGLLV LSRLTERKIH VCKAAGADVP |
|
| 201 | SENAANIETH EFGGPHPAGL SGTHIHFIEP VGANKTVWTI NYQDVIAIGR |
|
| 251 | LFVTGRLNTE RVVALGGLQV NKPRLLRTVL GAKVSQLTAG ELVDADNRVI |
|
| 301 | SGSVLNGAIA QGAHDYLGRY HN* |
[0485]Further work identified complete gonococcal gene <SEQ ID 131>:
[0000]| 1 | ATGATTAAAA TCAAAAAAGG TCTAAATCTG CCCATCGCGG GCAGACCGGA | |
|
| 51 | GCAAGTCATT TATGACGGCC CGGCCATTAC CGAAGTCGCG TTGCTTGGCG |
|
| 101 | AAGAATATGT CGGCATGCGC CCCTCGATGA AAATCAAGGA AGGTGAAGCC |
|
| 151 | GTCAAAAAAG GCCAAGTGCT GTTTGAAGAC AAAAAGAATC CGGGCGTAGT |
|
| 201 | ATTTACTGCG CCGGCTTCAG GCAAAATCGC CGCTATTCAC CGTGGCGAAA |
|
| 251 | AGCGCGTACT TCAGTCAGTC GTGATTGCCG TTGAAGGCAA CGACGAAATC |
|
| 301 | GAGTTCGAAC GCTACGTACC TGAAGCGCTG GCAAAATTGA GCAGCGAAAA |
|
| 351 | AGTGCGCCGC AACCTGATTC AATCAGGCTT ATGGACTGCG CTTCGCACCC |
|
| 401 | GTCCGTTCAG CAAAATCCCT GCCGTAGATG CCGAGCCGTT CGCCATCTTC |
|
| 451 | GTCAATGCGA TGGACACCAA TCCGCTGGCT GCCGACCCTA CGGTCATCAT |
|
| 501 | CAAAGAAGCC GCCGAAGACT TCAAACGCGG CCTGTTGGTA TTGAGCCGCC |
|
| 551 | TGACCGAACG TAAAATCCAT GTGTGTAAAG CAGCAGGCGC AGACGTGCCG |
|
| 601 | TCTGAAAATG CTGCCAATAT CGAAACACAT GAATTTGGCG GCCCGCATCC |
|
| 651 | TGCCGGCTTG AGTGGCACGC ACATTCATTT CATCGAGCCA GTCGGCGCGA |
|
| 701 | ATAAAACCGT GTGGACCATC AATTATCAAG ACGTGATTGC TATCGGACGT |
|
| 751 | TTGTTCGTAA CAGGCCGTCT GAATACCGAG CGCGTGGTTG CCTTGGGCGG |
|
| 801 | CCTGCAAGTC AACAAACCGC GCCTCTTGCG TACCGTTTTG GGTGCGAAGG |
|
| 851 | TGTCTCAACT TACCGCCGGC GAATTGGTTG ACGCGGACAA CCGCGTGATT |
|
| 901 | TCCGGTTCGG TATTGAACGG TGCGATTGCA CAAGGCGCGC ATGATTATTT |
|
| 951 | GGGACGCTAC CACAATCAGA TTTCCGTTAT CGAAGAAGGC CGCAGCAAAG |
|
| 1001 | AGCTGTTCGG CTGGGTTGCG CCGCAGCCGG ACAAATACTC CATCACGCGC |
|
| 1051 | ACCACTCTCG GCCATTTCCT AAAAAACAAA CTCTTCAAGT TCACGACAGC |
|
| 1101 | CGTCAACGGC GGCGACCGCG CCATGGTACC GATCGGCACT TATGAGCGCG |
|
| 1151 | TAATGCCGTT GGACATCCTG CCTACCTTGC TTTTGCGCGA TTTAATCGTC |
|
| 1201 | GGCGATACCG ACAGCGCGCA GGCTTTGGGT TGCTTGGAAT TGGACGAAGA |
|
| 1251 | AGACCTCGCT TTGTGCAGCT TCGTCTGCCC GGGCAAATAC GAATACGGCC |
|
| 1301 | CGCTGTTGCG CAAAGTGCTG GAAACCATTG AGAAGGAAGG CTGA |
[0486]This encodes a protein having amino acid sequence <SEQ ID 132; ORF22ng-1>:
[0000]| 1 | MIKIKKGLNL PIAGRPEQVI YDGPAITEVA LLGEEYVGMR PSMKIKEGEA | |
|
| 51 | VKKGQVLFED KKNPGVVFTA PASGKIAAIH RGEKRVLQSV VIAVEGNDEI |
|
| 101 | EFERYVPEAL AKLSSEKVRR NLIQSGLWTA LRTRPFSKIP AVDAEPFAIF |
|
| 151 | VNAMDTNPLA ADPTVIIKEA AEDFKRGLLV LSRLTERKIH VCKAAGADVP |
|
| 201 | SENAANIETH EFGGPHPAGL SGTHIHFIEP VGANKTVWTI NYQDVIAIGR |
|
| 251 | LFVTGRLNTE RVVALGGLQV NKPRLLRTVL GAKVSQLTAG ELVDADNRVI |
|
| 301 | SGSVLNGAIA QGAHDYLGRY HNQISVIEEG RSKELFGWVA PQPDKYSITR |
|
| 351 | TTLGHFLKNK LFKFTTAVNG GDRAMVPIGT YERVMPLDIL PTLLLRDLIV |
|
| 401 | GDTDSAQALG CLELDEEDLA LCSFVCPGKY EYGPLLRKVL ETIEKEG* |
[0487]The originally-identified partial strain B sequence (ORF22) shows 93.7% identity over a 158aa overlap with ORF22ng:
[0000]
[0488]The complete sequences from strain B (ORF22-1) and gonococcus (ORF22ng) show 96.2% identity in 447 aa overlap:
[0000]
[0489]Computer analysis of these sequences gave the following results:
[0000]Homology with 48 kDa Outer Membrane Protein of Actinobacillus pleuropneumoniae (Accession Number U24492).
[0490]ORF22 and this 48 kDa protein show 72% aa identity in 158aa overlap:
[0000]| Orf22 | 1 | MIKIKKGLNLPIAGRPEQAVYDGPAITEVALLGEEYAGMRPSMKVKEGDAVKKGQVLFED | 60 | |
| | MI IKKGL+LPIAG P Q +++G + EVA+LGEEY GMRPSMKV+EGD VKKGQVLFED | |
| 48 kDa | 1 | MITIKKGLDLPIAGTPAQVIHNGNTVNEVAMLGEEYVGMRPSMKVREGDVVKKGQVLFED | 60 |
|
| orf22 | 61 | KKNPGVVFTAPASGKIAAIHRGEKRVLQSVVIAVEXNDEIEFERYAPEALANLSGEEVRR | 120 |
| | KKNPGVVFTAPASG + I+RGEKRVLQSVVI VE +++I F RY LA+LS E+V++ | |
| 48 kDa | 61 | KKNPGVVFTAPASGTVVTINRGEKRVLQSVVIKVEGDEQITFTRYEAAQLASLSAEQVKQ | 120 |
|
| orf22 | 121 | NLIQSGLWTALRTRPFSKIPAVDAEPFAIFVNAMDTNP | 158 |
| | NLI+SGLWTA RTRPFSK+PA+DA P +IFVNAMDTNP | |
| 48 kDa | 121 | NLIESGLWTAFRTRPFSKVPALDAIPSSIFVNAMDTNP | 158 |
[0491]ORF22a also shows homology to the 48 kDa Actinobacillus pleuropneumoniae protein:
[0000]| gi|1185395 (U24492) 48 kDa outer membrane protein | |
| [Actinobacillus pleuropneumoniae] |
| Length = 449 |
| Score = 530 bits (1351), Expect = e−150 |
| Identities = 274/450 (60%), Positives = 323/450 (70%), Gaps = 4/450 (0%) |
|
| Query: | 1 | MIKIKKGLNLPIAGRPEQVIYDGPVITEVALLGEEYAGMRPXMKVKEGDAVKKGQVLFED | 60 | |
| | MI IKKGL+LPIAG P QVI++G + EVA+LGEEY GMRP MKV+EGD VKKGQVLFED | |
| Sbjct: | 1 | MITIKKGLDLPIAGTPAQVIHNGNTVNEVAMLGEEYVGMRPSMKVREGDVVKKGQVLFED | 60 |
|
| Query: | 61 | KKXPGVVFTAPVSGKIAAIHRGEKRVLQSVVIAVEGNDEIEFERYAPEALANLSGXEXXX | 120 |
| | KK PGVVFTAP SG + I+RGEKRVLQSVVI VEG+++I F RY LA+LS + | |
| Sbjct: | 61 | KKNPGVVFTAPASGTVVTINRGEKRVLQSVVIKVEGDEQITFTRYEAAQLASLSAEQVKQ | 120 |
|
| Query: | 121 | NLIQSGLWTALRXRPFSKIPAVDAEPFAIFVNAMDTNPLAADPVVVIKEAXXDFRRXXLV | 180 |
| | NLI+SGLWTA R RPFSK+PA+DA P +IFVNAMDTNPLAADP VV+KE DF+ V | |
| Sbjct: | 121 | NLIESGLWTAFRTRPFSKVPALDAIPSSIFVNAMDTNPLAADPEVVLKEYETDFKDGLTV | 180 |
|
| Query: | 181 | LSRL--TERKIHVCKAAGADVP-SENAANIETHEFGGPHPAGLSGTHIHFIEPVGANKTV | 237 |
| | L+RL ++ +++CK A +++P S I F G HPAGL GTHIHF++PVGA K V | |
| Sbjct: | 181 | LTRLFNGQKPVYLCKDADSNIPLSPAIEGITIKSFSGVHPAGLVGTHIHFVDPVGATKQV | 240 |
|
| Query: | 238 | WTINYQDVIAIGRLFATGRLNTERVIALGGSQVNKPRLLRTVLGAKVSQITAGELVDADN | 297 |
| | W +NYQDVIAIG+LF TG L T+R+I+L G QV PRL+RT LGA +SQ+TA EL +N | |
| Sbjct: | 241 | WHLNYQDVIAIGKLFTTGELFTDRIISLAGPQVKNPRLVRTRLGANLSQLTANELNAGEN | 300 |
|
| Query: | 298 | RVISGSVLNGAITQGAHDYLGRYHNQISVIEEGRSKELFGWVAPQPDKYSITRTTLGHFL | 357 |
| | RVISGSVL+GA G DYLGRY Q+SV+ EGR KELFGW+ P DK+SITRT LGHF | |
| Sbjct: | 301 | RVISGSVLSGATAAGPVDYLGRYALQVSVLAEGREKELFGWIMPGSDKFSITRTVLGHFG | 360 |
|
| Query: | 358 | KNKLFKFTTAVNGGDRAMVPIGTYERVMXXXXXXXXXXXXXXVGDTDSAQXXXXXXXXXX | 417 |
| | K KLF FTTAV+GG+RAMVPIG YERVM GDTDSAQ | |
| Sbjct: | 361 | K-KLFNFTTAVHGGERAMVPIGAYERVMPLDIIPTLLLRDLAAGDTDSAQNLGCLELDEE | 419 |
|
| Query: | 418 | XXXXXSFVCPGKYEXGPLLRKVLETXEKEG | 447 |
| | ++VCPGK GP+LR LE EKEG | |
[0492]ORF22ng-1 also shows homology with the OMP from A. pleuropneumoniae:
[0000]| gi|1185395 (U24492) 48 kDa outer membrane protein [Actinobacillus | |
| pleuropneumoniae] Length = 449 |
| Score = 555 bits (1414), Expect = e−157 |
| Identities = 284/450 (63%), Positives = 337/450 (74%), Gaps = 4/450 (0%) |
|
| Query: | 27 | MIKIKKGLNLPIAGRPEQVIYDGPAITEVALLGEEYVGMRPSMKIKEGEAVKKGQVLFED | 86 | |
| | MI IKKGL+LPIAG P QVI++G + EVA+LGEEYVGMRPSMK++EG+ VKKGQVLFED | |
| Sbjct: | 1 | MITIKKGLDLPIAGTPAQVIHNGNTVNEVAMLGEEYVGMRPSMKVREGDVVKKGQVLFED | 60 |
|
| Query: | 87 | KKNPGVVFTAPASGKIAAIHRGEKRVLQSVVIAVEGNDEIEFERYVPEALAKLSSEKVRR | 146 |
| | KKNPGVVFTAPASG + I+RGEKRVLQSVVI VEG+++I F RY LA LS+E+V++ | |
| Sbjct: | 61 | KKNPGVVFTAPASGTVVTINRGEKRVLOSVVIKVEGDEQITFTRYEAAQLASLSAEQVKQ | 120 |
|
| Query: | 147 | NLIQSGLWTALRTRPFSKIPAVDAEPFAIFVNAMDTNPLAADPTVIIKEAAEDFKRGLLV | 206 |
| | NLI+SGLWTA RTRPFSK+PA+DA P +IFVNAMDTNPLAADP V++KE DFK GL V | |
| Sbjct: | 121 | NLIESGLWTAFRTRPFSKVPALDAIPSSIFVNAMDTNPLAADPEVVLKEYETDFKDGLTV | 180 |
|
| Query: | 207 | LSRL--TERKIHVCKAAGADVP-SENAANIETHEFGGPHPAGLSGTHIHFIEPVGANKTV | 263 |
| | L+RL ++ +++CK A +++P S I F G HPAGL GTHIHF++PVGA K V | |
| Sbjct: | 181 | LTRLFNGQKPVYLCKDADSNIPLSPAIEGITIKSFSGVHPAGLVGTHIHFVDPVGATKQV | 240 |
|
| Query: | 264 | WTINYQDVIAIGRLFVTGRLNTERVVALGGLQVNKPRLLRTVLGAKVSQLTAGELVDADN | 323 |
| | W +NYQDVIAIG+LF TG L T+R+++L G QV PRL+RT LGA +SQLTA EL +N | |
| Sbjct: | 241 | WHLNYQDVIAIGKLFTTGELFTDRIISLAGPQVKNPRLVRTRLGANLSQLTANELNAGEN | 300 |
|
| Query: | 324 | RVISGSVLNGAIAQGAHDYLGRYHNQISVIEEGRSKELFGWVAPQPDKYSITRTTLGHFL | 383 |
| | RVISGSVL+GA A G DYLGRY Q+SV+EGR KELFGW+ P DK+SITRT LGHF | |
| Sbjct: | 301 | RVISGSVLSGATAAGPVDYLGRYALQVSVLAEGREKELFGWIMPGSDKFSITRTVLGHFG | 360 |
|
| Query: | 384 | KNKLFKFTTAVNGGDRAMVPIGTYERVMXXXXXXXXXXXXXXVGDIDSAQXXXXXXXXXX | 443 |
| | K KLF FTTAV+GG+RAMVPIG YERVM GDTDSAQ | |
| Sbjct: | 361 | K-KLFNFTTAVHGGERAMVPIGAYERVMPLDIIPTLLLRDLAAGDIDSAQNLGCLELDEE | 419 |
|
| Query: | 444 | XXXXXSFVCPGKYEYGPLLRKVLETIEKEG | 473 |
| | ++VCPGK YGP+LR LE IEKEG | |
| Sbjct: | 420 | DLALCTYVCPGKNNYGPMLRAALEKIEKEG | 449 |
[0493]Based on this analysis, including the homology with the outer membrane protein of Actinobacillus pleuropneumoniae, it was predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0494]ORF22-1 (35.4 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 5A shows the results of affinity purification of the GST-fusion protein, and FIG. 5B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for ELISA (positive result) and FACS analysis (FIG. 5C). These experiments confirm that ORF22-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 16
[0495]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 133>:
[0000]| 1 | ..GCGnCGnAAA TCATCCATCC CC..nACGTC GTAGGCCCTG AAGCCAACTG | |
|
| 51 | GTTTTTTATG GTAGCCAGTA CGTTTGTGAT TGCTTTGATT GGTTATTTTG |
|
| 101 | TTACTGAAAA AATCGTCGAA CCGCAATTGG GCCCTTATCA ATCAGATTTG |
|
| 151 | TCACAAGAAG AAAAAGACAT TCGGCATTCC AATGAAATCA CGCCTTTGGA |
|
| 201 | ATATAAAGGA TTAATTTGGG CTGGCGTGGT GTTTGTTGCC TTATCCGCCC |
|
| 251 | TATTGGCTTG GAGCATCGTC CCTGCCGACG GTATTTTGCG TCATCCTGAA |
|
| 301 | ACAGGATTGG TTTCCGGTTC GCCGTTTTTA AAATCGATTG TTGTTTTTAT |
|
| 351 | TTTCTTGTTG TTTGCACTGC CGGGCATTGT TTATGGCCGG GTAACCCGAA |
|
| 401 | GTTTGCGCGG CGAACAGGAA GTCGTTAATG CGmyGGCCGA ATCGATGAGT |
|
| 451 | ACTCTGGsGC TTTmTTTGsw CAkcATCTTT TTTGCCGCAC AGTTTGTCGC |
|
| 501 | ATTTTTTAAT TGGACGAATA TTGGGCAATA TATTGCCGTT AAAGGGGCGA |
|
| 551 | CGTTCTTAAA AGAAGTCGGC TTGGGCGGCA GCGTGTTGTT TATCGGTTTT |
|
| 601 | ATTTTAATTT GTGCTTTTAT CAATCTGATG ATAGGCTCCG CCTCCGCGCA |
|
| 651 | ATGGGCGGTA ACTGCGCCGA TTTTCGTCCC TATGCTGATG TTGGCCGGCT |
|
| 701 | ACGCGCCCGA AGTCATTCAA GCCGCTTACC GCATCGGTGA TTCCGTTACC |
|
| 751 | AATATTATTA CGCCGATGAT GAGTTATTTC GGGCTGATTA TGGCGACGGT |
|
| 801 | GrkCmmmTAC AAAAAAGATG CGGGCGTGGG TaCGcTGATT wCTATGATGT |
|
| 851 | TGCCGTATTC CGCTTTCTTC TTGATTGCgT GGATTGCCTT ATTCTGCATT |
|
| 901 | TGGGTATTTg TTTTGGGCCT GCCCGTCGGT CCCGGCGCGC CCACATTCTA |
|
| 951 | TCCCGCACCT TAA |
[0496]This corresponds to the amino acid sequence <SEQ ID 134; ORF12>:
[0000]| 1 | ..AXXIIHPXXVVGPEANWFFM VASTFVIALI GYFVTEKIVEPQLGPYQSDL | |
|
| 51 | SQEEKDIRHS NEITPLEYKG LIWAGVVFVA LSALLAWSIV PADGILRHPE |
|
| 101 | TGLVSGSPFL KSIVVFIFLL FALPGIVYGR VTRSLRGEQEVVNAXAESMS |
|
| 151 | TLXLXLXXIF FAAQFVAFFN WTNIGQYIAV KGATFLKEVG LGGSVLFIGF |
|
| 201 | ILICAFINLM IGSASAQWAV TAPIFVPMLM LAGYAPEVIQ AAYRIGDSVT |
|
| 251 | NIITPMMSYF GLIMATVXXY KKDAGVGTLI XMMLPYSAFF LIAWIALFCI |
|
| 301 | WVFVLGLPVG PGAPTFYPAP * |
[0497]Further sequence analysis revealed the complete DNA sequence <SEQ ID 135> to be:
[0000]| 1 | ATGAGTCAAA CCGATACGCA ACGGGACGGA CGATTTTTAC GCACAGTCGA | |
|
| 51 | ATGGCTGGGC AATATGTTGC CGCATCCGGT TACGCTTTTT ATTATTTTCA |
|
| 101 | TTGTGTTATT GCTGATTGCC TCTGCCGTCG GTGCGTATTT CGGACTATCC |
|
| 151 | GTCCCCGATC CGCGCCCTGT TGGTGCGAAA GGACGTGCCG ATGACGGTTT |
|
| 201 | GATTTACATT GTCAGCCTGC TCAATGCCGA CGGTTTTATC AAAATCCTGA |
|
| 251 | CGCATACCGT TAAAAATTTC ACCGGTTTCG CGCCGTTGGG AACGGTGTTG |
|
| 301 | GTTTCTTTAT TGGGCGTGGG GATTGCGGAA AAATCGGGCT TGATTTCCGC |
|
| 351 | ATTAATGCGC TTATTGCTCA CAAAATCGCC ACGCAAACTC ACTACTTTTA |
|
| 401 | TGGTTGTTTT TACAGGGATT TTATCTAATA CCGCTTCTGA ATTGGGCTAT |
|
| 451 | GTCGTCCTAA TCCCTTTGTC CGCCATCATC TTTCATTCCC TCGGCCGCCA |
|
| 501 | TCCGCTTGCC GGTCTGGCTG CGGCTTTCGC CGGCGTTTCG GGCGGTTATT |
|
| 551 | CGGCCAATCT GTTCTTAGGC ACAATCGATC CGCTCTTGGC AGGCATCACC |
|
| 601 | CAACAGGCGG CGCAAATCAT CCATCCCGAC TACGTCGTAG GCCCTGAAGC |
|
| 651 | CAACTGGTTT TTTATGGTAG CCAGTACGTT TGTGATTGCT TTGATTGGTT |
|
| 701 | ATTTTGTTAC TGAAAAAATC GTCGAACCGC AATTGGGCCC TTATCAATCA |
|
| 751 | GATTTGTCAC AAGAAGAAAA AGACATTCGG CATTCCAATG AAATCACGCC |
|
| 801 | TTTGGAATAT AAAGGATTAA TTTGGGCTGG CGTGGTGTTT GTTGCCTTAT |
|
| 851 | CCGCCCTATT GGCTTGGAGC ATCGTCCCTG CCGACGGTAT TTTGCGTCAT |
|
| 901 | CCTGAAACAG GATTGGTTTC CGGTTCGCCG TTTTTAAAAT CGATTGTTGT |
|
| 951 | TTTTATTTTC TTGTTGTTTG CACTGCCGGG CATTGTTTAT GGCCGGGTAA |
|
| 1001 | CCCGAAGTTT GCGCGGCGAA CAGGAAGTCG TTAATGCGAT GGCCGAATCG |
|
| 1051 | ATGAGTACTC TGGGGCTTTA TTTGGTCATC ATCTTTTTTG CCGCACAGTT |
|
| 1101 | TGTCGCATTT TTTAATTGGA CGAATATTGG GCAATATATT GCCGTTAAAG |
|
| 1151 | GGGCGACGTT CTTAAAAGAA GTCGGCTTGG GCGGCAGCGTGTTGTTTATC |
|
| 1201 | GGTTTTATTT TAATTTGTGC TTTTATCAAT CTGATGATAG GCTCCGCCTC |
|
| 1251 | CGCGCAATGG GCGGTAACTG CGCCGATTTT CGTCCCTATG CTGATGTTGG |
|
| 1301 | CCGGCTACGC GCCCGAAGTC ATTCAAGCCG CTTACCGCAT CGGTGATTCC |
|
| 1351 | GTTACCAATA TTATTACGCC GATGATGAGT TATTTCGGGC TGATTATGGC |
|
| 1401 | GACGGTGATC AAATACAAAA AAGATGCGGG CGTGGGTACG CTGATTTCTA |
|
| 1451 | TGATGTTGCC GTATTCCGCT TTCTTCTTGA TTGCGTGGAT TGCCTTATTC |
|
| 1501 | TGCATTTGGG TATTTGTTTT GGGCCTGCCC GTCGGTCCCG GCGCGCCCAC |
|
| 1551 | ATTCTATCCC GCACCTTAA |
[0498]This corresponds to the amino acid sequence <SEQ ID 136; ORF12-1>:
[0000]| 1 | MSQTDTQRDG RFLRTVEWLG NMLPHPVTLF IIFIVLLLIA SAVGAYFGLS | |
|
| 51 | VPDPRPVGAK GRADDGLIYI VSLLNADGFI KILTHTVKNF TGFAPLGTVL |
|
| 101 | VSLLGVGIAE KSGLISALMR LLLTKSPRKL TTFMVVFTGI LSNTASELGY |
|
| 151 | VVLIPLSAII FHSLGRHPLA GLAAAFAGVS GGYSANLFLG TIDPLLAGIT |
|
| 201 | QQAAQIIHPD YVVGPEANWF FMVASTFVIA LIGYFVTEKI VEPQLGPYQS |
|
| 251 | DLSQEEKDIR HSNEITPLEY KGLIWAGVVF VALSALLAWS IVPADGILRH |
|
| 301 | PETGLVSGSP FLKSIVVFIF LLFALPGIVY GRVTRSLRGE QEVVNAMAES |
|
| 351 | MSTLGLYLVI IFFAAQFVAF FNWTNIGQYI AVKGATFLKE VGLGGSVLFI |
|
| 401 | GFILICAFIN LMIGSASAQW AVTAPIFVPM LMLAGYAPEV IQAAYRIGDS |
|
| 451 | VTNIITPMMS YFGLIMATVIKYKKDAGVGT LISMMLPYSA FFLIAWIALF |
|
| 501 | CIWVFVLGLP VGPGAPTFYP AP* |
[0499]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0500]ORF12 shows 96.3% identity over a 320aa overlap with an ORF (ORF12a) from strain A of N. meningitidis.
[0000]
[0501]The complete length ORF12a nucleotide sequence <SEQ ID 137> is:
[0000]| 1 | ATGAGTCAAA CCGATACGCA ACGGGACGGA CGATTTTTAC GCACAGTCGA | |
|
| 51 | ATGGCTGGGC AATATGTTGC CGCACCCGGT TACGCTTTTT ATTATTTTCA |
|
| 101 | TTGTGTTATT GCTGATTGCC TCTGCCGCCG GTGCGTATTT CGGACTATCC |
|
| 151 | GTCCCCGATC CGCGCCCTGT TGGTGCGAAA GGACGTGCCG ATGACGGTTT |
|
| 201 | GATTCACGTT GTCAGCCTGC TCGATGCTGA CGGTTTGATC AAAATCCTGA |
|
| 251 | CGCATACCGT TAAAAATTTC ACCGGTTTCG CGCCGTTGGG AACGGTGTTG |
|
| 301 | GTTTCTTTAT TGGGCGTGGG GATTGCGGAA AAATCGGGCT TGATTTCCGC |
|
| 351 | ATTAATGCGC TTATTGCTCA CAAAATCTCC ACGCAAACTC ACTACTTTTA |
|
| 401 | TGGTTGTTTT TACAGGGATT TTATCTAATA CCGCTTCTGA ATTGGGCTAT |
|
| 451 | GTCGTCCTAA TCCCTTTGTC CGCCATCATC TTTCATTCCC TCGGCCGCCA |
|
| 501 | TCCGCTTGCC GGTCTGGCTG CGGCTTTCGC CGGCGTTTCG GGCGGTTATT |
|
| 551 | CGGCCAATCT GTTCTTAGGC ACAATCGATC CGCTCTTGGC AGGCATCACC |
|
| 601 | CAACAGGCGG CGCAAATCAT CCATCCCGAC TACGTCGTAG GCCCTGAAGC |
|
| 651 | CAACTGGTTT TTTATGGTAG CCAGTACGTT TGTGATTGCT TTGATTGGTT |
|
| 701 | ATTTTGTTAC TGAAAAAATC GTCGAACCGC AATTGGGCCC TTATCAATCA |
|
| 751 | GATTTGTCAC AAGAAGAAAA AGACATTCGA CATTCCAATG AAATCACGCC |
|
| 801 | TTTGGAATAT AAAGGATTAA TTTGGGCTGG CGTGGTGTTT GTTGCCTTAT |
|
| 851 | CCGCCCTATT GGCTTGGAGC ATCGTCCCTG CCGACGGTAT TTTGCGTCAT |
|
| 901 | CCTGAAACAG GATTGGTTTC CGGTTCGCCG TTTTTAAAAT CAATTGTTGT |
|
| 951 | TTTTATTTTC TTGTTGTTTG CACTGCCGGG CATTGTTTAT GGCCGGGTAA |
|
| 1001 | CCCGAAGTTT GCGCGGCGAA CAGGAAGTCG TTAATGCGAT GGCCGAATCG |
|
| 1051 | ATGAGTACTC TGGGGCTTTA TTTGGTCATC ATCTTTTTTG CCGCACAGTT |
|
| 1101 | TGTCGCATTT TTTAATTGGA CGAATATTGG GCAATATATT GCCGTTAAAG |
|
| 1151 | GGGCGACGTT CTTAAAAGAA GTCGGCTTGG GCGGCAGCGT GTTGTTTATC |
|
| 1201 | GGTTTTATTT TAATTTGTGC TTTTATCAAT CTGATGATAG GCTCCGCCTC |
|
| 1251 | CGCGCAATGG GCGGTAACTG CGCCGATTTT CGTCCCTATG CTGATGTTGG |
|
| 1301 | CCGGCTACGC GCCCGAAGTC ATTCAAGCCG CTTACCGCAT CGGTGATTCC |
|
| 1351 | GTTACCAATA TTATTACGCC GATGATGAGT TATTTCGGGC TGATTATGGC |
|
| 1401 | GACGGTGATC AAATACAAAA AAGATGCGGG CGTGGGTACG CTGATTTCTA |
|
| 1451 | TGATGTTGCC GTATTCCGCT TTCTTCTTGA TTGCGTGGAT TGCCTTATTC |
|
| 1501 | TGCATTTGGG TATTTGTTTT GGGCCTGCCC GTCGGTCCCG GCGCGCCCAC |
|
| 1551 | ATTCTATCCC GCACCTTAA |
[0502]This encodes a protein having amino acid sequence <SEQ ID 138>:
[0000]| 1 | MSQTDTQRDG RFLRTVEWLG NMLPHPVTLF IIFIVLLLIA SAAGAYFGLS | |
|
| 51 | VPDPRPVGAK GRADDGLIHV VSLLDADGLI KILTHTVKNF TGFAPLGTVL |
|
| 101 | VSLLGVGIAE KSGLISALMR LLLTKSPRKL TTFMVVFTGI LSNTASELGY |
|
| 151 | VVLIPLSAII FHSLGRHPLA GLAAAFAGVS GGYSANLFLG TIDPLLAGIT |
|
| 201 | QQAAQIIHPD YVVGPEANWF FMVASTFVIA LIGYFVTEKI VEPQLGPYQS |
|
| 251 | DLSQEEKDIR HSNEITPLEY KGLIWAGVVF VALSALLAWS IVPADGILRH |
|
| 301 | PETGLVSGSP FLKSIVVFIF LLFALPGIVY GRVTRSLRGE QEVVNAMAES |
|
| 351 | MSTLGLYLVI IFFAAQFVAF FNWTNIGQYI AVKGATFLKE VGLGGSVLFI |
|
| 401 | GFILICAFIN LMIGSASAQW AVTAPIFVPM LMLAGYAPEV IQAAYRIGDS |
|
| 451 | VTNIITPMMS YFGLIMATVI KYKKDAGVGT LISMMLPYSA FFLIAWIALF |
|
| 501 | CIWVFVLGLP VGPGAPTFYP AP* |
[0503]ORF12a and ORF12-1 show 99.0% identity in 522 aa overlap:
[0000]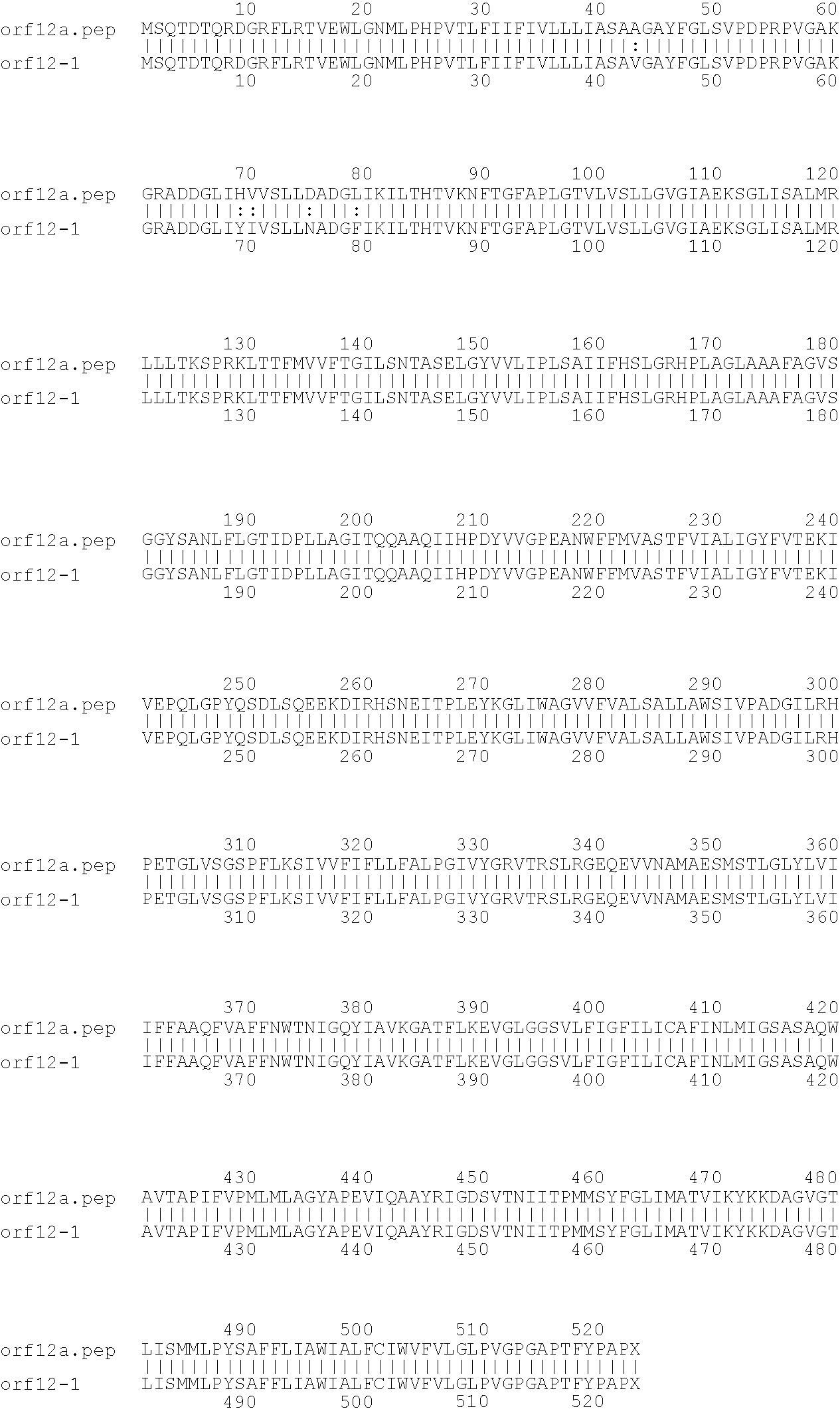
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0504]ORF12 shows 92.5% identity over a 320aa overlap with a predicted ORF (ORF12.ng) from N. gonorrhoeae.
[0000]
[0505]The complete length ORF12ng nucleotide sequence <SEQ ID 139> is:
[0000]| 1 | ATGAGTCAAA CCGACGCGCG TCGTAGCGGA CGATTTTTAC GCACAGTCGA | |
|
| 51 | ATGGCTGGGC AATATGTTGC CGCACCCGGT TACGCTTTTT ATTATTTTCA |
|
| 101 | TTGTGTTATT GCTGATTGcc tctgCCGTCG GTGCGTATTT CGGACTATCC |
|
| 151 | GTCCCCGATC CGCGTCCTGT TGGGGCGAAA GGACGTGCCG ATGACGGTTT |
|
| 201 | GATTCACGTT GTCAGCCTGC TCGATGCCGA CGGTTTGATC AAAATCCTGA |
|
| 251 | CGCATACCGT TAAAAATTTC ACCGGTTTCG CGCCGTTGGG AACGGTGTTG |
|
| 301 | GTTTCTTTAT TGGGCGTGGG GATTGCGGAA AAATCGGGCT TGATTTCCGC |
|
| 351 | ATTAATGCGC TTATTGCTCA CAAAATCCCC ACGCAAACTC ACTACTTTTA |
|
| 401 | TGGTTGTTTT TACAGGGATT TTATCCAATA CGGCTTCTGA ATTGGGCTAT |
|
| 451 | GTCGTCCTAA TCCCTTTGTC CGCCGTCATC TTTCATTCGC TCGGCCGCCA |
|
| 501 | TCCGCTTGCC GGTTTGGCTG CGGCTTTCGC CGGCGTTTCG GGCGGTTATT |
|
| 551 | CGGCCAATCT GTTCTTAGGC ACAATCGATC CGCTCTTGGC AGGCATCACC |
|
| 601 | CAACAGGCGG CGCAAATCAT CCATCCCGAC TACGTCGTAG GCCCTGAAGC |
|
| 651 | CAACTGGTTT TTTATGGCAG CCAGTACGTT TGTGATTGCT TTGATTGGTT |
|
| 701 | ATTTTGTTAC TGAAAAAATC GTCGAACCGC AATTGGGCCC TTATCAATCA |
|
| 751 | GATTTGTCAC AAGAAGAAAA AGACATTCGG CATTCCAATG AAATCACGCC |
|
| 801 | TTTGGAATAT AAAGGATTAA TTTGGGCAGG CGTGGTGTTT GTTGCCTTAT |
|
| 851 | CCGCCCTATT GGCTTGGAGC ATCGTCCCTG CCGACGGTAT TTTGCGTCAT |
|
| 901 | CCTGAAACAG GATTGGTTGC CGGTTCGCCG TTTTTAAAAT CGATTGTTGT |
|
| 951 | TTTTATTTTC TTGTTGTTTG CGCTGCCGGG CATTGTTTAT GGCCGGATAA |
|
| 1001 | CCCGAAGTTT GCGCGGCGAA CGGGAAGTCG TTAATGCGAT GGCCGAATCG |
|
| 1051 | ATGAGTACTT TGGGACTTTA TTTGGTCATC ATCTTTTTTG CCGCACAGTT |
|
| 1101 | TGTCGCATTT TTTAATTGGA CGAATATTGG GCAATATATT GCCGTTAAAG |
|
| 1151 | GGGCGGTGTT CTTAAAAGAA GTCGGCTTGG GCGGCAGTGT GTTGTTTATC |
|
| 1201 | GGTTTTATTT TAATTTGTGC TTTTATCAAT CTGATGATAG GCTCCGCCTC |
|
| 1251 | CGCGCAATGG GCGGTAACTG CGCCGATTTT CGTCCCTATG CTGATGTTGG |
|
| 1301 | CCGGCTACGC GCCCGAAGTC ATTCAAGCCG CTTACCGCAT CGGTGATTCC |
|
| 1351 | GTTACCAATA TTATTACGCC GATGATGAGT TATTTCGGGC TGATTATGGC |
|
| 1401 | GACGGTAATC AAATACAAAA AAGATGCGGG CGTAGGCACG CTGATTTCTA |
|
| 1451 | TGATGTTGCC GTATTCCGCT TTCTTCTTAA TTGCATGGAT CGCCTTATTC |
|
| 1501 | TGCATTTGGG TATTTGTTTT GGGTCTGCCC GTCGGTCCCG GCACACCCAC |
|
| 1551 | ATTCTATCCG GTGCCTTAA |
[0506]This encodes a protein having amino acid sequence <SEQ ID 140>:
[0000]| 1 | MSQTDARRSG RFLRTVEWLG NMLPHPVTLF IIFIVLLLIA SAVGAYFGLS | |
|
| 51 | VPDPRPVGAK GRADDGLIHV VSLLDADGLI KILTHTVKNF TGFAPLGTVL |
|
| 101 | VSLLGVGIAE KSGLISALMR LLLTKSPRKL TTFMVVFTGI LSNTASELGY |
|
| 151 | VVLIPLSAVI FHSLGRHPLA GLAAAFAGVS GGYSANLFLG TIDPLLAGIT |
|
| 201 | QQAAQIIHPD YVVGPEANWF FMAASTFVIA LIGYFVTEKI VEPQLGPYQS |
|
| 251 | DLSQEEKDIR HSNEITPLEY KGLIWAGVVF VALSALLAWS IVPADGILRH |
|
| 301 | PETGLVAGSP FLKSIVVFIF LLFALPGIVY GRITRSLRGE REVVNAMAES |
|
| 351 | MSTLGLYLVI IFFAAQFVAFFNWTNIGQYI AVKGAVFLKK FRLGGSVLFI |
|
| 401 | GFILICAFIN LMIGSASAQW AVTAPIFVPM LMLAGNAPQV IQAAYRIGDS |
|
| 451 | VTNIITPMMS YFGLIMATVI KYKKDAGVGT LISMMLPYSA FFLIAWIALF |
|
| 501 | CIWVFVLGLP VGPGTPTFYP VP* |
[0507]ORF12ng shows 97.1% identity in 522 aa overlap with ORF12-1:
[0000]
[0508]In addition, ORF12ng shows significant homology with a hypotehtical protein from E. coli:
[0000]| sp|P46133|YDAH_ECOLI HYPOTHETICAL 55.1 KD PROTEIN IN OGT-DBPA | |
| INTERGENIC REGION |
| >gi|1787597 (AE000231) hypothetical protein in ogt 5′region |
| [Escherichia coli] |
| Length = 510 |
| Score = 329 bits (835), Expect = 2e−89 |
| Identities = 178/507 (35%), Positives = 281/507 (55%), Gaps = 15/507 (2%) |
|
| Query: | 8 | RSGRFLRTVEWLGNMLPHPVTXXXXXXXXXXXASAVGAYFGLSVPDPRPVGAKGRADDGL | 67 | |
| | +SG+ VE +GN +PHP +A+ + FG+S +P D | |
| Sbjct: | 13 | QSGKLYGWVERIGNKVPHPFLLFIYLIIVLMVTTAILSAFGVSAKNP--------TDGTP | 64 |
|
| Query: | 68 | IHVVSLLDADGLIKILTHTVKNFTGFAPXXXXXXXXXXXXIAEKSGLISALMRLLLTKSP | 127 |
| | + V +LL +GL L + +KNF+GFAP +AE+ GL+ ALM + + | |
| Sbjct: | 65 | VVVKNLLSVEGLHWFLPNVIKNFSGFAPLGAILALVLGAGLAERVGLLPALMVKMASHVN | 124 |
|
| Query: | 128 | RKLTTFMVVFTGILSNTASELGYVVLIPLSAVIFHSLGRHPLAGLAAAFAGVSGGYSANL | 187 |
| | + ++MV+F S+ +S+ V++ P+ A+IF ++GRHP+AGL AA AGV G++ANL | |
| Sbjct: | 125 | ARYASYMVLFIAFFSHISSDAALVIMPPMGALIFLAVGRHPVAGLLAAIAGVGCGFTANL | 184 |
|
| Query: | 188 | FLGTIDPLLAGITQQAAQIIHPDYVVGPEANWFFMAASTFVIALIGYFVTEKIVEPQLGP | 247 |
| | + T D LL+GI+ +AA +P V NW+FMA+S V+ ++G +T+KI+EP+LG | |
| Sbjct: | 185 | LIVTTDVLLSGISTEAAAAFNPQMHVSVIDNWYFMASSVVVLTIVGGLITDKIIEPRLGQ | 244 |
|
| Query: | 248 | YQSDLSQEEKDIRHSNEITPLEYKGLIWAGVVFVALSALLAWSIVPADGILRHPETGLVA | 307 |
| | +Q + ++ + + S GL AGVV + A +A ++P +GILR P V | |
| Sbjct: | 245 | WQGNSDEKLQTLTESQRF------GLRIAGVVSLLFIAAIALMVIPQNGILRDPINHTVM | 298 |
|
| Query: | 308 | GSPFLKSIVVFIFLLFALPGIVYGRITRSLRGEREVVNAMAESMSTLGLYLXXXXXXXXX | 367 |
| | SPF+K IV I L F + + YG TR++R + ++ + M E M + ++ | |
| Sbjct: | 299 | PSPFIKGIVPLIILFFFVVSLAYGIATRTIRRQADLPHLMIEPMKEMAGFIVMVFPLAQF | 358 |
|
| Query: | 368 | XXXXNWTNIGQYIAVKGAVFLKEVGLGGSVLFIGFILICAFINLMIGSASAQWAVTAPIF | 427 |
| | NW+N+G++IAV L+ GL G F+G L+ +F+ +I S SA W++ APIF | |
| Sbjct: | 359 | VAMFNWSNMGKFIAVGLTDILESSGLSGIPAFVGLALLSSFLCMFIASGSAIWSILAPIF | 418 |
|
| Query: | 428 | VPMLMLAGYAPEVIQAAYRIGDSVTNIITPMMSYFGLIMATVIKYKKDAGVGTLISMMLP | 487 |
| | VPM ML G+ P Q +RI DS + P+ + L + + +YK DA +GT S++LP | |
| Sbjct: | 419 | VPMFMLLGFHPAFAQILFRIADSSVLPLAPVSPFVPLFLGFLQRYKPDAKLGTYYSLVLP | 478 |
|
| Query: | 488 | YSAFFLIAWIALFCIWVFVLGLPVGPG | 514 |
| | Y FL+ W+ + W +++GLP+GPG | |
| Sbjct: | 479 | YPLIFLVVWLLMLLAW-YLVGLPIGPG | 504 |
[0509]Based on this analysis, including the presence of several putative transmembrane domains and the predicted actinin-type actin-binding domain signature (shown in bold) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 17
[0510]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 141>:
[0000]| 1 | ..ACAGCCGGCG CAGCAGGTTn CnCGGTCTTC GTTTTCGTAA CGGACAGTCA | |
|
| 51 | GGTGGAGGTG TTCGGGAACA TCCAGACCGC AGTGGAAACA GGTTTTTTTC |
|
| 101 | ATGGCATTTC GGTTTCGTCT GTGTTTGGTG CGGCGGCACA AGACTCGGCA |
|
| 151 | ATgGCTTCGC GCAGTGCGTC TATACCGGTA TTTTCAGCAA CGGAAATGCG |
|
| 201 | GACGGcGgCA ATTTTTCCCG CAGCGTCGCG CCATATGCCC GTGTTTTgTT |
|
| 251 | CTTCAGACGG CAGCAGGTCG GTTTTGTTGT ACACCTTgAT GCACGGAaTA |
|
| 301 | TCGCCGGCAT GGATTTCTTG CAGTACGTTT TCCACGTCTT CAATCTGCTG |
|
| 351 | TCCGCTGTTC GGAGCGGCGG CATCGACGAC GTGCAGCAGC ACATCgGcTT |
|
| 401 | gCGCGGTTTC TTCCAGCGTG GCgGAAAAGG CGGAAATCAG TTTgTGCGGC |
|
| 451 | agATyGCTnA CGAATCCGAC GGTATCGGTC AGGATAATGC TGCATTCGGG |
|
| 501 | ACT.. |
[0511]This corresponds to the amino acid sequence <SEQ ID 142; ORF14>:
[0000]| 1 | ..TAGAAGXXVF VFVTDSQVEV FGNIQTAVET GFFHGISVSS VFGAAAQDSA | |
|
| 51 | MASRSASIPV FSATEMRTAA IFPAASRHMP VFCSSDGSRS VLLYTLMHGI |
|
| 101 | SPAWISCSTF STSSICCPLF GAAASTTCSS TSACAVSSSV AEKAEISLCG |
|
| 151 | RXLTNPTVSV RIMLHSG.. |
[0512]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0513]ORF14 shows 94.0% identity over a 167aa overlap with an ORF (ORF14a) from strain A of N. meningitidis.
[0000]
[0514]The complete length ORF14a nucleotide sequence <SEQ ID 143> is:
[0000]| 1 | ATGGAGGATT TGCAGGAAAT CGGGTTCGAT GTCGCCGCCG TAAAGGTAGG | |
|
| 51 | TCGGCAGCGC GAACATCATC GTCTGCATCA TCCCCAGCCC GGCAACGGCG |
|
| 101 | AGGCGGACGA TGTATTGTTT GCGTTCTTTT TGGTTGGCGG CTTCGATTTT |
|
| 151 | TTGCGCGTCA TAGGGTGCGG CGGTGTAGCC TATCTGCCTG ATTTTCAACA |
|
| 201 | GAATGTCGGA AAGGCGGATT TTGCCGTCGT CCCAGACGAC GCGGCAGCGG |
|
| 251 | TGCGTGCTGT AATTGAGGTC GATGCGGACG ATGCCGTCTG TACGCAAAAG |
|
| 301 | CTGCTGTTCG ATCAGCCAGA CGCAGGCGGC GCAGGTGATG CCGCCGAGCA |
|
| 351 | TTAAAACCGC CTCGCGCGTG CCGCCGTGGG TTTCCACAAA GTCGGACTGG |
|
| 401 | ACTTCGGGCA GGTCGTACAG GCGGATTTGG TCGAGGATTT CTTGGGGCGG |
|
| 451 | CAGCTCGGTT TTTTGCGCGT CGGCGGTGCG TTGTTTGTAA TAACTGCCCA |
|
| 501 | AGCCCGCGTC AATAATGCTT TGTGCGACTG CCTGACAACC GGCGCAGCAG |
|
| 551 | GTTTCGCGGT CTTCGTTTTC GTAACGGACG GTCAGATGCA GGTTTTCGGG |
|
| 601 | AACGTCCAGC CCGCAGTGGA AACAGGTTTT TTTCATGGCA TTTCGGTTTC |
|
| 651 | GTCTGTGTTT GGTGCGGCGG CACAATACTC GGCAATGGCT TCGCGCAGTG |
|
| 701 | CGTCTATACC GGTATTTTCA GCAACGGAAA TGCGGACGGC GGCAATTTTT |
|
| 751 | CCCGCAGCGT CGCGCCATAT GCCCGTGTTT TGTTCTTCAG ACGGCAGCAG |
|
| 801 | GTCGGTTTTG TTGTACACCT TGATGCACGG AATATCGCCG GCATGGATTT |
|
| 851 | CTTGCAGTAC GTTTTCCACG TCTTCAATCT GCTGTCCGCT GTTCGGAGCG |
|
| 901 | GCGGCATCGA CGACGTGCAG CAGCACATCG GCTTGCGCGG TTTCTTCCAG |
|
| 951 | CGTGGCGGAA AAGGCGGAAA TCAGTTTGTG CGGCAGATCG CTGACGAATC |
|
| 1001 | CGACGGTATC GGTCAGGATA ATGCTGCATT CGGGACTGAT GTACAGCCGC |
|
| 1051 | CGCGCCGTCG TGTCGAGTGT GGCGAAAAGC TGGTCTTTCG CATATATGCC |
|
| 1101 | CGACTTGGTC AGCCGGTTGA ACAGACTGGA TTTGCCGACA TTGGTATAG |
[0515]This encodes a protein having amino acid sequence <SEQ ID 144>:
[0000]| 1 | MEDLQEIGFD VAAVKVGRQR EHHRLHHPQP GNGEADDVLF AFFLVGGFDF | |
|
| 51 | LRVIGCGGVA YLPDFQQNVG KADFAVVPDD AAAVRAVIEV DADDAVCTQK |
|
| 101 | LLFDQPDAGG AGDAAEH*NR LARAAVGFHK VGLDFGQVVQ ADLVEDFLGR |
|
| 151 | QLGFLRVGGA LFVITAQARV NNALCDCLTT GAAGFAVFVF VTDGQMQVFG |
|
| 201 | NVQPAVETGF FHGISVSSVF GAAAQYSAMA SRSASIPVFS ATEMRTAAIF |
|
| 251 | PAASRHMPVF CSSDGSRSVL LYTLMHGISP AWISCSTFST SSICCPLFGA |
|
| 301 | AASTTCSSTS ACAVSSSVAE KAEISLCGRS LTNPTVSVRI MLHSGLMYSR |
|
| 351 | RAVVSSVAKS WSFAYMPDLV SRLNRLDLPT LV* |
[0516]It should be noted that this sequence includes a stop codon at position 118.
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0517]ORF14 shows 89.8% identity over a 167aa overlap with a predicted ORF (ORF14.ng) from N. gonorrhoeae:
[0000]
[0518]The complete length ORF14ng nucleotide sequence <SEQ ID 145> is predicted to encode a protein having amino acid sequence <SEQ ID 146>:
[0000]| 1 | MEDLQEIGFD VAAVKVGRQR EHHRLHHTQS GNGKADDVLF AFFLVGGFDF | |
|
| 51 | LRVIGCGGVA CLPDFQQNVG EADFAVVPDD AAAVRAVIEV DADDAVCAQK |
|
| 101 | LLFDQPDAGG AGNAAEHQHC FVRAIMGFHK VGLDFGQVVQ ADLVEDFLGR |
|
| 151 | QFGFFRVGGA SFVITAQAGI DDALCDCLTA DAAGFAVFAF VADGQMQVFG |
|
| 201 | NVQPAVETGF FHGISVSSVF GAAAQYSAMA SRSASIPVFS ATEMRTAAIF |
|
| 251 | PAASRHMPVF CSSDGSRSVL LYTLMHGISW AWISCSTFST SSICCPLFRA |
|
| 301 | AASTTCSSTS ACTVSSKVAE KAEISLCGRS LTNPTVSVRI MLHAGLMYSR |
|
| 351 | RAVVSRVAKS WSFAYMPDLV SRLNRLDLPT LV* |
[0519]Based on the putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 18
[0520]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 147>:
[0000]| 1 | ..GGCCATTACT CCGACCGCAC TTGGAAGCCG CGTTTGGNCG GCCGCCGTCT | |
|
| 51 | GCCGTATCTG CTTTATGGCA CGCTGATTGC GGTTATTGTG ATGATTTTGA |
|
| 101 | TGCCGAACTC GGGCAGCTTC GGTTTCGGCT ATGCGTCGCT GGCGGCTTTG |
|
| 151 | TCGTTCGGCG CGCTGATGAT TGCGCTGTTA GACGTGTCGT CAAATATGGC |
|
| 201 | GATGCAGCCG TTTAAGATGA TGGTCGGCGA CATGGTCAAC GAGGAGCAGA |
|
| 251 | AAA.NTACGC CTACGGGATT CAAAGTTTCT TAGCAAATAC GGGCGCGGTC |
|
| 301 | GTGGCGGCGA TTCTGCCGTT TGTGTTTGCG TATATCGGTT TGGCGAACAC |
|
| 351 | CGCCGANAAA GGCGTTGTGC CGCAGACCGT GGTCGTGGCG TTTTATGTGG |
|
| 401 | GTGCGGCGTT GCTGGTGATT ACCAGCGCGT TCACGATTTT CAAAGTGAAG |
|
| 451 | GAATACGANC CGGAAACCTA CGCCCGTTAC CACGGCATCG ATGTCGCCGC |
|
| 501 | GAATCAGGAA AAAGCCAACT GGATCGCACT CTTAAAA.CC GCGC.. |
[0521]This corresponds to the amino acid sequence <SEQ ID 148; ORF16>:
[0000]| 1 | ..GHYSDRTWKP RLXGRRLPYL LYGTLIAVIV MILMPNSGSF GFGYASLAAL | |
|
| 51 | SFGALMIALL DVSSNMAMQP FKMMVGDMVN EEQKXYAYGI QSFLANTGAV |
|
| 101 | VAAILPFVFA YIGLANTAXK GVVPQTVVVA FYVGAALLVI TSAFTIFKVK |
|
| 151 | EYXPETYARY HGIDVAANQE KANWIALLKX A.. |
[0522]Further work revealed the complete nucleotide sequence <SEQ ID 149>:
[0000]| 1 | ATGTCGGAAT ATACGCCTCA AACAGCAAAA CAAGGTTTGC CCGCGCTGGC | |
|
| 51 | AAAAAGCACG ATTTGGATGC TCAGTTTCGG CTTTCTCGGC GTTCAGACGG |
|
| 101 | CCTTTACCCT GCAAAGCTCG CAAATGAGCC GCATTTTTCA AACGCTAGGC |
|
| 151 | GCAGACCCGC ACAATTTGGG CTGGTTTTTC ATCCTGCCGC CGCTGGCGGG |
|
| 201 | GATGCTGGTG CAGCCGATTG TCGGCCATTA CTCCGACCGC ACTTGGAAGC |
|
| 251 | CGCGTTTGGG CGGCCGCCGT CTGCCGTATC TGCTTTATGG CACGCTGATT |
|
| 301 | GCGGTTATTG TGATGATTTT GATGCCGAAC TCGGGCAGCT TCGGTTTCGG |
|
| 351 | CTATGCGTCG CTGGCGGCTT TGTCGTTCGG CGCGCTGATG ATTGCGCTGT |
|
| 401 | TAGACGTGTC GTCAAATATG GCGATGCAGC CGTTTAAGAT GATGGTCGGC |
|
| 451 | GACATGGTCA ACGAGGAGCA GAAAGGCTAC GCCTACGGGA TTCAAAGTTT |
|
| 501 | CTTAGCAAAT ACGGGCGCGG TCGTGGCGGC GATTCTGCCG TTTGTGTTTG |
|
| 551 | CGTATATCGG TTTGGCGAAC ACCGCCGAGA AAGGCGTTGT GCCGCAGACC |
|
| 601 | GTGGTCGTGG CGTTTTATGT GGGTGCGGCG TTGCTGGTGA TTACCAGCGC |
|
| 651 | GTTCACGATT TTCAAAGTGA AGGAATACGA TCCGGAAACC TACGCCCGTT |
|
| 701 | ACCACGGCAT CGATGTCGCC GCGAATCAGG AAAAAGCCAA CTGGATCGAA |
|
| 751 | CTCTTGAAAA CCGCGCCTAA GGCGTTTTGG ACGGTTACTT TGGTGCAATT |
|
| 801 | CTTCTGCTGG TTCGCCTTCC AATATATGTG GACTTACTCG GCAGGCGCGA |
|
| 851 | TTGCGGAAAA CGTCTGGCAC ACCACCGATG CGTCTTCCGT AGGTTATCAG |
|
| 901 | GAGGCGGGTA ACTGGTACGG CGTTTTGGCG GCGGTGCAGT CGGTTGCGGC |
|
| 951 | GGTGATTTGT TCGTTTGTAT TGGCGAAAGT GCCGAATAAA TACCATAAGG |
|
| 1001 | CGGGTTATTT CGGCTGTTTG GCTTTGGGCG CGCTCGGCTT TTTCTCCGTT |
|
| 1051 | TTCTTCATCG GCAACCAATA CGCGCTGGTG TTGTCTTATA CCTTAATCGG |
|
| 1101 | CATCGCTTGG GCGGGCATTA TCACTTATCC GCTGACGATT GTGACCAACG |
|
| 1151 | CCTTGTCGGG CAAGCATATG GGCACTTACT TGGGCTTGTT TAACGGCTCT |
|
| 1201 | ATCTGTATGC CTCAAATCGT CGCTTCGCTG TTGAGTTTCG TGCTTTTCCC |
|
| 1251 | TATGCTGGGC GGCTTGCAGG CCACTATGTT CTTGGTAGGG GGCGTCGTCC |
|
| 1301 | TGCTGCTGGG CGCGTTTTCC GTGTTCCTGA TTAAAGAAAC ACACGGCGGG |
|
| 1351 | GTTTGA |
[0523]This corresponds to the amino acid sequence <SEQ ID 150; ORF16-1>:
[0000]| 1 | MSEYTPQTAK QGLPALAKST IWMLSFGFLG VQTAFTLQSS QMSRIFQTLG | |
|
| 51 | ADPHNLGWFF ILPPLAGMLV QPIVGHYSDR TWKPRLGGRR LPYLLYGTLI |
|
| 101 | AVIVMILMPN SGSFGFGYAS LAALSFGALM IALLDVSSNM AMQPFKMMVG |
|
| 151 | DMVNEEQKGY AYGIQSFLAN TGAVVAAILP FVFAYIGLAN TAEKGVVPQT |
|
| 201 | VVVAFYVGAA LLVITSAFTI FKVKEYDPET YARYHGIDVA ANQEKANWIE |
|
| 251 | LLKTAPKAFW TVTLVQFFCW FAFQYMWTYS AGAIAENVWH TTDASSVGYQ |
|
| 301 | EAGNWYGVLA AVQSVAAVIC SFVLAKVPNK YHKAGYFGCL ALGALGFFSV |
|
| 351 | FFIGNQYALV LSYTLIGIAW AGIITYPLTI VTNALSGKHM GTYLGLFNGS |
|
| 401 | ICMPQIVASL LSFVLFPMLG GLQATMFLVG GVVLLLGAFS VFLIKETHGG |
|
| 451 | V* |
[0524]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0525]ORF16 shows 96.7% identity over a 181aa overlap with an ORF (ORF16a) from strain A of N. meningitidis:
[0000]
[0526]The complete length ORF16a nucleotide sequence <SEQ ID 151> is:
[0000]| 1 | ATGTCGGAAT ATACGCCTCA AACAGCAAAA CAAGGTTTGC CCGCGCTGGC | |
|
| 51 | AAAAAGCACG ATTTGGATGC TCAGTTTCGG CTTTCTCGGC GTTCAGACGG |
|
| 101 | CCTTTACCCT GCAAAGCTCG CAGATGAGCC GCATCTTCCA GACGCTCGGT |
|
| 151 | GCCGATCCGC ACAGCCTCGG CTGGTTCTTT ATCCTGCCGC CGCTGGCGGG |
|
| 201 | GATGCTGGTG CAGCCGATTG TCGGCCATTA CTCCGACCGC ACTTGGAAGC |
|
| 251 | CGCGTTTGGG CGGCCGCCGT CTGCCGTATC TGCTTTATGG CACGCTGATT |
|
| 301 | GCGGTTATTG TGATGATTTT GATGCCGAAC TCGGGCAGCT TCGGTTTCGG |
|
| 351 | CTATGCGTCG CTGGCGGCTT TGTCGTTCGG CGCGCTGATG ATTGCGCTGT |
|
| 401 | TAGACGTGTC GTCAAATATG GCGATGCAGC CGTTTAAGAT GATGGTCGGC |
|
| 451 | GACATGGTCA ACGAGGAGCA GAAAGGCTAC GCCTACGGGA TTCAAAGTTT |
|
| 501 | CTTAGCGAAT ACGGGCGCGG TCGTGGCGGC GATTCTGCCG TTTGTGTTTG |
|
| 551 | CGTATATCGG TTTGGCGAAC ACCGCCGAGA AAGGCGTTGT GCCGCAGACC |
|
| 601 | GTGGTCGTGG CGTTTTATGT GGGTGCGGCG TTGCTGGTGA TTACCAGCGC |
|
| 651 | GTTCACGATT TTCAAAGTGA AGGAATACAA TCCGGAAACC TACGCCCGTT |
|
| 701 | ACCACGGCAT CGATGTCGCC GCGAATCAGG AAAAAGCCAA CTGGATCGAA |
|
| 751 | CTCTTGAAAA CCGCGCCTAA GGCGTTTTGG ACGGTTACTT TGGTGCAATT |
|
| 801 | CTTCTGCTGG TTCGCCTTCC AATATATGTG GACTTACTCG GCAGGCGCGA |
|
| 851 | TTGCGGAAAA CGTCTGGCAC ACCACCGATG CGTCTTCCGT AGGTTATCAG |
|
| 901 | GAGGCGGGTA ACTGGTACGG CGTTTTGGCG GCGGTGCAGT CGGTTGCGGC |
|
| 951 | GGTGATTTGT TCGTTTGTAT TGGCGAAAGT GCCGAATAAA TACCATAAGG |
|
| 1001 | CGGGTTATTT CGGCTGTTTG GCTTTGGGCG CGCTCGGCTT TTTCTCCGTT |
|
| 1051 | TTCTTCATCG GCAACCAATA CGCGCTGGTG TTGTCTTATA CCTTAATCGG |
|
| 1101 | CATCGCTTGG GCGGGCATTA TCACTTATCC GCTGACGATT GTGACCAACG |
|
| 1151 | CCTTGTCGGG CAAGCATATG GGCACTTACT TGGGCCTGTT TAACGGCTCT |
|
| 1201 | ATCTGTATGC CGCAAATCGT CGCTTCGCTG TTGAGTTTCG TGCTTTTCCC |
|
| 1251 | TATGCTGGGC GGCTTGCAGG CCACTATGTT CTTGGTAGGG GGCGTCGTCC |
|
| 1301 | TGCTGCTGGG CGCGTTTTCC GTGTTCCTGA TTAAAGAAAC ACACGGCGGG |
|
| 1351 | GTTTGA |
[0527]This encodes a protein having amino acid sequence <SEQ ID 152>:
[0000]| 1 | MSEYTPQTAK QGLPALAKST IWMLSFGFLG VQTAFTLQSS QMSRIFQTLG | |
|
| 51 | ADPHSLGWFF ILPPLAGMLV QPIVGHYSDR TWKPRLGGRR LPYLLYGTLI |
|
| 101 | AVIVMILMPN SGSFGFGYAS LAALSFGALM IALLDVSSNM AMQPFKMMVG |
|
| 151 | DMVNEEQKGY AYGIQSFLAN TGAVVAAILP FVFAYIGLAN TAEKGVVPQT |
|
| 201 | VVVAFYVGAA LLVITSAFTI FKVKEYNPET YARYHGIDVA ANQEKANWIE |
|
| 251 | LLKTAPKAFW TVTLVQFFCW FAFQYMWTYS AGAIAENVWH TTDASSVGYQ |
|
| 301 | EAGNWYGVLA AVQSVAAVIC SFVLAKVPNK YHKAGYFGCL ALGALGFFSV |
|
| 351 | FFIGNQYALV LSYTLIGIAW AGIITYPLTI VTNALSGKHM GTYLGLFNGS |
|
| 401 | ICMPQIVASL LSFVLFPMLG GLQATMFLVG GVVLLLGAFS VFLIKETHGG |
|
| 451 | V* |
[0528]ORF16a and ORF16-1 show 99.6% identity in 451 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0529]ORF16 shows 93.9% identity over a 181aa overlap with a predicted ORF (ORF16.ng) from N. gonorrhoeae.
[0000]
[0530]The complete length ORF16ng nucleotide sequence <SEQ ID 153> is:
[0000]| 1 | ATGATAGGGG ATCGCCGCGC CGGCAACCAT TTCGGATTTT CCAAAGCAAA | |
|
| 51 | TACTTTTCAA ATCAAAAAAA AGGATTTACT TTATGTCGGA ATATACGCCT |
|
| 101 | CAAACAGCAA AACAAGGTTT GCCCGCGCCG GCAAAAAGCA CGATTTGGAT |
|
| 151 | GTTGAGCTTC GGCTATCTCG GCGTTCAGAC GGCCTTTACC CTGCAAAGCT |
|
| 201 | CGCAGATGAG CCGCATTTTT CAAACGCTAG GCGCAGACCC GCACAATTTG |
|
| 251 | GGCTGGTTTT TCATCCTGCC GCCGCTGGCG GGGATGCTGG TTCAGCCGAT |
|
| 301 | AGTGGCTACT ACTCAGACCG CACTTGGAAG CCGCGCTTGG GCGGCCGCCG |
|
| 351 | CCTGCCGTAT CTGCTTTACG GCACGCTGAT TGCGGTCATC GTGATGATTT |
|
| 401 | TGATGCCGAA CTCGGGCAGC TTCGGTTTCG GCTATGCGTC GCTGGCGGCC |
|
| 451 | TTGTCGTTCG GCGCGCTGAT GATTGCGCTG TTGGACGTGT CGTCGAATAT |
|
| 501 | GGCGATGCAG CCGTTTAAGA TGATGGTCGG CGATATGGTC AACGAGGAGC |
|
| 551 | AGAAAAGCTA CGCCTACGGG ATTCAAAGTT TCTTAGCGAA TACGGACGCG |
|
| 601 | GTTGTGGCAG CGATTCTGCC GTTTGTGTTC GCGTATATCG GTTTGGCGAA |
|
| 651 | CACTGCCGAG AAAGGCGTTG TGCCACAAAC CGTGGTCGTA GCATTCTATG |
|
| 701 | TGGGTGCGGC GTTACTGATT ATTACCAGTG CGTTCACAAT CTCCAAAGTC |
|
| 751 | AAAGAATACG ACCCGGAAAC CTACGCCCGT TACCACGGCA TCGATGTCGC |
|
| 801 | CGCGAATCAG GAAAAAGCCA ACTGGTTCGA ACTCTTAAAA ACCGCGCCTA |
|
| 851 | AAGTGTTTTG GACGGTTACT CCGGTACAGT TTTTCTGCTG GTTCGCCTTC |
|
| 901 | CGGTATATGT GGACTTACTC GGCAGGCGCG ATTGCAGAAA ACGTCTGGCA |
|
| 951 | CACTACCGAT GCGTCTTCCG TAGGCCATCA GGAGGCGGGC AACCGGTACG |
|
| 1001 | GCGTTTTGGC GGCGGTGTAG |
[0531]This encodes a protein having amino acid sequence <SEQ ID 154>:
[0000]| 1 | MIGDRRAGNH FGFSKANTFQ IKKKDLLYVG IYASNSKTRF ARAGKKHDLD | |
|
| 51 | VELRLSRRSD GLYPAKLADE PHFSNARRRP AQFGLVFHPA AAGGDAGSAD |
|
| 101 | SGYYSDRTWK PRLGGRRLPY LLYGTLIAVI VMILMPNSGS FIGFGYASLAA |
|
| 151 | LSFGALMIAL LDVSSNMAMQ PFKMMVGDMV NEEQKSYAYG IQSFLANTDA |
|
| 201 | VVAAILPFVF AYIGLANTAE KGVVPQTVVV AFYVGAALLI ITSAFTISKV |
|
| 251 | KEYDPETYAR YHGIDVAANQ EKANWFELLK TAPKVFWTVT PVQFFCWFAF |
|
| 301 | RYMWTYSAGA IAENVWHTTD ASSVGHQEAG NRYGVLAAV* |
[0532]ORF16ng and ORF16-1 show 89.3% identity in 261 aa overlap:
[0000]
[0533]Based on this analysis, including the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 19
[0534]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 155>:
[0000]| 1 | ATGTTGTTCC GTAAAACGAC CGCCGCCGTT TTGGCGCATA CCTTGATGCT | |
|
| 51 | GAACGGCTGT ACGTTGATGT TGTGGGGAAT GAACAACCCG GTCAGCGAAA |
|
| 101 | CAATCACCCG NAAACACGTT GNCAAAGACC AAATCCGNGN CTTCGGTGTG |
|
| 151 | GTTGCCGAAG ACAATGCCCA ATTGGAAAAG GGCAGCCTGG TGATGATGGG |
|
| 201 | CGGAAAATAC TGGTTCGTCG TCAATCCCGA AGATTCGGCG AA.NTGACGG |
|
| 251 | GNATTTTGAN GGCAGGGCTG GACAAACCCT TCCAAATAGT TNAGGATACC |
|
| 301 | CCGAGCTATG C.TGCCACCA AGCCCTGCCG GTCAAACTCG GATCGNCTGG |
|
| 351 | CAGCCAGAAT... |
[0535]This corresponds to the amino acid sequence <SEQ ID 156; ORF28>:
[0000]| 1 | MLFRKTTAAV LAHTLMLNGC TLMLWGMNNP VSETITRKHV XKDQIRXFGV | |
|
| 51 | VAEDNAQLEK GSLVMMGGKY WFVVNPEDSA XXTGILXAGL DKPFQIVXDT |
|
| 101 | PSYXCHQALP VKLGSXGSQN... |
[0536]Further work revealed the complete nucleotide sequence <SEQ ID 157>:
[0000]| 1 | ATGTTGTTCC GTAAAACGAC CGCCGCCGTT TTGGCGGCAA CCTTGATGCT | |
|
| 51 | GAACGGCTGT ACGTTGATGT TGTGGGGAAT GAACAACCCG GTCAGCGAAA |
|
| 101 | CAATCACCCG CAAACACGTT GACAAAGACC AAATCCGCGC CTTCGGTGTG |
|
| 151 | GTTGCCGAAG ACAATGCCCA ATTGGAAAAG GGCAGCCTGG TGATGATGGG |
|
| 201 | CGGAAAATAC TGGTTCGTCG TCAATCCCGA AGATTCGGCG AAGCTGACGG |
|
| 251 | GCATTTTGAA GGCAGGGCTG GACAAACCCT TCCAAATAGT TGAGGATACC |
|
| 301 | CCGAGCTATG CTCGCCACCA AGCCCTGCCG GTCAAACTCG AATCGCCTGG |
|
| 351 | CAGCCAGAAT TTCAGTACCG AAGGCCTTTG CCTGCGCTAC GATACCGACA |
|
| 401 | AGCCTGCCGA CATCGCCAAG CTGAAACAGC TCGGGTTTGA AGCGGTCAAA |
|
| 451 | CTCGACAATC GGACCATTTA CACGCGCTGC GTATCCGCCA AAGGCAAATA |
|
| 501 | CTACGCCACA CCGCAAAAAC TGAACGCCGA TTACCATTTT GAGCAAAGTG |
|
| 551 | TGCCTGCCGA TATTTATTAC ACGGTTACTG AAGAACATAC CGACAAATCC |
|
| 601 | AAGCTGTTTG CAAATATCTT ATATACGCCC CCCTTTTTGA TACTGGATGC |
|
| 651 | GGCGGGCGCG GTACTGGCCT TGCCTGCGGC GGCTCTGGGT GCGGTCGTGG |
|
| 701 | ATGCCGCCCG CAAATGA |
[0537]This corresponds to the amino acid sequence <SEQ ID 158; ORF28-1>:
[0000]| 1 | MLFRKTTAAV LAATLMLNGC TLMLWGMNNP VSETITRKHV DKDQIRAFGV | |
|
| 51 | VAEDNAQLEK GSLVMMGGKY WFVVNPEDSA KLTGILKAGL DKPFQIVEDT |
|
| 101 | PSYARHQALP VKLESPGSQN FSTEGLCLRY DTDKPADIAK LKQLGFEAVK |
|
| 151 | LDNRTIYTRC VSAKGKYYAT PQKLNADYHF EQSVPADIYY TVTEEHTDKS |
|
| 201 | KLFANILYTP PFLILDAAGA VLALPAAALG AVVDAARK* |
[0538]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0539]ORF28 shows 79.2% identity over a 120aa overlap with an ORF (ORF28a) from strain A of N. meningitidis:
[0000]
[0540]The complete length ORF28a nucleotide sequence <SEQ ID 159> is:
[0000]| 1 | ATGTTGTTCC GTAAAACGAC CGCCGCCGTT TTGGCGGCAA CCTTGATGTT | |
|
| 51 | GAACGGCTGT ACGGTAATGA TGTGGGGTAT GAACAGCCCG TTCAGCGAAA |
|
| 101 | CGACCGCCCG CAAACACGTT GACAAGGACC AAATCCGCGC CTTCGGTGTG |
|
| 151 | GTTGCCGAAG ACAATGCCCA ATTGGAAAAG GGCAGCCTGG TGATGATGGG |
|
| 201 | CGGGAAATAC TGGTTCGTCG TCAATCCTGA AGATTCGGCG AAGCTGACGG |
|
| 251 | GCATTTTGAA GGCCGGGTTG GACAAGCAGT TTCAAATGGT TGAGCCCAAC |
|
| 301 | CCGCGCTTTG CCTACCAAGC CCTGCCGGTC AAACTCGAAT CGCCCGCCAG |
|
| 351 | CCAGAATTTC AGTACCGAAG GCCTTTGCCT GCGCTACGAT ACCGACAGAC |
|
| 401 | CTGCCGACAT CGCCAAGCTG AAACAGCTTG AGTTTGAAGC GGTCGAACTC |
|
| 451 | GACAATCGGA CCATTTACAC GCGCTGCGTC TCCGCCAAAG GCAAATACTA |
|
| 501 | CGCCACACCG CAAAAACTGA ACGCCGATTA TCATTTTGAG CAAAGTGTGC |
|
| 551 | CTGCCGATAT TTATTACACG GTTACGAAAA AACATACCGA CAAATCCAAG |
|
| 601 | TTGTTTGAAA ATATTGCATA TACGCCCACC ACGTTGATAC TGGATGCGGT |
|
| 651 | GGGCGCGGTG CTGGCCTTGC CTGTCGCGGC GTTGATTGCA GCCACGAATT |
|
| 701 | CCTCAGACAA ATGA |
[0541]This encodes a protein having amino acid sequence <SEQ ID 160>:
[0000]| 1 | MLFRKTTAAV LAATLMLNGC TVMMWGMNSP FSETTARKHV DKDQIRAFGV | |
|
| 51 | VAEDNAQLEK GSLVMMGGKY WFVVNPEDSA KLTGILKAGL DKQFQMVEPN |
|
| 101 | PRFAYQALPV KLESPASQNF STEGLCLRYD TDRPADIAKL KQLEFEAVEL |
|
| 151 | DNRTIYTRCV SAKGKYYATP QKLNADYHFE QSVPADIYYT VTKKHTDKSK |
|
| 201 | LFENIAYTPT TLILDAVGAV LALPVAALIA ATNSSDK* |
[0542]ORF28a and ORF28-1 show 86.1% identity in 238 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0543]ORF28 shows 84.2% identity over a 120aa overlap with a predicted ORF (ORF28.ng) from N. gonorrhoeae:
[0000]
[0544]The complete length ORF28ng nucleotide sequence <SEQ ID 161> is
[0000]| 1 | ATGTTGTTCC GTAAAACGAC CGCCGCCGTT TTGGCGGCAA CCTTGATACT | |
|
| 51 | GAACGGCTGT ACGATGATGT TGCGGGGGAT GAACAACCCG GTCAGCCAAA |
|
| 101 | CAATCACCCG CAAACACGTT GACAAAGACC AAATCCGCGC CTTCGGTGTG |
|
| 151 | GTTGCCGAAG ACAATGCCCA ATTGGAAAAG GGCAGCCTGG TGATGATGGG |
|
| 201 | CGGGAAATAC TGGTTCGCCG TCAATCCCGA AGATTCGGCG AAGCTGACGG |
|
| 251 | GCCTTTTGAA GGCCGGGTTG GACAAGCCCT TCCAAATAGT TGAGGATACC |
|
| 301 | CCGAGCTATG CCCGCCACCA AGCCCTGCCG GTCAAATTCG AAGCGCCCGG |
|
| 351 | CAGCCAGAAT TTCAGTACCG GAGGTCTTTG CCTGCGCTAT GATACCGGCA |
|
| 401 | GACCTGACGA CATCGCCAAG CTGAAACAGC TTGAGTTTAA AGCGGTCAAA |
|
| 451 | CTCGACAATC GGACCATTTA CACGCGCTGC GTATCCGCCA AAGGCAAATA |
|
| 501 | CTACGCCACG CCGCAAAAAC TGAACGCCGA TTATCATTTT GAGCAAAGTG |
|
| 551 | TGCCCGCCGA TATTTATTAT ACGGTTACTG AAAAACATAC CGACAAATCC |
|
| 601 | AAGCTGTTTG GAAATATCTT ATATACGCCC CCCTTGTTGA TATTGGATGC |
|
| 651 | GGCGGCCGCG GTGCTGGTCT TGCCTATGGC TCTGATTGCA GCCGCGAATT |
|
| 701 | CCTCAGACAA ATGA |
[0545]This encodes a protein having amino acid sequence <SEQ ID 162>:
[0000]| 1 | MLFRKTTAAV LAATLILNGC TMMLRGMNNP VSQTITRKHV DKDQIRAFGV | |
|
| 51 | VAEDNAQLEK GSLVMMGGKY WFAVNPEDSA KLTGLLKAGL DKPFQIVEDT |
|
| 101 | PSYARHQALP VKFEAPGSQN FSTGGLCLRY DTGRPDDIAK LKQLEFKAVK |
|
| 151 | LDNRTIYTRC VSAKGKYYAT PQKLNADYHF EQSVPADIYY TVTEKHTDKS |
|
| 201 | KLFGNILYTP PLLILDAAAA VLVLPMALIA AANSSDK* |
[0546]ORF28ng and ORF28-1 share 90.0% identity in 231 aa overlap:
[0000]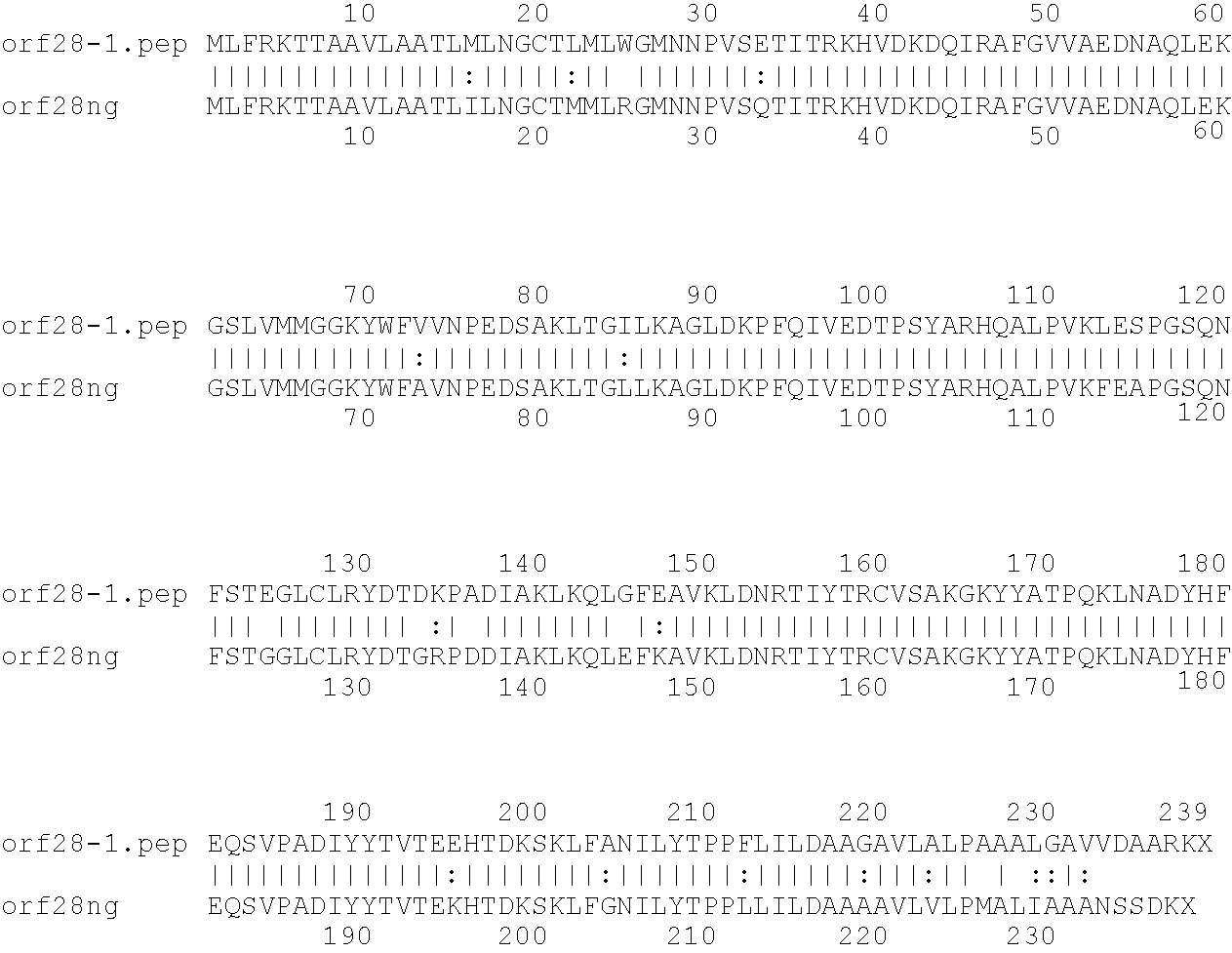
[0547]Based on this analysis, including the presence of a putative transmembrane domain in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0548]ORF28-1 (24 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 6A shows the results of affinity purification of the GST-fusion protein, and FIG. 6B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for ELISA, which gave a positive result. These experiments confirm that ORF28-1 is a surface-exposed protein, and that it may be a useful immunogen.
Example 20
[0549]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 163>:
[0000]| 1 | ..GTCAGTCCTG TACTGCCTAT TACACACGAA CGGACAGGGT TTGAAGGTGT | |
|
| 51 | TATCGGTTAT GAAACCCATT TTTCAGGGCA CGGACATGAA GTACACAGTC |
|
| 101 | CGTTCGATCA TCATGATTCA AAAAGCACTT CTGATTTCAG CGGCGGTGTA |
|
| 151 | GACGGCGGTT TTACTGTTTA CCAACTTCAT CGAACATGGT CGGAAATCCA |
|
| 201 | TCCGGAGGAT GAATATGACG GGCCGCAAGC AGCG.ATTAT CCGCCCCCCG |
|
| 251 | GAGGAGCAAG GGATATATAC AGCTATTATG TCAAAGGAAC TTCAACAAAA |
|
| 301 | ACAAAGACTA GTATTGTCCC TCAAGCCCCA TTTTCAGACC GTTGGCTAGA |
|
| 351 | AGAAAATGCC GGTGCCGCCT CTGGT.. |
[0550]This corresponds to the amino acid sequence <SEQ ID 164; ORF29>:
[0000]| 1 | ..VSPVLPITHE RTGFEGVIGY ETHFSGHGHE VHSPFDHHDS KSTSDFSGGV | |
|
| 51 | DGGFTVYQLH RTWSEIHPED EYDGPQAAXY PPPGGARDIY SYYVKGTSTK |
|
| 101 | TKTSIVPQAP FSDRWLEENA GAASG.. |
[0551]Further work revealed the complete nucleotide sequence <SEQ ID 165>:
[0000]| 1 | ATGAATTTGC CTATTCAAAA ATTCATGATG CTGTTTGCAG CAGCAATATC | |
|
| 51 | GTTGCTGCAA ATCCCCATTA GTCATGCGAA CGGTTTGGAT GCCCGTTTGC |
|
| 101 | GCGATGATAT GCAGGCAAAA CACTACGAAC CGGGTGGTAA ATACCATCTG |
|
| 151 | TTTGGTAATG CTCGCGGCAG TGTTAAAAAG CGGGTTTACG CCGTCCAGAC |
|
| 201 | ATTTGATGCA ACTGCGGTCA GTCCTGTACT GCCTATTACA CACGAACGGA |
|
| 251 | CAGGGTTTGA AGGTGTTATC GGTTATGAAA CCCATTTTTC AGGGCACGGA |
|
| 301 | CATGAAGTAC ACAGTCCGTT CGATCATCAT GATTCAAAAA GCACTTCTGA |
|
| 351 | TTTCAGCGGC GGTGTAGACG GCGGTTTTAC TGTTTACCAA CTTCATCGAA |
|
| 401 | CAGGGTCGGA AATCCATCCG GAGGATGGAT ATGACGGGCC GCAAGGCAGC |
|
| 451 | GATTATCCGC CCCCCGGAGG AGCAAGGGAT ATATACAGCT ATTATGTCAA |
|
| 501 | AGGAACTTCA ACAAAAACAA AGACTAATAT TGTCCCTCAA GCCCCATTTT |
|
| 551 | CAGACCGTTG GCTAAAAGAA AATGCCGGTG CCGCCTCTGG TTTTTTCAGC |
|
| 601 | CGTGCGGATG AAGCAGGAAA ACTGATATGG GAAAGCGACC CCAATAAAAA |
|
| 651 | TTGGTGGGCT AACCGTATGG ATGATGTTCG CGGCATCGTC CAAGGTGCGG |
|
| 701 | TTAATCCTTT TTTAATGGGT TTTCAAGGAG TAGGGATTGG GGCAATTACA |
|
| 751 | GACAGTGCAG TAAGCCCGGT CACAGATACA GCCGCGCAGC AGACTCTACA |
|
| 801 | AGGTATTAAT GATTTAGGAA AATTAAGTCC GGAAGCACAA CTTGCTGCCG |
|
| 851 | CGAGCCTATT ACAGGACAGT GCTTTTGCGG TAAAAGACGG TATCAACTCT |
|
| 901 | GCCAAACAAT GGGCTGATGC CCATCCAAAT ATAACAGCTA CTGCCCAAAC |
|
| 951 | TGCCCTTTCC GCAGCAGAGG CCGCAGGTAC GGTTTGGAGA GGTAAAAAAG |
|
| 1001 | TAGAACTTAA CCCGACTAAA TGGGATTGGG TTAAAAATAC CGGTTATAAA |
|
| 1051 | AAACCTGCTG CCCGCCATAT GCAGACTTTA GATGGGGAGA TGGCAGGTGG |
|
| 1101 | GAATAAACCT ATTAAATCTT TACCAAACAG TGCCGCTGAA AAAAGAAAAC |
|
| 1151 | AAAATTTTGA GAAGTTTAAT AGTAACTGGA GTTCAGCAAG TTTTGATTCA |
|
| 1201 | GTGCACAAAA CACTAACTCC CAATGCACCT GGTATTTTAA GTCCTGATAA |
|
| 1251 | AGTTAAAACT CGATACACTA GTTTAGATGG AAAAATTACA ATTATAAAAG |
|
| 1301 | ATAACGAAAA CAACTATTTT AGAATCCATG ATAATTCACG AAAACAGTAT |
|
| 1351 | CTTGATTCAA ATGGTAATGC TGTGAAAACC GGTAATTTAC AAGGTAAGCA |
|
| 1401 | AGCAAAAGAT TATTTACAAC AACAAACTCA TATCAGGAAC TTAGACAAAT |
|
| 1451 | GA |
[0552]This corresponds to the amino acid sequence <SEQ ID 166; ORF29-1>:
[0000]| 1 | MNLPIQKFMM LFAAAISLLQ IPISHANGLD ARLRDDMQAK HYEPGGKYHL | |
|
| 51 | FGNARGSVKK RVYAVQTFDA TAVSPVLPIT HERTGFEGVI GYETHFSGHG |
|
| 101 | HEVHSPFDHH DSKSTSDFSG GVDGGFTVYQ LHRTGSEIHP EDGYDGPQGS |
|
| 151 | DYPPPGGARD IYSYYVKGTS TKTKTNIVPQ APFSDRWLKE NAGAASGFFS |
|
| 201 | RADEAGKLIW ESDPNKNWWA NRMDDVRGIV QGAVNPFLMG FQGVGIGAIT |
|
| 251 | DSAVSPVTDT AAQQTLQGIN DLGKLSPEAQ LAAASLLQDS AFAVKDGINS |
|
| 301 | AKQWADAHPN ITATAQTALS AAEAAGTVWR GKKVELNPTK WDWVKNTGYK |
|
| 351 | KPAARHMQTL DGEMAGGNKP IKSLPNSAAE KRKQNFEKFN SNWSSASFDS |
|
| 401 | VHKTLTPNAP GILSPDKVKT RYTSLDGKIT IIKDNENNYF RIHDNSRKQY |
|
| 451 | LDSNGNAVKT GNLQGKQAKD YLQQQTHIRN LDK* |
[0553]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0554]ORF29 shows 88.0% identity over a 125aa overlap with an ORF (ORF29a) from strain A of N. meningitidis.
[0000]
[0555]The complete length ORF29a nucleotide sequence <SEQ ID 167> is:
[0000]| 1 | ATGAATTNGC CTATTCAAAA ATTCATGATG CTGTTTGCAG CAGCAATATC | |
|
| 51 | GTNGCTGCAA ATCCCNATTA GTCATGCGAA CGGTTTGGAT GCCCGTTTGC |
|
| 101 | GCGATGATAT GCAGGCAAAA CACTACGAAC CGGGTGGTAA ATACCATCTG |
|
| 151 | TTTGGTAATG CTCGCGGCAG TGTTAAAAAT CGGGTTTACG CCGTCCAAAC |
|
| 201 | ATTTGATGCA ACTGCGGTCG GCCCCATACT GCCTATTACA CACGAACGGA |
|
| 251 | CAGGATTTGA AGGCATTATC GGTTATGAAA CCCATTTTTC AGGACATGGA |
|
| 301 | CATGAAGTAC ACAGTCCGTT CGATAATCAT GATTCAAAAA GCACTTCTGA |
|
| 351 | TTTCAGCGGC GGCGTAGACG GTGGTTTTAC CGTTTACCAA CTTCATCGGA |
|
| 401 | CAGGGTCGGA AATCCATCCG GAGGATGGAT ATGACGGGCC GCAAGGCAGC |
|
| 451 | GATTATCCGC CCCCCGGAGG AGCAAGGGAT ATATACANNT ANTATGTCAA |
|
| 501 | AGGAACTTCA ACAAAAACAA AGAGTAATAT TGTTCCCCGA GCCCCATTTT |
|
| 551 | CAGACCGCTG GCTAAAAGAA AATGCCGGTG CCGCCTCTGG TTTTTTCAGC |
|
| 601 | CGTGCTGATG AAGCAGGAAA ACTGATATGG GAAAGCGACC CCAATAAAAA |
|
| 651 | TTGGTGGGCT AACCGTATGG ATGATATTCG CGGCATCGTC CAAGGTGCGG |
|
| 701 | TTAATCCTTT TTTAATGGGT TTTCAAGGAG TAGGGATTGG GGCAATTACA |
|
| 751 | GACAGTGCAG TAAGCCCGGT CACAGATACA GCCGCGCAGC AGACTCTACA |
|
| 801 | AGGTATNAAT CATTTAGGAA ANTTAAGTCC CGAAGCACAA CTTGCGGCTG |
|
| 851 | CAACCGCATT ACAAGACAGT GCTTTTGCGG TAAAAGACGG TATCAATTCC |
|
| 901 | GCCAGACAAT GGGCTGATGC CCATCCGAAT ATAACTGCAA CAGCCCAAAC |
|
| 951 | TGCCCTTGCC GTAGCAGANG CCGCAACTAC GGTTTGGGGC GGTAAAAAAG |
|
| 1001 | TAGAACTTAA CCCGACCAAA TGGGATTGGG TTAAAAATAC NGGCTATAAN |
|
| 1051 | ACACCTGCTG TTCGCACCAT GCATACTTTG GATGGGGAAA TGGCCGGTGG |
|
| 1101 | GAATAGACCG CCTAAATCTA TAACGTCCAA CAGCAAAGCA GATGCTTCCA |
|
| 1151 | CACAACCGTC TTTACAAGCG CAACTAATTG GAGAACAAAT TANNNNNGGG |
|
| 1201 | CATGCTTATA ACAAGCATGT CATAAGACAA CAAGAATTTA CGGATTTAAA |
|
| 1251 | TATCAATTCA CCAGCAGATT TTGCTCGGCA TATTGAAAAT ATTGTTAGCC |
|
| 1301 | ATCCANCAAA TATGAAAGAG TTACCTCGCG GTAGAACTGC GTATTGGGAT |
|
| 1351 | NATAAAACAG GGACNATAGT TATCCGAGAT AAAAATTCTG ACGATGGAGG |
|
| 1401 | TACAGCATTT AGACCAACAT CAGGTAAAAA ATATTATGAT GATTTATAG |
[0556]This encodes a protein having amino acid sequence <SEQ ID 168>:
[0000]| 1 | MNXPIQKFMM LFAAAISXLQ IPISHANGLD ARLRDDMQAK HYEPGGKYHL | |
|
| 51 | FGNARGSVKN RVYAVQTFDA TAVGPILPIT HERTGFEGII GYETHFSGHG |
|
| 101 | HEVHSPFDNH DSKSTSDFSG GVDGGFTVYQ LHRTGSEIHP EDGYDGPQGS |
|
| 151 | DYPPPGGARD IYXXYVKGTS TKTKSNIVPR APFSDRWLKE NAGAASGFFS |
|
| 201 | RADEAGKLIW ESDPNKNWWA NRMDDIRGIV QGAVNPFLMG FQGVGIGAIT |
|
| 251 | DSAVSPVTDT AAQQTLQGXN HLGXLSPEAQ LAAATALQDS AFAVKDGINS |
|
| 301 | ARQWADAHPN ITATAQTALA VAXAATTVWG GKKVELNPTK WDWVKNTGYX |
|
| 351 | TPAVRTMHTL DGEMAGGNRP PKSITSNSKA DASTQPSLQA QLIGEQIXXG |
|
| 401 | HAYNKHVIRQ QEFTDLNINS PADFARHIEN IVSHPXNMKE LPRGRTAYWD |
|
| 451 | XKTGTIVIRD KNSDDGGTAF RPTSGKKYYD DL* |
[0557]ORF29a and ORF29-1 show 90.1% identity in 385 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0558]ORF29 shows 88.8% identity over a 125aa overlap with a predicted ORF (ORF29.ng) from N. gonorrhoeae:
[0000]
[0559]The complete length ORF29ng nucleotide sequence <SEQ ID 169> is predicted to encode a protein having amino acid sequence <SEQ ID 170>:
[0000]| 1 | MNLPIQKFMM LFAAAISLLQ IPISHANGLD ARLRDDMQAK HYEPGGKYHL | |
|
| 51 | FGNARGSVKN RVCAVQTFDA TAVGPILPIT HERTGFEGVI GYETHFSGHG |
|
| 101 | HEVHSPFDNH DSKSTSDFSG GVDGGFTVYQ LHRTGSEIHP EDGYDGPQGG |
|
| 151 | GYPPPGGARD IYSYHIKGTS TKTKINTVPQ APFSDRWLKE NAGAASGFLS |
|
| 201 | RADEAGKLIW ENDPDKNWRA NRMDDIRGIV QGAVNPFLTG FQGLGVGAIT |
|
| 251 | DSAVSPVTYA AARKTLQGIH NLGNLSPEAQ LAAATALQDS AFAVKDSINS |
|
| 301 | ARQWADAHPN ITATAQTALA VTEAATTVWG GKKVELNPAK WDWVKNTGYK |
|
| 351 | KPAARHMQTV DGEMAGGNKP LESKNTVTTN NFFENTGYTE KVLRQASNGD |
|
| 401 | YHGFPQSVDA FSENGTVIQI VGGDNIVRHK LYIPGSYKGK DGNFEYIREA |
|
| 451 | DGKINHRLFV PNQQLPEK* |
[0560]In a second experiment, the following DNA sequence <SEQ ID 171> was identified:
[0000]| 1 | atgAATTTGC CTATTCAAAA ATTCATGATG ctgttggcAg cggcaatatc | |
|
| 51 | gatgctGCat ATCCCCATTA GTCATGCGAA CGGTTTGGAT GCCCGTTTGC |
|
| 101 | GCGATGATAT GCAGGCAAAA CACTACGAAC CGGGTGGCAA ATACCATCTG |
|
| 151 | TTTGGTAATG CTCGCGGCAG TGTTAAAAAT CGGGTTTGCG CCGTCCAAAC |
|
| 201 | ATTTGATGCA ACTGCGGTCG GCCCCATACT GCCTATTACA CACGAACGGA |
|
| 251 | CAGGATTTGA AGGTGTTATC GGCTATGAAA CCCATTTTTC AGGACACGGA |
|
| 301 | CACGAAGTAC ACAGTCCGTT CGATAATCAT GATTCAAAAA GCACTTCTGA |
|
| 351 | TTTCAGCGGC GGCGTAGACG GCGGTTTTAC CGTTTACCAA CTTCATCGGA |
|
| 401 | CAGGGTCGGA AATACATCCC GCAGACGGAT ATGACGGGCC TCAAGGCGGC |
|
| 451 | GGTTATCCGG AACCACAAGG GGCAAGGGAT ATATACAGCT ACCATATCAA |
|
| 501 | AGGAACTTCA ACCAAAACAA AGATAAACAC TGTTCCGCAA GCCCCTTTTT |
|
| 551 | CAGACCGCTG GCTAAAAGAA AATGCCGGTG CCGCTTCCGG TTTTCTCAGC |
|
| 601 | CGTGCGGATG AAGCAGGAAA ACTGATATGG GAAAACGACC CCGATAAAAA |
|
| 651 | TTGGCGGGCT AACCGTATGG ATGATATTCG CGGCATCGTC CAAGGTGCGG |
|
| 701 | TTAATCCTTT TTTAACGGGT TTTCAAGGGG TAGGGATTGG GGCAATTACA |
|
| 751 | GACAGTGCGG TAAGCCCGGT CACAGATACA GCCGCTCAGC AGACTCTACA |
|
| 801 | AGGTATTAAT GATTTAGGAA ATTTAAGTCC GGAAGCACAA CTTGCCGCCG |
|
| 851 | CGAGCCTATT ACAGGACAGT GCCTTTGCGG TAAAAGACGG CATCAATTCC |
|
| 901 | GCCAGACAAT GGGCTGATGC CCATCCGAAT ATAACAGCAA CAGCCCAAAC |
|
| 951 | TGCCCTTGCC GTAGCAGAGG CCGCAGGTAC GGTTTGGCGC GGTAAAAAAG |
|
| 1001 | TAGAACTTAA CCCGACCAAA TGGGATTGGG TTAAAAATAC CGGCTATAAA |
|
| 1051 | AAACCTGCTG CCCGCCATAT GCAGACTGTA GATGGGGAGA TGGCAGGGGG |
|
| 1101 | GAATAGACCG CCTAAATCTA TAACGTCGGA AGGAAAAGCT AATGCTGCAA |
|
| 1151 | CCTATCCTAA GTTGGTTAAT CAGCTAAATG AGCAAAACTT AAATAACATT |
|
| 1201 | GCGGCTCAAG ATCCAAGATT GAGTCTAGCT ATTCATGAGG GTAAAAAAAA |
|
| 1251 | TTTTCCAATA GGAACTGCAA CTTATGAAGA GGCAGATAGA CTAGGTAAAA |
|
| 1301 | TTTGGGTTGG TGAGGGTGCA AGACAAACTA GTGGAGGCGG ATGGTTAAGT |
|
| 1351 | AGAGATGGCA CTCGACAATA TCGGCCACCA ACAGAAAAAA AATCACAATT |
|
| 1401 | TGCAACTACA GGTATTCAAG CAAATTTTGA AACTTATACT ATTGATTCAA |
|
| 1451 | ATGAAAAAAG AAATAAAATT AAAAATGGAC ATTTAAATAT TAGGTAA |
[0561]This encodes a protein having amino acid sequence <SEQ ID 172; ORF29ng-1>:
[0000]| 1 | MNLPIQKFMM LLAAAISMLH IPISHANGLD ARLRDDMQAK HYEPGGKYHL | |
|
| 51 | FGNARGSVKN RVCAVQTFDA TAVGPILPIT HERTGFEGVI GYETHFSGHG |
|
| 101 | HEVHSPFDNH DSKSTSDFSG GVDGGFTVYQ LHRTGSEIHP ADGYDGPQGG |
|
| 151 | GYPEPQGARD IYSYHIKGTS TKTKINTVPQ APFSDRWLKE NAGAASGFLS |
|
| 201 | RADEAGKLIW ENDPDKNWRA NRMDDIRGIV QGAVNPFLTG FQGVGIGAIT |
|
| 251 | DSAVSPVTDT AAQQTLQGIN DLGNLSPEAQ LAAASLLQDS AFAVKDGINS |
|
| 301 | ARQWADAHPN ITATAQTALA VAEAAGTVWR GKKVELNPTK WDWVKNTGYK |
|
| 351 | KPAARHMQTV DGEMAGGNRP PKSITSEGKA NAATYPKLVN QLNEQNLNNI |
|
| 401 | AAQDPRLSLA IHEGKKNFPI GTATYEEADR LGKIWVGEGA RQTSGGGWLS |
|
| 451 | RDGTRQYRPP TEKKSQFATT GIQANFETYT IDSNEKRNKI KNGHLNIR* |
[0562]ORF29ng-1 and ORF29-1 show 86.0% identity in 401 aa overlap:
[0000]
[0563]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 21
[0564]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 173>:
[0000]| 1 | ATGAAAAAAC AAATCACCGC AGCCGTAATG ATGCTGTCTA TGATTGCCCC | |
|
| 51 | CGCAATGGCA AACGGCTTGG ACAATCAGGC ATTTGAAGAC CAAATGTTCC |
|
| 101 | ACACGCGGGC AGATGCACCG ATGCAG... |
[0565]This corresponds to the amino acid sequence <SEQ ID 174; ORF30>:
[0000]| 1 | MKKQITAAVM MLSMIAPAMA NGLDNQAFED QMFHTRADAP MQ.. |
[0566]Further work revealed the complete nucleotide sequence <SEQ ID 175>:
[0000]| 1 | ATGAAAAAAC AAATCACCGC AGCCGTAATG ATGCTGTCTA TGATTGCCCC | |
|
| 51 | CGCAATGGCA AACGGCTTGG ACAATCAGGC ATTTGAAGAC CAAGTGTTCC |
|
| 101 | ACACGCGGGC AGATGCACCG ATGCAGTTGG CGGAGCTTTC TCAAAAGGAG |
|
| 151 | ATGAAGGAGA CAGAGGGGGC GTTTCTTCCA TTGGCTATCT TGGGTGGTGC |
|
| 201 | TGCCATTGGT ATGTGGACAC AGCATGGTTT TAGTTATGCA ACGACAGGCA |
|
| 251 | GACCAGCTTC TGTTAGAGAT GTTGCTATTG CTGGCGGATT AGGCGCAATT |
|
| 301 | CCTGGTGGTG TAGGCGCCGC AGGAAAGGTT GTTTCCTTTG CTAAATATGG |
|
| 351 | ACGTGAGATT AAAATCGGCA ATAATATGCG GATAGCCCCT TTCGGTAATA |
|
| 401 | GAACAGGTCA TCCTATTGGA AAATTTCCCC ATTATCATCG TCGAGTTACG |
|
| 451 | GATAATACGG GCAAGACTTT GCCTGGACAG GGAATTGGTC GTCATCGCCC |
|
| 501 | TTGGGAATCA AAATCTACGG ACAGATCATG GAAAAACCGC TTCTAA |
[0567]This corresponds to the amino acid sequence <SEQ ID 176; ORF30-1>:
[0000]| 1 | MKKQITAAVM MLSMIAPAMA NGLDNQAFED QVFHTRADAP MQLAELSQKE | |
|
| 51 | MKETEGAFLP LAILGGAAIG MWTQHGFSYA TTGRPASVRD VAIAGGLGAI |
|
| 101 | PGGVGAAGKV VSFAKYGREI KIGNNMRIAP FGNRTGHPIG KFPHYHRRVT |
|
| 151 | DNTGKTLPGQ GIGRHRPWES KSTDRSWKNR F* |
[0568]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0569]ORF30 shows 97.6% identity over a 42aa overlap with an ORF (ORF30a) from strain A of N. meningitidis.
[0000]
[0570]The complete length ORF30a nucleotide sequence <SEQ ID 177> is:
[0000]| 1 | ATGAAAAAAC AAATCACCGC AGCCGTAATG ATGCTGTCTA TGATTGCCCC | |
|
| 51 | CGCAATGGCA AACGGCTTGG ACAATCAGGC ATTTGAAGAC CAAGTGTTCC |
|
| 101 | ACACGCGGGC AGATGCACCG ATGCAGTTGG CGGAGCTTTC TCAAAAGGAG |
|
| 151 | ATGAAGGANA CAGNGGGGGC GTTTCTTCCA TTGGNTATCT TGGGTGGTGC |
|
| 201 | TGCCATTGGT ATGTGGACAC AGCATGGTTT TAGTTATGCA ACGACAGGCA |
|
| 251 | GACCAGCTTC TGTTAGAGAT GTTGCTATTG CTGGCGGATT AGGCGCAATT |
|
| 301 | CCTGGTGNTG TAGGCGCCGC AGGAAAGGTT GTTTCCTTTG CTAAATATGG |
|
| 351 | ACGTGAGATT AAAATCGGCA ATAATATGCG GATAGCCCCT TTCGGTAATA |
|
| 401 | GAACAGGTCA TCCTATTGGN AAATTTCCCC ATTATCATCG TCGAGTTACG |
|
| 451 | GATAATACGG GCAAGACTTT GCCTGGACAG GGAATTGGTC GTCATCGCCC |
|
| 501 | TTGGGAATCA AAATCTACGG ACAGATCATG GAAAAACCGC TTCTAA |
[0571]This encodes a protein having amino acid sequence <SEQ ID 178>:
[0000]| 1 | MKKQITAAVM MLSMIAPAMA NGLDNQAFED QVFHTRADAP MQLAELSQKE | |
|
| 51 | MKXTXGAFLP LXILGGAAIG MWTQHGFSYA TTGRPASVRD VAIAGGLGAI |
|
| 101 | PGXVGAAGKV VSFAKYGREI KIGNNMRIAP FGNRTGHPIG KFPHYHRRVT |
|
| 151 | DNTGKTLPGQ GIGRHRPWES KSTDRSWKNR F* |
[0572]ORF30a and ORF30-1 show 97.8% identity in 181 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0573]ORF30 shows 97.6% identity over a 42aa overlap with a predicted ORF (ORF30.ng) from N. gonorrhoeae.
[0000]
[0574]The complete length ORF30ng nucleotide sequence <SEQ ID 179> is
[0000]| 1 | ATGAAAAAAC AAATCACCGC AGCCGTAATG ATGCTGTCTA TGATCGCCCC | |
|
| 51 | CGCAATGGCA AACGGATTGG ACAATCAGGC ATTTGAAGAC CAAGTGTTCC |
|
| 101 | ACACGCGGGC AGATGCGCCG ATGCAGTTGG CGGAGCTTTC TCAGAAGGAG |
|
| 151 | ATGAAGGAGA CTGAAGGGGC TTTTCTTCCA TTGGCTATCT TGGGTGGTGC |
|
| 201 | TGCCATTGGT ATGTGGACAC AGCATGGTTT TAGTTATGCA ACGACAGGCA |
|
| 251 | GACCAGCTTC TGTTAGAGAT GTTGCTGGCG GATTAGGCGC AATTCCTGGT |
|
| 301 | GATGTAGGTG CTGCAGGAAA GGTTGTTTCC TTTGCTAAAT ATGGACGTGA |
|
| 351 | GATTAAAATC GGCAATAATA TGCGGATAGC CCCTTTCGGT AATAGAACAG |
|
| 401 | GTCATCCTAT TGGAAAATTT CCCCATTATC ATCGTCGAGT TACGGATAAT |
|
| 451 | ACGGGCAAGA CTTTGCCTGG ACAGGGAATT GGTCGTCATC GCCCTTGGGA |
|
| 501 | ATCAAAATCT ACGGACAGAT CATGGAAAAA CCGCTTCTAA |
[0575]This encodes a protein having amino acid sequence <SEQ ID 180>:
[0000]| 1 | MKKQITAAVM MLSMIAPAMA NGLDNQAFED QVFHTRADAP MQLAELSQKE | |
|
| 51 | MKETEGAFLP LAILGGAAIG MWTQHGFSYA TTGRPASVRD VAGGLGAIPG |
|
| 101 | DVGAAGKVVS FAKYGREIKI GNNMRIAPFG NRTGHPIGKF PHYHRRVTDN |
|
| 151 | TGKTLPGQGI GRHRPWESKS TDRSWKNRF* |
[0576]ORF30ng and ORF30-1 show 98.3% identity in 181 aa overlap:
[0000]
[0577]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 22
[0578]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 181>:
[0000]| 1 | ATGAATAAAA CTCTCTATCG TGTAATTTTC AACCGCAAAC GTGGGGCTGT | |
|
| 51 | GrTAGCCGTT GCTGAAACTA CCAAGCGCGA AGGTAAAAGC TGTGCCGATA |
|
| 101 | GTGATTCAGG CAGCGCTCAT GTGAAATCTG TTCCTTTTGG TACTACTCAT |
|
| 151 | GCACCTGTTT GTg.CGTTaC AAATATCTTT TCTTTTTCTT TATTGGGCTT |
|
| 201 | TTCTTTATGT TTGGCTGTAG GtacGGyCAA TATTGCTTTT GCTGATGGCA |
|
| 251 | TT.. |
[0579]This corresponds to the amino acid sequence <SEQ ID 182; ORF31>:
[0000]| 1 | MNKTLYRVIF NRKRGAVXAV AETTKREGKS CADSDSGSAH VKSVPFGTTH | |
|
| 51 | APVCXVTNIF SFSLLGFSLC LAVGTXNIAF ADGI.. |
[0580]Further work revealed a further partial nucleotide sequence <SEQ ID 183>:
[0000]| 1 | ATGAATAAAA CTCTCTATCG TGTAATTTTC AACCGCAAAC GTGGGGCTGT | |
|
| 51 | GGTAGCCGTT GCTGAAACTA CCAAGCGCGA AGGTAAAAGC TGTGCCGATA |
|
| 101 | GTGATTCAGG CAGCGCTCAT GTGAAATCTG TTCCTTTTGG TACTACTCAT |
|
| 151 | GCACCTGTTT GTCGTTCAAA TATCTTTTCT TTTTCTTTAT TGGGCTTTTC |
|
| 201 | TTTATGTTTG GCTGTAGGTA CGGCCAATAT TGCTTTTGCT GATGGCATT.. |
[0581]This corresponds to the amino acid sequence <SEQ ID 184; ORF31-1>:
[0000]| 1 | MNKTLYRVIF NRKRGAVVAV AETTKREGKS CADSDSGSAH VKSVPFGTTH | |
|
| 51 | APVCRSNIFS FSLLGFSLCL AVGTANIAFA DGI.. |
[0582]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0583]ORF31 shows 76.2% identity over a 84aa overlap with a predicted ORF (ORF31.ng) from N. gonorrhoeae:
[0000]
[0584]The complete length ORF31ng nucleotide sequence <SEQ ID 185> is:
[0000]| 1 | ATGAACAAAA CCCTCTATCG TGTGATTTTC AACCGCAAAC GCGGTGCTGT | |
|
| 51 | GGTAGCTGTT GCCGAAACCA CCAAGCGCGA AGGTAAAAGC TGTGCCGATA |
|
| 101 | GTGGTTCGGG CAGCGTTTAT GTGAAATCCG TTTCTTTCAT TCCTACTCAT |
|
| 151 | TCCAAAGCCT TTTGTTTTTC TGCATTAGGC TTTTCTTTAT GTTTGGCTTT |
|
| 201 | GGGTACGGTC AATATTGCTT TTGCTGACGG CATTATTACT GATAAAGCTG |
|
| 251 | CTCCTAAAAC CCAACAAGCC ACGATTCTGC AAACAGGTaa cGGCATACCG |
|
| 301 | CAAGTCAATA TTCAAACCCC TACTTCGGCA GGGGTTTCTG TTAATCAATA |
|
| 351 | TGCCCAGTTT GATGTGGGTA ATCGCGGGGC GATTTTAAAC AACAGTCGCA |
|
| 401 | GCAACACCCA AACACAGCTA GGCGGTTGGA TTCAAGGCAA TCCTTGGTTG |
|
| 451 | ACAAGGGGCG AAGCACGTGT GGTTGTAAAC CAAATCAACA GCAGCCATCC |
|
| 501 | TTCACAACTG AATGGCTATA TTGAAGTGGG TGGACGACGT GCAGAAGTCG |
|
| 551 | TTATTGCCAA TCCGGCAGGG ATTGCAGTCA ATGGTGGTGG TTTTATCAAT |
|
| 601 | GCTTCCCGTG CCACTTTGAC GACAGGCCAA CCGCAATATC AAGCAGGAGA |
|
| 651 | CTTTAGCGGC TTTAAGATAA GGCAAGGCAA TGCTGTAATC GCCGGACACG |
|
| 701 | GTTTGGATGC CCGTGATACC GATTTCACAC GTATTCTTGT ATGCCAACAA |
|
| 751 | AATCACCTTG ATCAGTACGG CCGAACAAGC AGGCATTCGT AA |
[0585]This encodes a protein having amino acid sequence <SEQ ID 186>:
[0000]| 1 | MNKTLYRVIF NRKRGAVVAV AETTKREGKS CADSGSGSVY VKSVSFIPTH | |
|
| 51 | SKAFCFSALG FSLCLALGTV NIAFADGIIT DKAAPKTQQA TILQTGNGIP |
|
| 101 | QVNIQTPTSA GVSVNQYAQF DVGNRGAILN NSRSNTQTQL GGWIQGNPWL |
|
| 151 | TRGEARVVVN QINSSHPSQL NGYIEVGGRR AEVVIANPAG IAVNGGGFIN |
|
| 201 | ASRATLTTGQ PQYQAGDFSG FKIRQGNAVI AGHGLDARDT DFTRILVCQQ |
|
| 251 | NHLDQYGRTS RHS* |
[0586]This gonococcal protein shares 50% identity over a 149aa overlap with the pore-forming hemolysins-like HecA protein from Erwinia chrysanthemi (accession number L39897):
[0000]| orf31ng | 96 | GNGIPQVNIQTPTSAGVSVNQYAQFDVGNRGAILNNSRSN-TQTQLGGWIQGNPWLTRGE | 154 | |
| | GNG+P VNI TP ++G+S N+Y F+V NRG ILNN + T +QLGG IQ NP L |
| HecA | 45 | GNGVPVVNIATPDASGLSHNRYHDFNVDNRGLILNNGTARLTPSQLGGLIQNNPNLNGRA | 104 |
|
| Orf31ng | 155 | ARVVVNQINSSHPSQLNGYIEVGGRRAEVVIANPAGIAVNGGGFINASRATLTTGQPQYQ | 214 |
| | A ++N++ S + S+L GY+EV G+ A VV+ANP GI +G GF+N R TLTTG PQ+ |
| HecA | 105 | AAAILNEVVSPNRSRLAGYLEVAGQAANVVVANPYGITCSGCGFLNTPRLTLTTGTPQFD | 164 |
|
| Orf31ng | 215 | -AGDFSGFKIRQGNAVIAGHGLDARDTDF | 242 |
| | AG SG +R G+ +I G GLDA +D+ |
| HecA | 165 | AAGGLSGLDVRGGDILIDGAGLDASRSDY | 193 |
[0587]Furthermore, ORF31ng and ORF31-1 show 79.5% identity in 83 aa overlap:
[0000]
[0588]On this basis, including the homology with hemolysins, and also with adhesins, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 23
[0589]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 187>:
[0000]| 1 | ATGAATACTC CTCCTTTTGT CTGTTGGATT TTTTGCAAGG TCATCGACAA | |
|
| 51 | TTTCGGCGAC ATCGGCGTTT CGTGGCGGCT CGCCCGTGTT TTGCACCGCG |
|
| 101 | AACTCGGTTG GCAGGTGCAT TTGTGGACGG ACGATGTGTC CGCCTTGCGT |
|
| 151 | GCGCTTTGCC CTGATTTGCC CGATGTTCCC TGCGTTCATC AGGATATTCA |
|
| 201 | TGTCCGCACT TGGCATTCCG ATGCGGCAGA TATTGATACC GCG.. |
[0590]This corresponds to the amino acid sequence <SEQ ID 188; ORF32>:
[0000]| 1 | MNTPPFVCWI FCKVIDNFGD IGVSWRLARV LHRELGWQVH LWTDDVSALR | |
|
| 51 | ALCPDLPDVP CVHQDIHVRT WHSDAADIDT A.. |
[0591]Further work revealed the complete nucleotide sequence <SEQ ID 189>:
[0000]| 1 | ATGAATACTC CTCCTTTTGT CTGTTGGATT TTTTGCAAGG TCATCGACAA | |
|
| 51 | TTTCGGCGAC ATCGGCGTTT CGTGGCGGCT CGCCCGTGTT TTGCACCGCG |
|
| 101 | AACTCGGTTG GCAGGTGCAT TTGTGGACGG ACGATGTGTC CGCCTTGCGT |
|
| 151 | GCGCTTTGCC CTGATTTGCC CGATGTTCCC TGCGTTCATC AGGATATTCA |
|
| 201 | TGTCCGCACT TGGCATTCCG ATGCGGCAGA TATTGATACC GCGCCTGTTC |
|
| 251 | CCGATGTCGT CATCGAAACT TTTGCCTGCG ACCTGCCCGA AAATGTGCTG |
|
| 301 | CACATTATCC GCCGACACAA GCCGCTTTGG CTGAATTGGG AATATTTGAG |
|
| 351 | CGCGGAGGAA AGCAATGAAA GGCTGCATCT GATGCCTTCG CCGCAGGAGG |
|
| 401 | GTGTTCAAAA ATATTTTTGG TTTATGGGTT TCAGCGAAAA AAGCGGCGGG |
|
| 451 | TTGATACGCG AACGTGATTA CTGCGAAGCC GTCCGTTTCG ATACTGAAGC |
|
| 501 | CCTGCGAGAG CGGCTGATGC TGCCCGAAAA AAACGCCTCC GAATGGCTGC |
|
| 551 | TTTTCGGCTA TCGGAGCGAT GTTTGGGCAA AGTGGCTGGA AATGTGGCGA |
|
| 601 | CAGGCAGGCA GCCCGATGAC ACTGTTGCTG GCGGGGACGC AAATCATCGA |
|
| 651 | CAGCCTCAAA CAAAGCGGCG TTATTCCGCA AGATGCCCTG CAAAACGACG |
|
| 701 | GCGATGTTTT TCAGACGGCA TCCGTCCGCC TCGTCAAAAT CCCTTTCGTG |
|
| 751 | CCGCAACAGG ACTTCGACCA ACTGCTGCAC CTTGCCGACT GCGCCGTCAT |
|
| 801 | CCGCGGCGAA GACAGTTTCG TGCGCGCCCA GCTTGCGGGC AAACCCTTCT |
|
| 851 | TTTGGCACAT CTACCCGCAA GACGAGAATG TCCATCTCGA CAAACTCCAC |
|
| 901 | GCCTTTTGGG ATAAGGCACA CGGTTTCTAC ACGCCCGAAA CCGTGTCGGC |
|
| 951 | ACACCGCCGT CTTTCGGACG ACCTCAACGG CGGAGAGGCT TTATCCGCAA |
|
| 1001 | CACAACGCCT CGAATGTTGG CAAACCCTGC AACAACATCA AAACGGCTGG |
|
| 1051 | CGGCAAGGCG CGGAGGATTG GAGCCGTTAT CTTTTCGGGC AGCCGTCAGC |
|
| 1101 | TCCTGAAAAA CTCGCTGCCT TTGTTTCAAA GCATCAAAAA ATACGCTAG |
[0592]This corresponds to the amino acid sequence <SEQ ID 190; ORF32-1>:
[0000]| 1 | MNTPPFVCWI FCKVIDNFGD IGVSWRLARV LHRELGWQVH LWTDDVSALR | |
|
| 51 | ALCPDLPDVP CVHQDIHVRT WHSDAADIDT APVPDVVIET FACDLPENVL |
|
| 101 | HIIRRHKPLW LNWEYLSAEE SNERLHLMPS PQEGVQKYFW FMGFSEKSGG |
|
| 151 | LIRERDYCEA VRFDTEALRE RLMLPEKNAS EWLLFGYRSD VWAKWLEMWR |
|
| 201 | QAGSPMTLLL AGTQIIDSLK QSGVIPQDAL QNDGDVFQTA SVRLVKIPFV |
|
| 251 | PQQDFDQLLH LADCAVIRGE DSFVRAQLAG KPFFWHIYPQ DENVHLDKLH |
|
| 301 | AFWDKAHGFY TPETVSAHRR LSDDLNGGEA LSATQRLECW QTLQQHQNGW |
|
| 351 | RQGAEDWSRY LFGQPSAPEK LAAFVSKHQK IR*w |
[0593]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0594]ORF32 shows 93.8% identity over a 81aa overlap with an ORF (ORF32a) from strain A of N. meningitidis.
[0000]
[0595]The complete length ORF32a nucleotide sequence <SEQ ID 191> is:
[0000]| 1 | ATGAATACTC CTCCTTTTTC TGCTGGANTT TTTTGCAAGG TCATCGACAA | |
|
| 51 | TTTCGGCGAC ATCGGCGTTT CGTGGCGGCT TGCCCGTGTT TTGCACCGCG |
|
| 101 | AACTCGGTTG GCAGGTGCAT TTGTGGACGG ACGATGTGTC CGCCTTGCGT |
|
| 151 | GCGCTTTGCC CTGATTTGCC CGATGTTCNC TGCGTTCATC AGGATATTCA |
|
| 201 | TGTCCGCACT TGGCATTCCG ATGCGGCAGA TATTGATACC GCGCCTGTTC |
|
| 251 | NCGATGTCGT CATCGAAACT TTTGCCTGCG ACCTGCCCGA AAATGTGCTG |
|
| 301 | CACATCATCC GCCGACACAA GCCGCTTTGG CTGAANTGGG AATATTTGAG |
|
| 351 | CGCGGAGGAN AGCAATGAAA GGCTGCACNT GATGCCTTCG CCGCAGGAGA |
|
| 401 | GTGTTCNAAA ATANTTTTGG TTTATGGGTT TCAGCGAANN NAGCGGCGGA |
|
| 451 | CTGATACGCG AACGCGATTA CTGCGAAGCC GTCCGTTTCG ATAGCGGAGC |
|
| 501 | CTTGCGCAAG AGGCTGATGC TTCCCGAAAA AAACGNCCCC GAATGGCTGC |
|
| 551 | TTTTCGGCTA TCGGAGCGAT GTTTGGGCAA AGTGGCTGGA AATGTGGCGA |
|
| 601 | CAGGCAGGCA GTCCGTTGAC ACTTTTGCTG GCNGGGGCGC ANATTATCGA |
|
| 651 | CAGCCTCAAA CAAAACGGCG TTATTCCGCA AGATGCCCTG CAAAACGACG |
|
| 701 | GCGATGTTTT TCAGACGGCA TCCGTCCGCC TCGTCAAAAT CCCTTTCGTG |
|
| 751 | CCGCAACAGG ACTTCGACAA ACTGCTGCAC CTTGCCGACT GCGCCGTCAT |
|
| 801 | CCGCGGCGAA GACAGTTTCG TGCGCGCCCA GCTTGCGGGC AAACCCTTCT |
|
| 851 | TTTGGCACAT CTACCCGCAA GATGAGAATG TCCATCTCGA CAAACTCCAC |
|
| 901 | GCCTTTTGGG ATAAGGCACA CGGTTTCTAC ACGCCCGAAA CCGCATCGGC |
|
| 951 | ACACCGCCGC CTTTCAGACG ACCTCAACGG CGGAGAGGCT TTATCCGCAA |
|
| 1001 | CACAACGCCT CGAATGTTGG CAAATCCTGC AACAACATCA AAACGGCTGG |
|
| 1051 | CGGCAAGGCG CGGAGGATTG GAGCCGTTAT CTTTTTGGGC AGCCTTCCGC |
|
| 1101 | ATCCGAAAAA CTCGCCGCCT TTGTTTCAAA GCATCAAAAA ATACGCTAG |
[0596]This encodes a protein having amino acid sequence <SEQ ID 192>:
[0000]| 1 | MNTPPFSAGX FCKVIDNFGD IGVSWRLARV LHRELGWQVH LWTDDVSALR | |
|
| 51 | ALCPDLPDVX CVHQDIHVRT WHSDAADIDT APVXDVVIET FACDLPENVL |
|
| 101 | HIIRRHKPLW LXWEYLSAEX SNERLHXMPS PQESVXKXFW FMGFSEXSGG |
|
| 151 | LIRERDYCEA VRFDSGALRK RLMLPEKNXP EWLLFGYRSD VWAKWLEMWR |
|
| 201 | QAGSPLTLLL AGAXIIDSLK QNGVIPQDAL QNDGDVFQTA SVRLVKIPFV |
|
| 251 | PQQDFDKLLH LADCAVIRGE DSFVRAQLAG KPFFWHIYPQ DENVHLDKLH |
|
| 301 | AFWDKAHGFY TPETASAHRR LSDDLNGGEA LSATQRLECW QILQQHQNGW |
|
| 351 | RQGAEDWSRY LFGQPSASEK LAAFVSKHQK IR* |
[0597]ORF32a and ORF32-1 show 93.2% identity in 382 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0598]ORF32 shows 95.1% identity over a 82aa overlap with a predicted ORF (ORF32.ng) from N. gonorrhoeae:
[0000]
[0599]An ORF32ng nucleotide sequence <SEQ ID 193> was predicted to encode a protein having amino acid sequence <SEQ ID 194>:
[0000]| 1 | MVMNTYAFPV CWIFCKVIDN FGDIGVSWRL ARVLHRELGW QVHLWTDDVS | |
|
| 51 | ALRALCPDLP DVPFVHQDIH VRTWHSDAAD IDTAPVPDAV IETFACDLPE |
|
| 101 | NVLNIIRRHK PLWLNWEYLS AEESNERLHL MPSPQEGVQK YFWFMGFSEK |
|
| 151 | SGGLIRERDY REAVRFDTEA LRRRLVLPEK NAPEWLLFGY RGDVWAKWLD |
|
| 201 | MWQQAGSLMT LLLAGAQIID SLKQSGVIPQ NALQNEGGVF QTASVRLVKI |
|
| 251 | PFVPQQDFDK LLHLADCAVI RGEDSFVRTQ LAGKPFFWHI YPQDENVHLD |
|
| 301 | KLHAFWDKAY GFYTPETASV HRLLSDDLNG GEALSATQRL ECGVL* |
[0600]Further sequencing revealed the following DNA sequence <SEQ ID 195>:
[0000]| 1 | ATGAATACAT ACGCTTTTCC TGTCTGTTGG ATTTTTTGCA AGGTCATCGA | |
|
| 51 | CAATTTCGGC GACATCGGCG TTTCGTGGCG GCTCGCCCGT GTTTTGCACC |
|
| 101 | GCGAACTCGG TTGGCAGGTG CATTTGTGGA CGGACGACGT GTCCGCCTTG |
|
| 151 | CGCGCGCTTT GTCCCGATTT GCCCGATGTT CCCTTCGTTC ATCAGGATAT |
|
| 201 | TCATGTCCGC ACTTGGCATT CCGATGCGGC AGACATTGAT ACCGCGCCCG |
|
| 251 | TTCCCGATGC CGTTATCGAA ACTTTTGCCT GCGACCTGCC CGAAAATGTG |
|
| 301 | CTGAACATCA TCCGCCGACA CAAACCGCTT TGGCTGAATT GGGAATATTT |
|
| 351 | GAGCGCGGAG GAAAGCAATG AAAGGCTGCA CCTGATGCCT TCGCCGCAGG |
|
| 401 | AGGGCGTTCA AAAATATTTT TGGTTTATGG GTTTCAGCGA AAAAAGCGGC |
|
| 451 | GGGTTGATAC GCGAACGCGA TTACCGCGAA GCCGTCCGTT TCGATACCGA |
|
| 501 | AGCCCTGCGC CGGCGGCTGG TGCTGCCCGA AAAAAACGCC CCCGAATGGC |
|
| 551 | TGCTTTTCGG CTATCGGGGC GATGTTTGGG CAAAGTGGCT GGACATGTGG |
|
| 601 | CAACAGGCAG GCAGCCTGAT GACCCTACTG CTGGCGGGGG CGCAAATTAT |
|
| 651 | CGACAGCCTC AAACAAAGCG GCGTTATTCC GCAAAACGCC CTGCAAAAtg |
|
| 701 | aaggcgGTGT CTTTCagacG gcatccgTcC gccttGTCAA AAtcCCGTTC |
|
| 751 | GTGCcGCAAC AGGAcTTCGA CAAATTGCTG CAcctcgcCG ACTGCGCCGT |
|
| 801 | GATACGCGGC GAAGACAGTT TCGTGCGTAC CCAGCTTGCC GGAAAACCCT |
|
| 851 | TTTTTTGGCA CATCTACCCG CAAGACGAGA ATGTCCATCT CGACAAACTC |
|
| 901 | CACGCCTTTT GGGATAAGGC ATACGGCTTC TACACGCCCG AAACCGCATC |
|
| 951 | GGTGCACCGC CTCCTTTCGG ACGACCTCAA CGGCGGAGAG GCTTTATCCG |
|
| 1001 | CAACACAACG CCTCGAATGT TGGCAAACCC TGCAACAACA TCAAAACGGC |
|
| 1051 | TGGCGGCAAG GCGCGGAGGA TTGGAGCCGT TATCTTTTCG GGCAGCCTTC |
|
| 1101 | CGCATCCGAA AAACTCGCCG CCTTTGTTTC AAAGCATCAA AAAATACGCT |
|
| 1151 | AG |
[0601]This encodes a protein having amino acid sequence <SEQ ID 196; ORF32ng-1>:
[0000]| 1 | MNTYAFPVCW IFCKVIDNFG DIGVSWRLAR VLHRELGWQV HLWTDDVSAL | |
|
| 51 | RALCPDLPDV PFVHQDIHVR TWHSDAADID TAPVPDAVIE TFACDLPENV |
|
| 101 | LNIIRRHKPL WLNWEYLSAE ESNERLHLMP SPQEGVQKYF WFMGFSEKSG |
|
| 151 | GLIRERDYRE AVRFDTEALR RRLVLPEKNA PEWLLFGYRG DVWAKWLDMW |
|
| 201 | QQAGSLMTLL LAGAQIIDSL KQSGVIPQNA LQNEGGVFQT ASVRLVKIPF |
|
| 251 | VPQQDFDKLL HLADCAVIRG EDSFVRTQLA GKPFFWHIYP QDENVHLDKL |
|
| 301 | HAFWDKAYGF YTPETASVHR LLSDDLNGGE ALSATQRLEC WQTLQQHQNG |
|
| 351 | WRQGAEDWSR YLFGQPSASE KLAAFVSKHQ KIR* |
[0602]ORF32ng-1 and ORF32-1 show 93.5% identity in 383 aa overlap:
[0000]
[0603]On this basis, including the RGD sequence in the gonococcal protein, characteristic of adhesins, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0604]ORF32-1 (42 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 7A shows the results of affinity purification of the His-fusion protein, and FIG. 7B shows the results of expression of the GST-fusion in E. coli. Purified His-fusion protein was used to immunise mice, whose sera were used for ELISA, giving a positive result. These experiments confirm that ORF32-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 24
[0605]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 197>:
[0000]| 1 | ..TTGTTCCTGC GTGTNAAAGT GGGGCGTTTT TTCAGCAGTC CGGCGACGTG | |
|
| 51 | GTTTCGGGNC AAAGACCCTG TAAATCAGGC GGTGTTGCGG CTGTATNCGG |
|
| 101 | ACGAGTGGCG GCA.ACTTCG GTACGTTGGA AAATAGNCGC AACGTCGCAC |
|
| 151 | AGCCTGTGGC TCTGCACGCT GCTCGGAATG CTGGTGTCGG TATTGTTGCT |
|
| 201 | GCTTTTGGTG CGGCAATATA CGTTCAACTG GGAAAGCACG CTGTTGAGCA |
|
| 251 | ATGCCGCTTC GGTACGCGCG GTGGAAATGT TGGCATGGCT GCCGTCGAAA |
|
| 301 | CTCGGTTTCC CTGTCCCCGA TGCGCGGTCG GTCATCGAAG GCCGTCTGAA |
|
| 351 | CGGCAATATT GCCGATGCGC GGGCTTGGTC GGGGCTGCTG GTCGNCAGTA |
|
| 401 | TCGCCTGCTA NGGCATCCTG CCGCGCCTG.. |
[0606]This corresponds to the amino acid sequence <SEQ ID 198; ORF33>:
[0000]| 1 | ..LFLRVKVGRF FSSPATWFRX KDPVNQAVLR LYXDEWRXTS VRWKIXATSH | |
|
| 51 | SLWLCTLLGM LVSVLLLLLV RQYTFNWEST LLSNAASVRA VEMLAWLPSK |
|
| 101 | LGFPVPDARS VIEGRLNGNI ADARAWSGLL VXSIACXGIL PRL.. |
[0607]Further work revealed the complete nucleotide sequence <SEQ ID 199>:
[0000]| 1 | ATGTTGAATC CATCCCGAAA ACTGGTTGAG CTGGTCCGTA TTTTGGACGA | |
|
| 51 | AGGCGGTTTT ATTTTCAGCG GCGATCCCGT ACAGGCGACG GAGGCTTTGC |
|
| 101 | GCCGCGTGGA CGGCAGTACG GAGGAAAAAA TCATCCGTCG GGCGGAGATG |
|
| 151 | ATTGACAGGA ACCGTATGCT GCGGGAGACG TTGGAACGTG TGCGTGCGGG |
|
| 201 | GTCGTTCTGG TTGTGGGTGG TGGCGGCGAC GTTTGCATTT TTTACCGGTT |
|
| 251 | TTTCAGTCAC TTATCTTCTA ATGGACAATC AGGGTCTGAA TTTCTTTTTG |
|
| 301 | GTTTTGGCGG GCGTGTTGGG CATGAATACG CTGATGCTGG CAGTATGGTT |
|
| 351 | GGCAATGTTG TTCCTGCGTG TGAAAGTGGG GCGTTTTTTC AGCAGTCCGG |
|
| 401 | CGACGTGGTT TCGGGGCAAA GACCCTGTAA ATCAGGCGGT GTTGCGGCTG |
|
| 451 | TATGCGGACG AGTGGCGGCA ACCTTCGGTA CGTTGGAAAA TAGGCGCAAC |
|
| 501 | GTCGCACAGC CTGTGGCTCT GCACGCTGCT CGGAATGCTG GTGTCGGTAT |
|
| 551 | TGTTGCTGCT TTTGGTGCGG CAATATACGT TCAACTGGGA AAGCACGCTG |
|
| 601 | TTGAGCAATG CCGCTTCGGT ACGCGCGGTG GAAATGTTGG CATGGCTGCC |
|
| 651 | GTCGAAACTC GGTTTCCCTG TCCCCGATGC GCGGGCGGTC ATCGAAGGCC |
|
| 701 | GTCTGAACGG CAATATTGCC GATGCGCGGG CTTGGTCGGG GCTGCTGGTC |
|
| 751 | GGCAGTATCG CCTGCTACGG CATCCTGCCG CGCCTGCTGG CTTGGGTAGT |
|
| 801 | GTGTAAAATC CTTTTGAAAA CAAGCGAAAA CGGATTGGAT TTGGAAAAGC |
|
| 851 | CCTATTATCA GGCGGTCATC CGCCGCTGGC AGAACAAAAT CACCGATGCG |
|
| 901 | GATACGCGTC GGGAAACCGT GTCCGCCGTT TCACCGAAAA TCATCTTGAA |
|
| 951 | CGATGCGCCG AAATGGGCGG TCATGCTGGA GACCGAGTGG CAGGACGGCG |
|
| 1001 | AATGGTTCGA GGGCAGGCTG GCGCAGGAAT GGCTGGATAA GGGCGTTGCC |
|
| 1051 | ACCAATCGGG AACAGGTTGC CGCGCTGGAG ACAGAGCTGA AGCAGAAACC |
|
| 1101 | GGCGCAACTG CTTATCGGCG TGCGCGCCCA AACTGTGCCG GACCGCGGCG |
|
| 1151 | TGTTGCGGCA GATTGTCCGA CTCTCGGAAG CGGCGCAGGG CGGCGCGGTG |
|
| 1201 | GTGCAGCTTT TGGCGGAACA GGGGCTTTCA GACGACCTTT CGGAAAAGCT |
|
| 1251 | GGAACATTGG CGTAACGCGC TGGCCGAATG CGGCGCGGCG TGGCTTGAGC |
|
| 1301 | CTGACAGGGC GGCGCAGGAA GGGCGTTTGA AAGACCAATA A |
[0608]This corresponds to the amino acid sequence <SEQ ID 200; ORF33-1>:
[0000]| 1 | MLNPSRKLVE LVRILDEGGF IFSGDPVQAT EALRRVDGST EEKIIRRAEM | |
|
| 51 | IDRNRMLRET LERVRAGSFW LWVVAATFAF FTGFSVTYLL MDNQGLNFFL |
|
| 101 | VLAGVLGMNT LMLAVWLAML FLRVKVGRFF SSPATWFRGK DPVNQAVLRL |
|
| 151 | YADEWRQPSV RWKIGATSHS LWLCTLLGML VSVLLLLLVR QYTFNWESTL |
|
| 201 | LSNAASVRAV EMLAWLPSKL GFPVPDARAV IEGRLNGNIA DARAWSGLLV |
|
| 251 | GSIACYGILP RLLAWVVCKI LLKTSENGLD LEKPYYQAVI RRWQNKITDA |
|
| 301 | DTRRETVSAV SPKIILNDAP KWAVMLETEW QDGEWFEGRL AQEWLDKGVA |
|
| 351 | TNREQVAALE TELKQKPAQL LIGVRAQTVP DRGVLRQIVR LSEAAQGGAV |
|
| 401 | VQLLAEQGLS DDLSEKLEHW RNALAECGAA WLEPDRAAQE GRLKDQ* |
[0609]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0610]ORF33 shows 90.9% identity over a 143aa overlap with an ORF (ORF33a) from strain A of N. meningitidis.
[0000]
[0611]The complete length ORF33a nucleotide sequence <SEQ ID 201> is:
[0000]| 1 | ATGTTGAATC CATCCCGAAA ACTGGTTGAG CTGGTCCGTA TTTTGGAAGA | |
|
| 51 | AGGCGGCTTT ATTTTCAGCG GCGATCCCGT GCAGGCGACG GAGGCTTTGC |
|
| 101 | GCCGCGTGGA CGGCAGTACG GAGGAAAAAA TCATCCGTCG GGCGAAGATG |
|
| 151 | ATCGACAGGA ACCGTATGCT GCGGGAGACG TTGGAACGTG TGCGTGCGGG |
|
| 201 | GTCGTTCTGG TTGTGGGTGG CGGCGGCGAC GTTTGCGTTT NTTACCGNTT |
|
| 251 | TTTCAGTTAC TTATCTTCTA ATGGACAATC AGGGTCTGAA TTTCTTTTTG |
|
| 301 | GTTTTGGCGG GCGTGNTGGG CATGAATACG CTGATGCTGG CAGTATGGTT |
|
| 351 | GGCAATGTTG TTCCTGCGCG TGAAAGTGGG GCGTTTTTTC AGCAGTCCGG |
|
| 401 | CGACGTGGTT TCGGGGCAAA GACCCTGTCA ATCAGGCGGT GTTGCGGCTG |
|
| 451 | TATGCGGACG AGTGGCGGCN ACCTTCGGTA CGTTGGAAAA TAGGCGCAAC |
|
| 501 | GTCGCACAGC CTGTGGCTCT GCACGCTGCT CGGAATGCTG GTGTCGGTAT |
|
| 551 | TGTTGCTGCT TTTGGTGCGG CAATATACGT TCAACTGGGA AAGCACGCTG |
|
| 601 | TTGGGCGATT CGTCTTCGGT ACGGCTGGTG GAAATGTTGG CATGGCTGCC |
|
| 651 | TGCGAAACTG GGTTTTCCCG TGCCTGATGC GCGGGCGGTC ATCGAAGGTC |
|
| 701 | GTCTGAACGG CAATATTGCC GATGCGCGGG CTTGGTCGGG GCTGCTGGTC |
|
| 751 | GGCAGTATCG CCTGCTACGG CATCCTGCCG CGCCTCTTGG CTTGGGCGGT |
|
| 801 | ATGCAAAATC CTTNTGNAAA CAAGCGAAAA CGGCTTGGAT TTGGAAAAGC |
|
| 851 | NCNNNNNTCN NNCGNTCATC CGCCGCTGGC AGAACAAAAT CACCGATGCG |
|
| 901 | GATACGCGTC GGGAAACCGT GTCCGCCGTT TCGCCGAAAA TCGTCTTGAA |
|
| 951 | CGATGCGCCG AAATGGGCGG TCATGCTGGA GACCGAATGG CAGGACGGCG |
|
| 1001 | AATGGTTCGA GGGCAGGCTG GCGCAGGAAT GGCTGGATAA GGGCGTTGCC |
|
| 1051 | GCCAATCGGG AACAGGTTGC CGCGCTGGAG ACAGAGCTGA AGCAGAAACC |
|
| 1101 | GGCGCAACTG CTTATCGGCG TGCGCGCCCA AACTGTGCCC GACCGCGGCG |
|
| 1151 | TGTTGCGGCA GATCGTCCGA CTTTCGGAAG CGGCGCAGGG CGGCGCGGTG |
|
| 1201 | GTGCANCTTT TGGCGGAACA GGGGCTTTCA GACGACCTTT CGGAAAAGCT |
|
| 1251 | GGAACATTGG CGTAACGCGC TGACCGAATG CGGCGCGGCG TGGCTGGAAC |
|
| 1301 | CCGACAGAGC GGCGCAGGAA GGCCGTCTGA AAACCAACGA CCGCACTTGA |
[0612]This encodes a protein having amino acid sequence <SEQ ID 202>:
[0000]| 1 | MLNPSRKLVE LVRILEEGGF IFSGDPVQAT EALRRVDGST EEKIIRRAKM | |
|
| 51 | IDRNRMLRET LERVRAGSFW LWVAAATFAF XTXFSVTYLL MDNQGLNFFL |
|
| 101 | VLAGVXGMNT LMLAVWLAML FLRVKVGRFF SSPATWFRGK DPVNQAVLRL |
|
| 151 | YADEWRXPSV RWKIGATSHS LWLCTLLGML VSVLLLLLVR QYTFNWESTL |
|
| 201 | LGDSSSVRLV EMLAWLPAKL GFPVPDARAV IEGRLNGNIA DARAWSGLLV |
|
| 251 | GSIACYGILP RLLAWAVCKI LXXTSENGLD LEKXXXXXXI RRWQNKITDA |
|
| 301 | DTRRETVSAV SPKIVLNDAP KWAVMLETEW QDGEWFEGRL AQEWLDKGVA |
|
| 351 | ANREQVAALE TELKQKPAQL LIGVRAQTVP DRGVLRQIVR LSEAAQGGAV |
|
| 401 | VXLLAEQGLS DDLSEKLEHW RNALTECGAA WLEPDRAAQE GRLKTNDRT* |
[0613]ORF33a and ORF33-1 show 94.1% identity in 444 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0614]ORF33 shows 91.6% identity over a 143aa overlap with a predicted ORF (ORF33.ng) from N. gonorrhoeae:
[0000]
[0615]An ORF33ng nucleotide sequence <SEQ ID 203> was predicted to encode a protein having amino acid sequence <SEQ ID 204>:
[0000]| 1 | MIDRDRMLRD TLERVRAGSF WLWVVVASMM FTAGFSGTYL LMDNQGLNFF | |
|
| 51 | LVLAGVLGMN TLMLAVWLAT LFLRVKVGRF FSSPATWFRG KGPVNQAVLR |
|
| 101 | LYADQWRQPS VRWKIGATAH SLWLCTLLGM LVSVLLLLLV RQYTFNWEST |
|
| 151 | LLSNAASVRA VEMLAWLPSK LGFPVPDARA VIEGRLNGNI ADARAWSGLL |
|
| 201 | VGSIVCYGIL PRLLAWVVCK ILLKTSENGL DLEKTYYQAV IRRWQNKITD |
|
| 251 | ADTRRETVSA VSPKIVLNDA PKWALMLETE WQDGQWFEGR LAQEWLDKGV |
|
| 301 | AANREQVAAL ETELKQKPAQ LLIGVRAQTV PDRGVLRQIV RLSEAAQGGA |
|
| 351 | VVQLLAEQGL SDDLSEKLEH WRNALTECGA AWLEPDRVAQ EGRLKDQ* |
[0616]Further sequence analysis revealed the following DNA sequence <SEQ ID 205>:
[0000]| 1 | ATGTTGaatC CATCCCgaAA ACTGgttgag ctGgTCCgtA Ttttgaataa | |
|
| 51 | agggggtTTT attttcagcg gcgatcctgt gcaggcgacg gaggctttgc |
|
| 101 | gccgcgtgga cggcAGTACG GAggAaaaaa tcttccgtcg GGCGGAGAtg |
|
| 151 | atcgACAGGg accgtatgtt gcgggACaCg TtggaacGTG TGCGTGCggg |
|
| 201 | gtcgtTctgG TTATGGGTGG TggtggCAtC gATGATGTtt aCCGCCGGAT |
|
| 251 | TTTCAGgcac ttatCttCTG ATGGACaatC AGGGGCtGAA TtTCTTTTTA |
|
| 301 | GTTTTggcgG GAGTGTtggG CATGaatacG ctgATGCTGG CAGTATGGtt |
|
| 351 | gGCAACGTTG TTCCTGCGCG TGAAAGTGGG ACGGTTTTTC AGCAGTCCGG |
|
| 401 | CGACGTGGTT TCGGGGCAAA GGCCCTGTAA ATCAGGCGGT GTTGCGGCTG |
|
| 451 | TATGCGGACC AGTGGCGGCA ACCTTCGGTA CGATGGAAAA TAGGCGCAAC |
|
| 501 | GGCGCACAGC TTGTGGCTCT GCACGCTGCT CGGAATGCTG GTGTCGGTAT |
|
| 551 | TGCTGCTGCT TTTGGTGCGG CAATATACGT TCAACTGGGA AAGCACGCTG |
|
| 601 | TTGAGCAATG CCGCTTCGGT ACGCGCGGTG GAAATGTTGG CATGGCTGCC |
|
| 651 | GTCGAAACTC GGTTTCCCTG TCCCCGATGC GCGGGCGGTC ATCGAAGGTC |
|
| 701 | GTCTGAACGG CAATATTGCC GATGCGCGGG CTTGGTCGGG GCTGCTGGTC |
|
| 751 | GGCAGTATCG TCTGCTACGG CATCCTGCCG CGCCTCTTGG CTTGGGTAGT |
|
| 801 | GTGTAAAATC CTTTTGAAAA CAAGCGAAAA CGGattgGAT TTGGAAAAAA |
|
| 851 | CCTATTATCA GGCGGTCATC CGCCGCTGGC AGAACAAAAT CACCGATGCG |
|
| 901 | GATACGCGTC GGGAAACCGT GTCCGCCGTT TCGCcgaAAA TCGTCTTGAA |
|
| 951 | CGATGCGCCG AAATGGGCGC TCATGCTGGA GACCGAGTGG CAGGACGGCC |
|
| 1001 | AATGGTTCGA GGGCAGGCTG GCGCAGGAAT GGCTGGATAA GGGCGTTGCC |
|
| 1051 | GCCAATCGGG AACAGGTTGC CGCGCTGGAG ACAGAGCTGA AGCAGAAACC |
|
| 1101 | GGCGCAACTG CTTATCGGCG TACGCGCCCA AACTGTGCCG GACCGGGGCG |
|
| 1151 | TGCTGCGGCA GATTGTGCGG CTTTCGGAAG CGGCGCAGGG CGGCGCGGTG |
|
| 1201 | GTGCAGCTTT TGGCGGAACA GGGGCTTTCA GACGACCTTT CGGAAAAGCT |
|
| 1251 | GGAACATTGG CGTAACGCGC TGACCGAATG CGGCGCGGCG TGGCTTGAGC |
|
| 1301 | CTGACAGGGT GGCGCAGGAA GGCCGTTTGA AAGACCAATA A |
[0617]This encodes a protein having amino acid sequence <SEQ ID 206; ORF33ng-1>:
[0000]| 1 | MLNPSRKLVE LVRILNKGGF IFSGDPVQAT EALRRVDGST EEKIFRRAEM | |
|
| 51 | IDRDRMLRDT LERVRAGSFW LWVVVASMMF TAGFSGTYLL MDNQGLNFFL |
|
| 101 | VLAGVLGMNT LMLAVWLATL FLRVKVGRFF SSPATWFRGK GPVNQAVLRL |
|
| 151 | YADQWRQPSV RWKIGATAHS LWLCTLLGML VSVLLLLLVR QYTFNWESTL |
|
| 201 | LSNAASVRAV EMLAWLPSKL GFPVPDARAV IEGRLNGNIA DARAWSGLLV |
|
| 251 | GSIVCYGILP RLLAWVVCKI LLKTSENGLD LEKTYYQAVI RRWQNKITDA |
|
| 301 | DTRRETVSAV SPKIVLNDAP KWALMLETEW QDGQWFEGRL AQEWLDKGVA |
|
| 351 | ANREQVAALE TELKQKPAQL LIGVRAQTVP DRGVLRQIVR LSEAAQGGAV |
|
| 401 | VQLLAEQGLS DDLSEKLEHW RNALTECGAA WLEPDRVAQE GRLKDQ* |
[0618]ORF33ng-1 and ORF33-1 show 94.6% identity in 446 aa overlap:
[0000]
[0619]Based on the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 25
[0620]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 207>:
[0000]| 1 | ..CAGAAGAGTT TGTCGAGAAT TTCTTTATGG GGTTTGGGCG GCGTGTTTTT | |
|
| 51 | CGGGGTGTCC GGTCTGGTAT GGTTTTCTTT GGGCGTTTCT TT.GAGTGCG |
|
| 101 | CCTGTTTTTC GGGTGTTTCT TTTCGGGGTT CGGGACGGGG GACGTTTGTG |
|
| 151 | GGCAGTACGG GGGTTTCTTT GAGTGTGTTT TCAGCTTGTG TTCC.GGCGT |
|
| 201 | CGTCCGGCTG CCTGTCGGTT TGAGCTGTGT CGGCAGGTTG CG..GTTTGA |
|
| 251 | CCCGGTTTTT CTTGGGTGCG GCAGGGGACG TCATTCTCCT GCCGCTTTCG |
|
| 301 | TCTGTGCCGT CCGGCTGTGC GGGTTCGGAT GAGGCGGCGT GGTGGTGTTC |
|
| 351 | GGGTTGGGCG GCATCTTGTT CCGACTACGC CGTTTGGCAG CCAGAATTCG |
|
| 401 | GTTTCGCGGG GGCTGTCGGT GTGTTGCGGT TCGGCTTGAA GGGTTTTGTC |
|
| 451 | GTCC.. |
[0621]This corresponds to the amino acid sequence <SEQ ID 208; ORF34>:
[0000]| 1 | ..QKSLSRISLW GLGGVFFGVS GLVWFSLGVS XECACFSGVS FRGSGRGTFV | |
|
| 51 | GSTGVSLSVF SACVXGVVRL PVGLSCVGRL XXLTRFFLGA AGDVILLPLS |
|
| 101 | SVPSGCAGSD EAAWWCSGWA ASCPTTPFGS QNSVSRGLSV CCGSA*RVLS |
|
| 151 | S.. |
[0622]Further work revealed the complete nucleotide sequence <SEQ ID 209>:
[0000]| 1 | ATGATGATGC CGTTCATAAT GCTTCCTTGG ATTGCkGGTG TGCCTGCCGT | |
|
| 51 | GCCGGGTCAG AATAGGTTGT CCAGAATTTC TTTATGGGGT TTGGGCGGCG |
|
| 101 | TGTTTTTCGG GGTGTCCGGT TTGGTATGGT TTTCTTTGGG CGTTTCTTTG |
|
| 151 | GGCTGCGCCT GTTTTTCGGG TGTTTCTTTT CGGGGTTCGG GACGGGGGAC |
|
| 201 | GTTTGTGGGC AGTACGGGGG TTTCTTTGAG TGTGTTTTCA GCTTGTGTTC |
|
| 251 | CGGCGTCGTC CGGCTGCCTG TCGGTTTGAG CTGTGTCGGC AGGTTGCGGT |
|
| 301 | TTGACCCGGT TTTTCTTGGG TGCGGCAGGG GACGGCAGTC CGCTGCCGCT |
|
| 351 | TTCGTCTGTG CCGTCCGGCT GTGCGGGTTC GGATGAGGCG GCGTGGTGGT |
|
| 401 | GTTCGGGTTG GGCGGCATCT TGTCCGACTA CGCCGTTTGG CAGCCAGAAT |
|
| 451 | TCGGTTTCGC GGGGGCTGTC GGTGTGTTGC GGTTCGGCTT GAAGGGTTTT |
|
| 501 | GTCGCCGTTC GGGTTGAATG TGCTGACGAT GCCTATTGCC AATGCGCCGA |
|
| 551 | TGGCGGCGAT ACAGATGAGC AATACGGCGC GTATCAGGAG TTTGGGGGTC |
|
| 601 | AGCCTGAAGG GTTTGTTCGG TTTTTTTGCC ATTTTGATTG TGCTTTTGGG |
|
| 651 | GTGTCGGGCA ATGCCGTCTG AAGGCGGTTC AGACGGCATT GCCGAGTCAG |
|
| 701 | CGTTGGACGT AGTTTTGGTA GAGGGTGATG ACTTTTTGTA CGCCGACGGT |
|
| 751 | GGTGCTGACT TTTTGGGTAA TCTGCGCCTG TTCTTCGGGG GTGAGGATGC |
|
| 801 | CCATAACGTA GGTTACGTTG CCGTAGGTAA CGATTTTGAC GCGCGCCTGT |
|
| 851 | GTGGCGGGGC TGATGCCCAA CAGCGTGGCG CGGACTTTGG ATGTGTTCCA |
|
| 901 | AGTGTCGCCG GCGATGTCGC CGGCAGTGCG CGGCAGGGAG GCGACGGTAA |
|
| 951 | TATAGTTGTA CACGCCTTCG GCGGCCTGTT CGGAACGTGC AATCTGACCG |
|
| 1001 | ACGAACTGTT TTTCGCCTTC GGTGGCGACT TGTCCGAGCA GCAGCAGGTG |
|
| 1051 | GCGGTTGTAG CCGACGACGG AGATTTGGGG CGTGTAGCCT TTGGTTTGGT |
|
| 1101 | TGTTTTGGCG CAGATAGGAA CGGGCGGTGG TTTCGATACG CAACGCCATA |
|
| 1151 | ACGTTGTCGT CGGTTTGCGC GCCGGTGGTT CGGCGGTCGA CGGCGGATTT |
|
| 1201 | CGCGCCGACG GCGGCGCTTC CGATTACTGC GCTGACGCAG CCGCTAAGGG |
|
| 1251 | CAAGGCTGAA AATGGCGGCA ATCAGGGTGC GGACGGTGTG CGGTTTGGGT |
|
| 1301 | TTCATCGGGT GCTTCCTTTC TTGGGCGTTT CAGACGGCAT TGCTTTGCGC |
|
| 1351 | CATGCCGTCT GA |
[0623]This corresponds to the amino acid sequence <SEQ ID 210; ORF34-1>:
[0000]| 1 | MMMPFIMLPW IAGVPAVPGQ NRLSRISLWG LGGVFFGVSG LVWFSLGVSL | |
|
| 51 | GCACFSGVSF RGSGRGTFVG STGVSLSVFS ACVPASSGCL SV*AVSAGCG |
|
| 101 | LTRFFLGAAG DGSPLPLSSV PSGCAGSDEA AWWCSGWAAS CPTTPFGSQN |
|
| 151 | SVSRGLSVCC GSA*RVLSPF GLNVLTMPIA NAPMAAIQMS NTARIRSLGV |
|
| 201 | SLKGLFGFFA ILIVLLGCRA MPSEGGSDGI AESALDVVLV EGDDFLYADG |
|
| 251 | GADFLGNLRL FFGGEDAHNV GYVAVGNDFD ARLCGGADAQ QRGADFGCVP |
|
| 301 | SVAGDVAGSA RQGGDGNIVV HAFGGLFGTC NLTDELFFAF GGDLSEQQQV |
|
| 351 | AVVADDGDLG RVAFGLVVLA QIGTGGGFDT QRHNVVVGLR AGGSAVDGGF |
|
| 401 | RADGGASDYC ADAAAKGKAE NGGNQGADGV RFGFHRVLPF LGVSDGIALR |
|
| 451 | HAV* |
[0624]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0625]ORF34 shows 73.3% identity over a 161aa overlap with an ORF (ORF34a) from strain A of N. meningitidis:
[0000]
[0626]The complete length ORF34a nucleotide sequence <SEQ ID 211> is:
[0000]| 1 | ATGATGATNC CGTTNATAAT GCTTCCTTGG ATTGCGGGTG TGCCTGCCGT | |
|
| 51 | GCCGGGTCAG AAGAGGTTGT CGAGAANTTC TTTATGGGGT TTAGGCGGCN |
|
| 101 | TGTTTTTCGG GGTGTCCGGT TTGGTATGGT TTTCTTTGGG CGTTTCTNTT |
|
| 151 | TCTTTGGGTG TTTCTNTGGG CTGTGCCTGT TTTTCGGGTG TTTCTTTTCG |
|
| 201 | GGGTTCGGGA CGGGGGACGT TTGTGGGCAG TACNGGGGTT TCTTTGAGTG |
|
| 251 | TGTTTTCAGC TTGTGCTCCG GCGTCGTCCG GCTGCCTGTC GGTTTNAGCT |
|
| 301 | GTGTCGGCAG GTTGCGGTTT GACCCGGNTT TTCTTNGGTG CGGCAGGGGA |
|
| 351 | CGGCAGTCCG CTGCCGCTTT CGTCTGTGCC GTCCGGCTGT GCGGGTGCGG |
|
| 401 | ATGAGGAGGC GTNGTNGTGT TCGGGTTGGG CGGCATCTTG TCCGACTACG |
|
| 451 | CCGTTTGGCA GCCAGAATTC GGTTTCGCGG GGGCTGTCGG TGTGTTGCGG |
|
| 501 | TTCGGTNTGG AGGGTTTTGT CNCCGTTCGG GTNGAATGTG CTGACGATGC |
|
| 551 | CTATTGCCAA TGCGCCGATG GCGGTGATAC AGATGAGCAA TACGGCGCGT |
|
| 601 | ATCAGGAGTT TGGGGGTCAG CCTGAAGGGT TTGTTCNGTT TTTTTGCCAT |
|
| 651 | TTTGATTGTG CTTTTGGGGT GTCGGGCAAT GCCGTCTGAA GGCGGTTCAG |
|
| 701 | ACGGCATTGC CGAGTCAGCG TTGGACGTAG TTTNGGTAGA GGGTGATGAC |
|
| 751 | TTTTTGTACG CCGACGGTGG TGCTGACTTT TTGGGTAATC TGCGCCTGTT |
|
| 801 | CTTCGGGGGT GAGGATGCCC ATAACGTAGG TTACGTTGCC GTAGGTAACG |
|
| 851 | ATTTTGACGC GCGCCTGTGT GGCGGGGCTG ATGCCCAACA GCGTGGCGCG |
|
| 901 | GACTTTGGAT GTGTTCCAAG TGTCGCCGGC GATGTCGCCG GCAGTGCGCG |
|
| 951 | GCAGGGAGGC GACGGTAATG TANTTGTACA CGCCTTCGGC GGCCTGTTCG |
|
| 1001 | GAACGTGCAA TCTGACCGAC GAACTGTTTC TCGCCTTCGG TGGCGACTTG |
|
| 1051 | TCCGAGCAGC AGCAGGTGGC GGTTGTAGCC GACAACGGAG ATTTGGGGCG |
|
| 1101 | TGTANCCTTT GGTTTGGTTG TTTTGGCGCA GATAGGAGCG GGCGGTGGTT |
|
| 1151 | TCGATACGCA GCGCCATTAC GTTGTCGTCG GTTNGCGCGC CGGTGGTTCG |
|
| 1201 | GCGGTCGACG GCGGATTTCG CGCCGACCGC CGCGCCGCCG ACGACTGCGC |
|
| 1251 | TGACGCAGCC GCCGAGGGCA AGGCTGAGGA CGGCGGCAGT CAGGGTGCGG |
|
| 1301 | ACGGTGTGCG GTTTGGGTTT CATCGGGTGC TTCCTTTCTT GGGCGTTTCA |
|
| 1351 | GACGGCATTG CTTTGCGCCA TGCCGTCTGA |
[0627]This encodes a protein having amino acid sequence <SEQ ID 212>:
[0000]| 1 | MMXPXIMLPW IAGVPAVPGQ KRLSRXSLWG LGGXFFGVSG LVWFSLGVSX | |
|
| 51 | SLGVSXGCAC FSGVSFRGSG RGTFVGSTGV SLSVFSACAP ASSGCLSVXA |
|
| 101 | VSAGCGLTRX FXGAAGDGSP LPLSSVPSGC AGADEEAXXC SGWAASCPTT |
|
| 151 | PFGSQNSVSR GLSVCCGSVW RVLSPFGXNV LTMPIANAPM AVIQMSNTAR |
|
| 201 | IRSLGVSLKG LFXFFAILIV LLGCRAMPSE GGSDGIAESA LDVVXVEGDD |
|
| 251 | FLYADGGADF LGNLRLFFGG EDAHNVGYVA VGNDFDARLC GGADAQQRGA |
|
| 301 | DFGCVPSVAG DVAGSARQGG DGNVXVHAFG GLFGTCNLTD ELFLAFGGDL |
|
| 351 | SEQQQVAVVA DNGDLGRVXF GLVVLAQIGA GGGFDTQRHY VVVGXRAGGS |
|
| 401 | AVDGGFRADR RAADDCADAA AEGKAEDGGS QGADGVRFGF HRVLPFLGVS |
|
| 451 | DGIALRHAV* |
[0628]ORF34a and ORF34-1 show 91.3% identity in 459 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0629]ORF34 shows 77.6% identity over a 161aa overlap with a predicted ORF (ORF34.ng) from N. gonorrhoeae.
[0000]
[0630]The complete length ORF34ng nucleotide sequence <SEQ ID 213> is:
[0000]| 1 | ATGATGATGC CGTTCATAAT GCTTCCTTGG ATTGCGGGTG TGCCTGCCGT | |
|
| 51 | GCCGGGTCAA AAGAGGTTGT CGAGAATCTC TTTATGGGGT TTGGCCGGCG |
|
| 101 | TGTTTTTCGG GGTGTCCGGT TTGGTATGGT TTTCTTTGGG CGTTTCTTTT |
|
| 151 | TCTTTGGGTG TTTCTTTGGG CTGCGCCTGT TTTTCGGGTG TTTCTTTTCG |
|
| 201 | GGGTTCGGGA TGGGGGGCGT TTGTGGGCAG TACGGGGGTT TCTTTGAGTG |
|
| 251 | TGTTTTCAGC TTGTGTTCCG GTGCCGGTTA ACGAATCGGC TGCCCGGGCC |
|
| 301 | GCATCCGAAG GGCGCGGTTT gACCCGGTTT TTCTTGGGTG CGGCAGGGGA |
|
| 351 | CGGCAGTCCG CTGCCGCTTT CTTCTGTGCC GTCCGGCTGT GCGGGTTCGG |
|
| 401 | ATGAGGCGGC GTGGTGGTGT TCGGGTTGGG CGGCATCTTG TCCGACGGCG |
|
| 451 | CCGTTTGGCA GCCAGAATTC GGTTTCGCGG GGGCTGTCGG TGTGTTGCGG |
|
| 501 | TTCGGTTTGG AGGGTTTTGT CGCCGTTCGG GTTGAATGTG CTGACGATGC |
|
| 551 | CTACTGCCAA TGCGCCGATG GCGGTGATAC AGATGAGCAA TACGGCGCGT |
|
| 601 | ATCAGGAGTT TGGGGGTCAG CCTGAAGGGT TTGTTCGGTT TTTTTGCCAT |
|
| 651 | TTTGATTGTG CTTTTGGGGT GTCGGGCAAT GCCGTCTGAA GGCGGTTCAG |
|
| 701 | ACGGCATTGC CGAGTCAGCG TTGGACGTAG TTTTGGTAGA GGGTAATGAC |
|
| 751 | TTTTTGTACG CCGAcggTGG TGCTGACTTT TTGGGTAATC TGCGCCTGTT |
|
| 801 | CTTCGGGGGT GAGGATGCCC ATAACGTAGG TTACATTGCC GTAGGTAATG |
|
| 851 | ATTTTGACGC GCGCCTGTGT AGCGGGGCTG ATGCCCAGCA GcgtgGCGCG |
|
| 901 | GACTTTGGAC GTGTTCCAAG TGTCGCCGGC GATGTCGCCC GCAGTGCGCG |
|
| 951 | GCAGGGAGGC GACGGTAATG TAGTTGTATA CGCCTTCGGC GGCCTGTTCG |
|
| 1001 | GAACGTGCAA TCTGACCGAC GAACTGTTTT TCGCCTTCGG TGGCGACTTG |
|
| 1051 | TCCGAGCAGC AGCAGGTGGC GGTTGTAGCC GACGACGGAG ATTTGGGGCG |
|
| 1101 | TGTAGCCTTT GGTTTGGTTG TTTTGGCGCA GGTAGGAACG GGCGGTGGTT |
|
| 1151 | TCGATACGCA ACGCCATAAC GTtgtCATCG GTTtgcgcgc CGGTGGTTcg |
|
| 1201 | gCGGTCGATG ACGGATTTTG CGCCGACGGC GGCCCCGCCG ACGACTGCGC |
|
| 1251 | TGAAGCAGCC GCCGAGGGCA AGGCTGAGGA CGGCGGCAAT CAGGGTGCGG |
|
| 1301 | ACGGTGTGTG GTTTGGGTTT CATCGGGGAC TTCCTTTCTT GGGCGTTTCA |
|
| 1351 | GACGGCATTG CTTTGCGCCA TGCCGTCTGA |
[0631]This encodes a protein having amino acid sequence <SEQ ID 214>:
[0000]| 1 | MMMPFIMLPW IAGVPAVPGQ KRLSRISLWG LAGVFFGVSG LVWFSLGVSF | |
|
| 51 | SLGVSLGCAC FSGVSFRGSG WGAFVGSTGV SLSVFSACVP VPVNESAARA |
|
| 101 | ASEGRGLTRF FLGAAGDGSP LPLSSVPSGC AGSDEAAWWC SGWAASCPTA |
|
| 151 | PFGSQNSVSR GLSVCCGSVW RVLSPFGLNV LTMPTANAPM AVIQMSNTAR |
|
| 201 | IRSLGVSLKG LFGFFAILIV LLGCRAMPSE GGSDGIAESA LDVVLVEGND |
|
| 251 | FLYADGGADF LGNLRLFFGG EDAHNVGYIA VGNDFDARLC SGADAQQRGA |
|
| 301 | DFGRVPSVAG DVARSARQGG DGNVVVYAFG GLFGTCNLTD ELFFAFGGDL |
|
| 351 | SEQQQVAVVA DDGDLGRVAF GLVVLAQVGT GGGFDTQRHN VVIGLRAGGS |
|
| 401 | AVDDGFCADG GPADDCAEAA AEGKAEDGGN QGADGVWFGF HRGLPFLGVS |
|
| 451 | DGIALRHAV* |
[0632]ORF34ng and ORF34-1 show 90.0% identity in 459 aa overlap:
[0000]
[0633]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 26
[0634]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 215>:
[0000]| 1 | ATGAAAACCT TCTTCAAAAC CCTTTCCGCC GCCGCACTCG CGCTCATCCT | |
|
| 51 | CGCCGCCTGC GGATT.CAAA AAGACAGCGC GCCCGCCGCA TCCGCTTCTG |
|
| 101 | CCGCCGCCGA CAACGGCGCG GCGTAAAAAA GAAATCGTCT TCGGCACGAC |
|
| 151 | CGTCGGCGAC TTCGGCGATA TGGTCAAAGA ACAAATCCAA GCCGAGCTGG |
|
| 201 | AGAAAAAAGG CTACACCGTC AAACTGGTCG AGTTTACCGA CTATGTACGC |
|
| 251 | CCGAATCTGG CATTGGCTGA GGGCGAGTTG |
[0635]This corresponds to the amino acid sequence <SEQ ID 216; ORF4>:
[0000]| 1 | MKTFFKTLSA AALALILAAC G.QKDSAPAA SASAAADNGA AKKEIVFGTT | |
|
| 51 | VGDFGDMVKE QIQAELEKKG YTVKLVEFTD YVRPNLALAE GEL |
[0636]Further sequence analysis revealed the complete nucleotide sequence <SEQ ID 217>:
[0000]| 1 | ATGAAAACCT TCTTCAAAAC CCTTTCCGCC GCCGCACTCG CGCTCATCCT | |
|
| 51 | CGCCGCCTGC GGCGGTCAAA AAGACAGCGC GCCCGCCGCA TCCGCTTCTG |
|
| 101 | CCGCCGCCGA CAACGGCGCG GCGAAAAAAG AAATCGTCTT CGGCACGACC |
|
| 151 | GTCGGCGACT TCGGCGATAT GGTCAAAGAA CAAATCCAAG CCGAGCTGGA |
|
| 201 | GAAAAAAGGC TACACCGTCA AACTGGTCGA GTTTACCGAC TATGTACGCC |
|
| 251 | CGAATCTGGC ATTGGCTGAG GGCGAGTTGG ACATCAACGT CTTCCAACAC |
|
| 301 | AAACCCTATC TTGACGACTT CAAAAAAGAA CACAATCTGG ACATCACCGA |
|
| 351 | AGTCTTCCAA GTGCCGACCG CGCCTTTGGG ACTGTACCCG GGCAAGCTGA |
|
| 401 | AATCGCTGGA AGAAGTCAAA GACGGCAGCA CCGTATCCGC GCCCAACGAC |
|
| 451 | CCGTCCAACT TCGCCCGCGT CTTGGTGATG CTCGACGAAC TGGGTTGGAT |
|
| 501 | CAAACTCAAA GACGGCATCA ATCCGTTGAC CGCATCCAAA GCGGACATCG |
|
| 551 | CCGAGAACCT GAAAAACATC AAAATCGTCG AGCTTGAAGC CGCGCAACTG |
|
| 601 | CCGCGTAGCC GCGCCGACGT GGATTTTGCC GTCGTCAACG GCAACTACGC |
|
| 651 | CATAAGCAGC GGCATGAAGC TGACCGAAGC CCTGTTCCAA GAACCGAGCT |
|
| 701 | TTGCCTATGT CAACTGGTCT GCCGTCAAAA CCGCCGACAA AGACAGCCAA |
|
| 751 | TGGCTTAAAG ACGTAACCGA GGCCTATAAC TCCGACGCGT TCAAAGCCTA |
|
| 801 | CGCGCACAAA CGCTTCGAGG GCTACAAATC CCCTGCCGCA TGGAATGAAG |
|
| 851 | GCGCAGCCAA ATAA |
[0637]This corresponds to the amino acid sequence <SEQ ID 218; ORF4-1>:
[0000]| 1 | MKTFFKTLSA AALALILAAC GGQKDSAPAA SASAAADNGA AKKEIVFGTT | |
|
| 51 | VGDFGDMVKE QIQAELEKKG YTVKLVEFTD YVRPNLALAE GELDINVFQH |
|
| 101 | KPYLDDFKKE HNLDITEVFQ VPTAPLGLYP GKLKSLEEVK DGSTVSAPND |
|
| 151 | PSNFARVLVM LDELGWIKLK DGINPLTASK ADIAENLKNI KIVELEAAQL |
|
| 201 | PRSRADVDFA VVNGNYAISS GMKLTEALFQ EPSFAYVNWS AVKTADKDSQ |
|
| 251 | WLKDVTEAYN SDAFKAYAHK RFEGYKSPAA WNEGAAK* |
[0638]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0639]ORF4 shows 93.5% identity over a 93aa overlap with an ORF (ORF4a) from strain A of N. meningitidis:
[0000]
[0640]The complete length ORF4a nucleotide sequence <SEQ ID 219> is:
[0000]| 1 | ATGAAAACCT TCTTCAAAAC CCTTTCCGCC GCCGCACTCG CGCTCATCCT | |
|
| 51 | CGCCGCCTGC GGCGGTCAAA AAGATAGCGC GCCCGCCGCA TCCGCTTCTG |
|
| 101 | CCGCCGCCGA CAACGGCGCG GCGAANAAAG AAATCGTCTT CGGCACGACC |
|
| 151 | GTCGGCGACT TCGGCGATAT GGTCAAAGAA CANATCCAAC CCGAGCTGGA |
|
| 201 | GAAAAAAGGC TACACCGTCA AACTGGTCGA GTNTACCGAC TATGTGCGCN |
|
| 251 | CGAATCTGGC ATTGGCTGAG GGCGAGTTGG ACATCAACGT CTTNCAACAC |
|
| 301 | ANACNCTATC TTGACGACTN CAAAAAANAA CACAATCTGG ACATCACCNN |
|
| 351 | AGTCTTNCAA GTGCCGACCG CGCCTTTGGG ACTGTACCCG GGCAAGCTGA |
|
| 401 | AATCGCTGGA NNAAGTCAAA GANGGCAGCA CCGTATCCGC GCCCAACGAC |
|
| 451 | CCGTNNNACT TCGNCCGCGT CTTGGTGATG CTCGACGAAC TGGGTTNGAT |
|
| 501 | CAAACTCAAA GACNGCATCA NNNNGNNGNN NNNANCNANA NNNGANANNN |
|
| 551 | NNNNANNNNT NNNNNNNNNN NNNNNCNNCG NNNNNNNANN NNNNNNNNNN |
|
| 601 | NCGNNTNNNN NNGCNNNNNT NNANNNTNNN NNCNNCNNNN NNNNNTNNNN |
|
| 651 | NANNANNAGC GGCATGAAGC TGACCGAAGC CCTGTTCCAA GAACCGAGCT |
|
| 701 | TTGCCTATGT CAACTGGTCT GCCGTCAAAA CCGCCGACAA AGACAGCCAA |
|
| 751 | TGGCTTAAAG ACGTAACCGA GGCCTATAAC TCCGACGCGT TCAAAGCCTA |
|
| 801 | CGCGCACAAA CGCTTCGAGG GCTACAAATC CCCTGCCGCA TGGAATGAAG |
|
| 851 | GCGCAGCCAA ATAA |
[0641]This is predicted to encode a protein having amino acid sequence <SEQ ID 220>:
[0000]| 1 | MKTFFKTLSA AALALILAAC GGQKDSAPAA SASAAADNGA AXKEIVFGTT | |
|
| 51 | VGDFGDMVKE XIQPELEKKG YTVKLVEXTD YVRXNLALAE GELDINVXQH |
|
| 101 | XXYLDDXKKX HNLDITXVXQ VPTAPLGLYP GKLKSLXXVK XGSTVSAPND |
|
| 151 | PXXFXRVLVM LDELGXIKLK DXIXXXXXXX XXXXXXXXXX XXXXXXXXXX |
|
| 201 | XXXXAXXXXX XXXXXXXXXS GMKLTEALFQ EPSFAYVNWS AVKTADKDSQ |
|
| 251 | WLKDVTEAYN SDAFKAYAHK RFEGYKSPAA WNEGAAK* |
[0642]A leader peptide is underlined.
[0643]Further analysis of these strain A sequences revealed the complete DNA sequence <SEQ ID 221>:
[0000]| 1 | ATGAAAACCT TCTTCAAAAC CCTTTCCGCC GCCGCACTCG CGCTCATCCT | |
|
| 51 | CGCCGCCTGC GGCGGTCAAA AAGATAGCGC GCCCGCCGCA TCCGCTTCTG |
|
| 101 | CCGCCGCCGA CAACGGCGCG GCGAAAAAAG AAATCGTCTT CGGCACGACC |
|
| 151 | GTCGGCGACT TCGGCGATAT GGTCAAAGAA CAAATCCAAC CCGAGCTGGA |
|
| 201 | GAAAAAAGGC TACACCGTCA AACTGGTCGA GTTTACCGAC TATGTGCGCC |
|
| 251 | CGAATCTGGC ATTGGCTGAG GGCGAGTTGG ACATCAACGT CTTCCAACAC |
|
| 301 | AAACCCTATC TTGACGACTT CAAAAAAGAA CACAATCTGG ACATCACCGA |
|
| 351 | AGTCTTCCAA GTGCCGACCG CGCCTTTGGG ACTGTACCCG GGCAAGCTGA |
|
| 401 | AATCGCTGGA AGAAGTCAAA GACGGCAGCA CCGTATCCGC GCCCAACGAC |
|
| 451 | CCGTCCAACT TCGCCCGCGT CTTGGTGATG CTCGACGAAC TGGGTTGGAT |
|
| 501 | CAAACTCAAA GACGGCATCA ATCCGCTGAC CGCATCCAAA GCGGACATTG |
|
| 551 | CCGAAAACCT GAAAAACATC AAAATCGTCG AGCTTGAAGC CGCGCAACTG |
|
| 601 | CCGCGTAGCC GCGCCGACGT GGATTTTGCC GTCGTCAACG GCAACTACGC |
|
| 651 | CATAAGCAGC GGCATGAAGC TGACCGAAGC CCTGTTCCAA GAACCGAGCT |
|
| 701 | TTGCCTATGT CAACTGGTCT GCCGTCAAAA CCGCCGACAA AGACAGCCAA |
|
| 751 | TGGCTTAAAG ACGTAACCGA GGCCTATAAC TCCGACGCGT TCAAAGCCTA |
|
| 801 | CGCGCACAAA CGCTTCGAGG GCTACAAATC CCCTGCCGCA TGGAATGAAG |
|
| 851 | GCGCAGCCAA ATAA |
[0644]This encodes a protein having amino acid sequence <SEQ ID 222; ORF4a-1>:
[0000]| 1 | MKTFFKTLSA AALALILAAC GGQKDSAPAA SASAAADNGA AKKEIVFGTT | |
|
| 51 | VGDFGDMVKE QIQPELEKKG YTVKLVEFTD YVRPNLALAE GELDINVFQH |
|
| 101 | KPYLDDFKKE HNLDITEVFQ VPTAPLGLYP GKLKSLEEVK DGSTVSAPND |
|
| 151 | PSNFARVLVM LDELGWIKLK DGINPLTASK ADIAENLKNI KIVELEAAQL |
|
| 201 | PRSRADVDFA VVNGNYAISS GMKLTEALFQ EPSFAYVNWS AVKTADKDSQ |
|
| 251 | WLKDVTEAYN SDAFKAYAHK RFEGYKSPAA WNEGAAK* |
[0645]ORF4a-1 and ORF4-1 show 99.7% identity in 287 aa overlap:
[0000]
[0000]Homology with an Outer Membrane Protein of Pasteurella haemolitica (Accession q08869).
[0646]ORF4 and this outer membrane protein show 33% aa identity in 91aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0647]ORF4 shows 93.6% identity over a 94aa overlap with a predicted ORF (ORF4.ng) from N. gonorrhoeae:
[0000]
[0648]The complete length ORF4ng nucleotide sequence <SEQ ID 223> was predicted to encode a protein having amino acid sequence <SEQ ID 224>:
[0000]| 1 | MKTFFKTLST ASLALILAAC GGQKDSAPAA SAAAPSADNG AAKKEIVFGT | |
|
| 51 | TVGDFGDMVK EQIQAELEKK GYTVKLVEFT DYVRPNLALA EGELDINVFQ |
|
| 101 | HKPYLDDFKK EHNLDITEAF QVPTAPLGLY PGKLKSLEEV KDGSTVSAPN |
|
| 151 | DPSNFARALV MLNELGWIKL KDGINPLTAS KADIAENLKN IKIVELEAAQ |
|
| 201 | LPRSRADVDF AVVNGNYAIS SGMKLTEALF QEPSFAYVNW SAVKTADKDS |
|
| 251 | QWLKDVTEAY NSDAFKAYAH KRFEGYKYPA AWNEGAAK* |
[0649]Further analysis revealed the complete length ORF4ng DNA sequence <SEQ ID 225> to be:
[0000]| 1 | atgAAAACCT TCTTCAAAAC cctttccgcc gccgcaCTCG CGCTCATCCT | |
|
| 51 | CGCAGCCTGc ggCggtcaAA AAGACAGCGC GCCCgcagcc tctgcCGCCG |
|
| 101 | CCCCTTCTGC CGATAACGgc gCgGCGAAAA AAGAAAtcgt ctTCGGCACG |
|
| 151 | Accgtgggcg acttcggcgA TAtggTCAAA GAACAAATCC AagcCGAgct |
|
| 201 | gGAGAAAAAA GgctACACcg tcAAattggt cgaatttacc gactatgtGC |
|
| 251 | gCCCGAATCT GGCATTGGCG GAGGGCGAGT TGGACATCAA CGTCTTCCAA |
|
| 301 | CACAAACCCT ATCTTGACGA TTTCAAAAAA GAACACAACC TGGACATCAC |
|
| 351 | CGAAGCCTTC CAAGTGCCGA CCGCGCCTTT GGGACTGTAT CCGGGCAAAC |
|
| 401 | TGAAATCGCT GGAAGAAGTC AAAGACGGCA GCACCGTATC CGCGCCCAac |
|
| 451 | gACccgTCCA ACTTCGCACG CGCCTTGGTG ATGCTGAACG AACTGGGTTG |
|
| 501 | GATCAAACTC AAAGACGGCA TCAATCCGCT GACCGCATCC AAAGCCGACA |
|
| 551 | TCGCGGAAAA CCTGAAAAAC ATCAAAATCG TCGAGCTTGA AGCCGCACAA |
|
| 601 | CTGCCGCGCA GCCGCGCCGA CGTGGATTTT GCCGTCGTCA ACGGCAACTA |
|
| 651 | CGCCATAAGC AGCGGCATGA AGCTGACCGA AGCCCTGTTC CAAGAGCCGA |
|
| 701 | GCTTTGCCTA TGTCAACTGG TCTGCCgtcA AAACCGCCGA CAAAGACAGC |
|
| 751 | CAATGGCTTA AAGACGTAAC CGAGGCCTAT AACTCCGACG CGTTCAAAGC |
|
| 801 | CTACGCGCAC AAACGCTTCG AGGGCTACAA ATACCCTGCC GCATGGAATG |
|
| 851 | AAGGCGCAGC CAAATAA |
[0650]This encodes a protein having amino acid sequence <SEQ ID 226; ORF4ng-1>:
[0000]| 1 | MKTFFKTLSA AALALILAAC GGQKDSAPAA SAAAPSADNG AAKKEIVFGT | |
|
| 51 | TVGDFGDMVK EQIQAELEKK GYTVKLVEFT DYVRPNLALA EGELDINVFQ |
|
| 101 | HKPYLDDFKK EHNLDITEAF QVPTAPLGLY PGKLKSLEEV KDGSTVSAPN |
|
| 151 | DPSNFARALV MLNELGWIKL KDGINPLTAS KADIAENLKN IKIVELEAAQ |
|
| 201 | LPRSRADVDF AVVNGNYAIS SGMKLTEALF QEPSFAYVNW SAVKTADKDS |
|
| 251 | QWLKDVTEAY NSDAFKAYAH KRFEGYKYPA AWNEGAAK* |
[0651]This shows 97.6% identity in 288 aa overlap with ORF4-1:
[0000]
[0652]In addition, ORF4ng-1 shows significant homology with an outer membrane protein from the database:
[0000]
[0653]Based on this analysis, including the homology with the outer membrane protein of Pasteurella haemolitica, and on the presence of a putative prokaryotic membrane lipoprotein lipid attachment site in the gonococcal protein, it was predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0654]ORF4-1 (30 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIGS. 8A and 8B show, respectively, the results of affinity purification of the His-fusion and GST-fusion proteins. Purified His-fusion protein was used to immunise mice, whose sera were used for ELISA (positive result), Western blot (FIG. 8C), FACS analysis (FIG. 8D), and a bactericidal assay (FIG. 8E). These experiments confirm that ORF4-1 is a surface-exposed protein, and that it is a useful immunogen.
[0655]FIG. 8F shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF4-1.
Example 27
[0656]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 227>:
[0000]| 1 | CCTCGTCGTC CTCGGCATGC TCCAGTTTCA AGGGGCGATT TACTCCAAGG | |
|
| 51 | CGGTGGAACG TATGCTCGGC ACGGTCATCG GGCTGGGCGC GGGTTTGGGC |
|
| 101 | GTTTTATGGC TGAACCAGCA TTATTTCCAC GGCAACCTCC TCTTCTACCT |
|
| 151 | CACCGTCGGC ACGGCAAGCG CACTGGCCGG CTGGGCGGCG GTCGGCAAAA |
|
| 201 | ACGGCTACGT CCCTmTGCTG GCAGGGCTGA CGATGTGTAT GCTCATCGGC |
|
| 251 | GACAACGGCA GCGAATGGCT CGACAGCGGA CTCATGCGCG CCATGAACGT |
|
| 301 | CCTCATCGGC GyGGCCATCG CCATCGCCGC CGCCAAACTG CTGCCGCTGA |
|
| 351 | AATCCACACT GATGTGGCGT TTCATGCTTG CCGACAACCT GGCCGACTGC |
|
| 401 | AGCAAAATGA TTGCCGAAAT CAGCAACGGC AGGCGCATGA CCCGCGAACG |
|
| 451 | CCTCGAGGAG AACATGGCGA AAATGCGCCA AATCAACGCA CGCATGGTCA |
|
| 501 | AAAGCCGCAG CCATCTCGCC GCCACATCGG GCGAAAGCTG CATCAGCCCC |
|
| 551 | GCCATGATGG AAGCCATGCA GCACGCCCAC CGTAAAATCG TCAACACCAC |
|
| 601 | CGAGCTGCTC CTGACCACCG CCGCCAAGCT GCAATCTCCC AAACTCAACG |
|
| 651 | GCAGCGAAAT CCGGCTGCTT GACCGCCACT TCACACTGCT CCAAAC.... |
|
| 701 | ............................. GC AGACACGCCC GCCGCATCCG |
|
| 751 | CATCGACACC GCCATCAACC CCGAACTGGA AGCCCTCGCC GAACACCTCC |
|
| 801 | ACTACCAATG GCAGGGCTTC CTCTGGCTCA GCACCGATAT GCGTCAGGAA |
|
| 851 | ATTTCCGCCC TCGTCATCCT GCTGCAACGC ACCCGCCGCA AATGGCTGGA |
|
| 901 | TGCCCACGAA CGCCAACACC TGCGCCAAAG CCTGCTTGA |
[0657]This corresponds to the amino acid sequence <SEQ ID 228; ORF8>:
[0000]| 1 | ......PRRP RHAPVSRGDL LQGGGTYARH GHRAGRGFGR FMAEPALFPR | |
|
| 51 | QPPLLPHRRH GKRTGRLGGG RQKRLRPXAG RADDVYAHRR QRQRMARQRT |
|
| 101 | HARHERPHRR GHRHRRRQTA AAEIHTDVAF HACRQPGRLQ QNDCRNQQRQ |
|
| 151 | AHDPRTPRGE HGENAPNQRT HGQKPQPSRR HIGRKLHQPR HDGSHAARPP |
|
| 201 | XNRQHHRAAP DHRRQAAISQ TQRQRNPAAX PPLHTAPN.. .........Q |
|
| 251 | TRPPHPHRHR HQPRTGSPRR TPPLPMAGLP LAQHRYASGN FRPRHPAATH |
|
| 301 | PPQMAGCPRT PTPAPKPA* |
[0658]Computer analysis of this amino acid sequence gave the following results:
Sequence Motifs
[0659]ORF8 is proline-rich and has a distribution of proline residues consistent with a surface localization. Furthermore the presence of an RGD motif may indicate a possible role in bacterial adhesion events.
[0660]Homology with a Predicted ORF from N. gonorrhoeae
[0661]ORF8 shows 86.5% identity over a 312aa overlap with a predicted ORF (ORF8.ng) from N. gonorrhoeae.
[0000]
[0662]The complete length ORF8ng nucleotide sequence <SEQ ID 229> is predicted to encode a protein having amino acid sequence <SEQ ID 230>:
[0000]| 1 | MDRDDRLRRP RHAPVPRRDL LQRGGTYARY GHRAGRGFGR FMAEPALFPR | |
|
| 51 | QPPLLPDHRH GKRTGRLGGG RQKRLRPYVG GADDVHAHRR QRQRMARQRP |
|
| 101 | DARDERPHRR RHRHCRRQTA AAEIHTDVAF HACRQPGRLQ QNDCRNQQRQ |
|
| 151 | AYDARTFGAE YGQNAPNQRT HGQKPQPPRR HIGRKPHQPL HDGSHAARPP |
|
| 201 | QNRQHHRAAP DHRRQAAISQ TQRQRNPAAR PPLHTAPNRP ATNRRPHQRQ |
|
| 251 | TRPPHPHRHR HQPRTGSPRR TPPLPMAGFP LAQHQYASGN FRPRHPPATH |
|
| 301 | PPQMAGCPRT PTPAPKPA* |
[0663]Based on the sequence motifs in these proteins, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 28
[0664]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 231>:
[0000]| 1 | ..GAAATCAGCC TGCGGTCCGA CNACAGGCCG GTTTCCGTGN CGAAGCGGCG | |
|
| 51 | GGATTCGGAA CGTTTTCTGC TGTTGGACGG CGGCAACAGC CGGCTCAAGT |
|
| 101 | GGGCGTGGGT GGAAAACGGC ACGTTCGCAA CCGTCGGTAG CGCGCCGTAC |
|
| 151 | CGCGATTTGT CGCCTTTGGG CGCGGAGTGG GCGGAAAAGG CGGATGGAAA |
|
| 201 | TGTCCGCATC GTCGGTTGCG CTGTGTGCGG AGAATTCAAA AAGGCACAAG |
|
| 251 | TGCAGGAACA GCTCGCCCGA AAAATCGAGT GGCTGCCGTC TTCCGCACAG |
|
| 301 | GCTTT.GGCA TACGCAACCA CTACCGCCAC CCCGAAGAAC ACGGTTCCGA |
|
| 351 | CCGCTGGTTC AACGCCTTGG GCAGCCGCCG CTTCAGCCGC AACGCCTGCG |
|
| 401 | TCGTCGTCAG TTGCGGCACG GCGGTAACGG TTGACGCGCT CACCGATGAC |
|
| 451 | GGACATTATC TCGGAGA.GG AACCATCATG CCCGGTTTCC ACCTGATGAA |
|
| 501 | AGAATCGCTC GCCGTCCGAA CCGCCAACCT CAACCGGCAC GCCGGTAAGC |
|
| 551 | GTTATCCTTT CCCGACCGG.. |
[0665]This corresponds to the amino acid sequence <SEQ ID 232; ORF61>:
[0000]| 1 | ..EISLRSDXRP VSVXKRRDSE RFLLLDGGNS RLKWAWVENG TFATVGSAPY | |
|
| 51 | RDLSPLGAEW AEKADGNVRI VGCAVCGEFK KAQVQEQLAR KIEWLPSSAQ |
|
| 101 | AXGIRNHYRH PEEHGSDRWF NALGSRRFSR NACVVVSCGT AVTVDALTDD |
|
| 151 | GHYLGXGTIM PGFHLMKESL AVRTANLNRH AGKRYPFPT.. |
[0666]Further work revealed the complete nucleotide sequence <SEQ ID 233>:
[0000]| 1 | ATGACGGTTT TGAAGCTTTC GCACTGGCGG GTGTTGGCGG AGCTTGCCGA | |
|
| 51 | CGGTTTGCCG CAACACGTCT CGCAACTGGC GCGTATGGCG GATATGAAGC |
|
| 101 | CGCAGCAGCT CAACGGTTTT TGGCAGCAGA TGCCGGCGCA CATACGCGGG |
|
| 151 | CTGTTGCGCC AACACGACGG CTATTGGCGG CTGGTGCGCC CATTGGCGGT |
|
| 201 | TTTCGATGCC GAAGGTTTGC GCGAGCTGGG GGAAAGGTCG GGTTTTCAGA |
|
| 251 | CGGCATTGAA GCACGAGTGC GCGTCCAGCA ACGACGAGAT ACTGGAATTG |
|
| 301 | GCGCGGATTG CGCCGGACAA GGCGCACAAA ACCATATGCG TGACCCACCT |
|
| 351 | GCAAAGTAAG GGCAGGGGGC GGCAGGGGCG GAAGTGGTCG CACCGTTTGG |
|
| 401 | GCGAGTGTCT GATGTTCAGT TTTGGCTGGG TGTTTGACCG GCCGCAGTAT |
|
| 451 | GAGTTGGGTT CGCTGTCGCC TGTTGCGGCA GTGGCGTGTC GGCGCGCCTT |
|
| 501 | GTCGCGTTTA GGTTTGGATG TGCAGATTAA GTGGCCCAAT GATTTGGTTG |
|
| 551 | TCGGACGCGA CAAATTGGGC GGCATTCTGA TTGAAACGGT CAGGACGGGC |
|
| 601 | GGCAAAACGG TTGCCGTGGT CGGTATCGGC ATCAATTTTG TCCTGCCCAA |
|
| 651 | GGAAGTAGAA AATGCCGCTT CCGTGCAATC GCTGTTTCAG ACGGCATCGC |
|
| 701 | GGCGGGGCAA TGCCGATGCC GCCGTGCTGC TGGAAACGCT GTTGGTGGAA |
|
| 751 | CTGGACGCGG TGTTGTTGCA ATATGCGCGG GACGGATTTG CGCCTTTTGT |
|
| 801 | GGCGGAATAT CAGGCTGCCA ACCGCGACCA CGGCAAGGCG GTATTGCTGT |
|
| 851 | TGCGCGACGG CGAAACCGTG TTCGAAGGCA CGGTTAAAGG CGTGGACGGA |
|
| 901 | CAAGGCGTTT TGCACTTGGA AACGGCAGAG GGCAAACAGA CGGTCGTCAG |
|
| 951 | CGGCGAAATC AGCCTGCGGT CCGACGACAG GCCGGTTTCC GTGCCGAAGC |
|
| 1001 | GGCGGGATTC GGAACGTTTT CTGCTGTTGG ACGGCGGCAA CAGCCGGCTC |
|
| 1051 | AAGTGGGCGT GGGTGGAAAA CGGCACGTTC GCAACCGTCG GTAGCGCGCC |
|
| 1101 | GTACCGCGAT TTGTCGCCTT TGGGCGCGGA GTGGGCGGAA AAGGCGGATG |
|
| 1151 | GAAATGTCCG CATCGTCGGT TGCGCTGTGT GCGGAGAATT CAAAAAGGCA |
|
| 1201 | CAAGTGCAGG AACAGCTCGC CCGAAAAATC GAGTGGCTGC CGTCTTCCGC |
|
| 1251 | ACAGGCTTTG GGCATACGCA ACCACTACCG CCACCCCGAA GAACACGGTT |
|
| 1301 | CCGACCGCTG GTTCAACGCC TTGGGCAGCC GCCGCTTCAG CCGCAACGCC |
|
| 1351 | TGCGTCGTCG TCAGTTGCGG CACGGCGGTA ACGGTTGACG CGCTCACCGA |
|
| 1401 | TGACGGACAT TATCTCGGGG GAACCATCAT GCCCGGTTTC CACCTGATGA |
|
| 1451 | AAGAATCGCT CGCCGTCCGA ACCGCCAACC TCAACCGGCA CGCCGGTAAG |
|
| 1501 | CGTTATCCTT TCCCGACCAC AACGGGCAAT GCCGTCGCCA GCGGCATGAT |
|
| 1551 | GGATGCGGTT TGCGGCTCGG TTATGATGAT GCACGGGCGT TTGAAAGAAA |
|
| 1601 | AAACCGGGGC GGGCAAGCCT GTCGATGTCA TCATTACCGG CGGCGGCGCG |
|
| 1651 | GCAAAAGTTG CCGAAGCCCT GCCGCCTGCA TTTTTGGCGG AAAATACCGT |
|
| 1701 | GCGCGTGGCG GACAACCTCG TCATTTACGG GTTGTTGAAC ATGATTGCCG |
|
| 1751 | CCGAAGGCAG GGAATATGAA CATATTTAA |
[0667]This corresponds to the amino acid sequence <SEQ ID 234; ORF61-1>:
[0000]| 1 | MTVLKLSHWR VLAELADGLP QHVSQLARMA DMKPQQLNGF WQQMPAHIRG | |
|
| 51 | LLRQHDGYWR LVRPLAVFDA EGLRELGERS GFQTALKHEC ASSNDEILEL |
|
| 101 | ARIAPDKAHK TICVTHLQSK GRGRQGRKWS HRLGECLMFS FGWVFDRPQY |
|
| 151 | ELGSLSPVAA VACRRALSRL GLDVQIKWPN DLVVGRDKLG GILIETVRTG |
|
| 201 | GKTVAVVGIG INFVLPKEVE NAASVQSLFQ TASRRGNADA AVLLETLLVE |
|
| 251 | LDAVLLQYAR DGFAPFVAEY QAANRDHGKA VLLLRDGETV FEGTVKGVDG |
|
| 301 | QGVLHLETAE GKQTVVSGEI SLRSDDRPVS VPKRRDSERF LLLDGGNSRL |
|
| 351 | KWAWVENGTF ATVGSAPYRD LSPLGAEWAE KADGNVRIVG CAVCGEFKKA |
|
| 401 | QVQEQLARKI EWLPSSAQAL GIRNHYRHPE EHGSDRWFNA LGSRRFSRNA |
|
| 451 | CVVVSCGTAV TVDALTDDGH YLGGTIMPGF HLMKESLAVR TANLNRHAGK |
|
| 501 | RYPFPTTTGN AVASGMMDAV CGSVMMMHGR LKEKTGAGKP VDVIITGGGA |
|
| 551 | AKVAEALPPA FLAENTVRVA DNLVIYGLLN MIAAEGREYE HI* |
[0668]FIG. 9 shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF61-1. Further computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the baf Protein of B. pertussis (Accession Number U12020).
[0669]ORF61 and baf protein show 33% aa identity in 166aa overlap:
[0000]| orf61 | 23 | LLLDGGNSRLKWAWVE-NGTFATVGSAPYR----DLSPLGAEWAEKADGNVRIVGCAVCG | 77 | |
| | +L+D GNSRLK W + + A AP DL LG A R +G V G |
| baf | 3 | ILIDSGNSRLKVGWFDPDAPQAAREPAPVAFDNLDLDALGRWLATLPRRPQRALGVNVAG | 62 |
|
| orf61 | 78 | EFKKAQVQEQLAR---KIEWLPSSAQAXGIRNHYRHPEEHGSDRW---FNALGSRRFSRN | 131 |
| | + + L I WL + A G+RN YR+P++ G+DRW L + |
| baf | 63 | LARGEAIAATLRAGGCDIRWLRAQPLAMGLRNGYRNPDQLGADRWACMVGVLARQPSVHP | 122 |
|
| orf61 | 132 | ACVVVSCGTAVTVDALTDDGHYLGXGTIMPGFHLMKESLAVRTANL | 177 |
| | +V S GTA T+D + D + G G I+PG +M+ +LA TA+L |
| baf | 123 | PLLVASFGTATTLDTIGPDNVFPG-GLILPGPAMMRGALAYGTAHL | 167 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0670]ORF61 shows 97.4% identity over a 189aa overlap with an ORF (ORF61a) from strain A of N. meningitidis.
[0000]
[0671]The complete length ORF61a nucleotide sequence <SEQ ID 235> is:
[0000]| 1 | ATGACGGTTT TGAAGCCTTC GCACTGGCGG GTGTTGGCGG AGCTTGCCGA | |
|
| 51 | CGGTTTGCCG CAACACGTCT CGCAACTGGC GCGTATGGCG GATATGAAGC |
|
| 101 | CGCAGCAGCT CAACGGTTTT TGGCAGCAGA TGCCGGCGCA CATACGCGGG |
|
| 151 | CTGTTGCGCC AACACGACGG CTATTGGCGG CTGGTGCGCC CATTGGCGGT |
|
| 201 | TTTCGATGCC GAAGGTTTGC GCGAGCTGGG GGAAAGGTCG GGTTTTCAGA |
|
| 251 | CGGCATTGAA GCACGAGTGC GCGTCCAGCA ACGACGAGAT ACTGGAATTG |
|
| 301 | GCGCGGATTG CGCCGGACAA GGCGCACAAA ACCATATGTG TGACCCACCT |
|
| 351 | GCAAAGTAAG GGCAGGGGGC GGCAGGGGCG GAAGTGGTCG CACCGTTTGG |
|
| 401 | GCGAGTGTCT GATGTTCAGT TTTGGCTGGG TGTTTGACCG GCCGCAGTAT |
|
| 451 | GAGTTGGGTT CGCTGTCGCC TGTTGCGGCA GTGGCGTGCC GGCGCGCCTT |
|
| 501 | GTCGCGTTTG GGTTTGAAAA CGCAAATCAA GTGGCCAAAC GATTTGGTCG |
|
| 551 | TCGGACGCGA CAAATTGGGC GGCATTCTGA TTGAAACGGT CAGGACGGGC |
|
| 601 | GGCAAAACGG TTGCCGTGGT CGGTATCGGC ATCAATTTCG TGCTGCCCAA |
|
| 651 | GGAAGTGGAA AACGCCGCTT CCGTGCAATC GCTGTTTCAG ACGGCATCGC |
|
| 701 | GGCGGGGAAA TGCCGATGCC GCCGTGTTGC TGGAAACGCT GTTGGCGGAA |
|
| 751 | CTTGATGCGG TGTTGTTGCA ATATGCGCGG GACGGATTTG CGCCTTTTGT |
|
| 801 | GGCGGAATAT CAGGCTGCCA ACCGCGACCA CGGCAAGGCG GTATTGCTGT |
|
| 851 | TGCGCGACGG CGAAACCGTG TTCGAAGGCA CGGTTAAAGG CGTGGACGGA |
|
| 901 | CAAGGCGTTC TGCACTTGGA AACGGCAGAG GGCAAACAGA CGGTCGTCAG |
|
| 951 | CGGCGAAATC AGCCTGCGGT CCGACGACAG GCCGGTTTCC GTGCCGAAGC |
|
| 1001 | GGCGGGATTC GGAACGTTTT CTGCTGTTGG ACGGCGGCAA CAGCCGGCTC |
|
| 1051 | AAGTGGGCGT GGGTGGAAAA CGGCACGTTC GCAACCGTCG GTAGCGCGCC |
|
| 1101 | GTACCGCGAT TTGTCGCCTT TGGGCGCGGA GTGGGCGGAA AAGGTGGATG |
|
| 1151 | GAAATGTCCG CATCGTCGGT TGCGCCGTGT GCGGAGAATT CAAAAAGGCA |
|
| 1201 | CAAGTGCAGG AACAGCTCGC CCGAAAAATC GAGTGGCTGC CGTCTTCCGC |
|
| 1251 | ACAGGCTTTG GGCATACGCA ACCACTACCG CCACCCCGAA GAACACGGTT |
|
| 1301 | CCGACCGCTG GTTCAACGCC TTGGGCAGCC GCCGCTTCAG CCGCAACGCC |
|
| 1351 | TGCGTCGTCG TCAGTTGCGG CACGGCGGTA ACGGTTGACG CGCTCACCGA |
|
| 1401 | TGACGGACAT TATCTCGGGG GAACCATCAT GCCCGGTTTC CACCTGATGA |
|
| 1451 | AAGAATCGCT CGCCGTCCGA ACCGCCAACC TCAACCGGCA CGCCGGTAAG |
|
| 1501 | CGTTATCCTT TCCCGACCAC AACGGGCAAT GCCGTCGCCA GCGGCATGAT |
|
| 1551 | GGATGCGGTT TGCGGCTCGG TTATGATGAT GCACGGGCGT TTGAAAGAAA |
|
| 1601 | AAACCGGGGC GGGCAAGCCT GTCGATGTCA TCATTACCGG CGGCGGCGCG |
|
| 1651 | GCAAAAGTTG CCGAAGCCCT GCCGCCTGCA TTTTTGGCGG AAAATACCGT |
|
| 1701 | GCGCGTGGCG GACAACCTCG TCATTCACGG GCTGCTGAAC CTGATTGCCG |
|
| 1751 | CCGAAGGCGG GGAATCGGAA CATACTTAA |
[0672]This encodes a protein having amino acid sequence <SEQ ID 236>:
[0000]| 1 | MTVLKPSHWR VLAELADGLP QHVSQLARMA DMKPQQLNGF WQQMPAHIRG | |
|
| 51 | LLRQHDGYWR LVRPLAVFDA EGLRELGERS GFQTALKHEC ASSNDEILEL |
|
| 101 | ARIAPDKAHK TICVTHLQSK GRGRQGRKWS HRLGECLMFS FGWVFDRPQY |
|
| 151 | ELGSLSPVAA VACRRALSRL GLKTQIKWPN DLVVGRDKLG GILIETVRTG |
|
| 201 | GKTVAVVGIG INFVLPKEVE NAASVQSLFQ TASRRGNADA AVLLETLLAE |
|
| 251 | LDAVLLQYAR DGFAPFVAEY QAANRDHGKA VLLLRDGETV FEGTVKGVDG |
|
| 301 | QGVLHLETAE GKQTVVSGEI SLRSDDRPVS VPKRRDSERF LLLDGGNSRL |
|
| 351 | KWAWVENGTF ATVGSAPYRD LSPLGAEWAE KVDGNVRIVG CAVCGEFKKA |
|
| 401 | QVQEQLARKI EWLPSSAQAL GIRNHYRHPE EHGSDRWFNA LGSRRFSRNA |
|
| 451 | CVVVSCGTAV TVDALTDDGH YLGGTIMPGF HLMKESLAVR TANLNRHAGK |
|
| 501 | RYPFPTTTGN AVASGMMDAV CGSVMMMHGR LKEKTGAGKP VDVIITGGGA |
|
| 551 | AKVAEALPPA FLAENTVRVA DNLVIHGLLN LIAAEGGESE HT* |
[0673]ORF61a and ORF61-1 show 98.5% identity in 591 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0674]ORF61 shows 94.2% identity over a 189aa overlap with a predicted ORF (ORF61.ng) from N. gonorrhoeae.
[0000]
[0675]An ORF61ng nucleotide sequence <SEQ ID 237> was predicted to encode a protein having amino acid sequence <SEQ ID 238>:
[0000]| 1 | MFSFGWAFDR PQYELGSLSP VAALACRRAL GCLGLETQIK WPNDLVVGRD | |
|
| 51 | KLGGILIETV RAGGKTVAVV GIGINFVLPK EVENAASVQS LFQTASRRGN |
|
| 101 | ADAAVLLETL LAELGAVLEQ YAEEGFAPFL NEYETANRDH GKAVLLLRDG |
|
| 151 | ETVCEGTVKG VDGRGVLHLE TAEGEQTVVS GEISLRPDNR SVSVPKRPDS |
|
| 201 | ERFLLLEGGN SRLKWAWVEN GTFATVGSAP YRDLSPLGAE WAEKADGNVR |
|
| 251 | IVGCAVCGES KKAQVKEQLA RKIEWLPSSA QALGIRNHYR HPEEHGSDRW |
|
| 301 | FNALGSRRFS RNACVVVSCG TAVTVDALTD DGHYLGGTIM PGFHLMKESL |
|
| 351 | AVRTANLNRP AGKRYPFPTT TGNAVASGMM DAVCGSIMMM HGRLKEKNGA |
|
| 401 | GKPVDVIITG GGAAKVAEAL PPAFLAENTV RVADNLVIHG LLNLIAAEGG |
|
| 451 | ESEHA* |
[0676]Further analysis revealed the complete gonococcal DNA sequence <SEQ ID 239> to be:
[0000]| 1 | ATGACGGTTT TGAAGCCTTC GCATTGGCGG GTGTTGGCGG AGCTTGCCGA | |
|
| 51 | CGGTTTGCCG CAACACGTAT CGCAATTGGC GCGTGAGGCG GACATGAAGC |
|
| 101 | CGCAGCAGCT CAACGGTTTT TGGCAGCAGA TGCCGGCGCA TATACGCGGG |
|
| 151 | CTGTTGCGCC AACACGACGG CTATTGGCGG CTGGTGCGCC CCTTGGCGGT |
|
| 201 | TTTCGATGCC GAAGGTTTGC GCGATCTGGG GGAAAGGTCG GGTTTTCAGA |
|
| 251 | CGGCATTGAA GCACGAGTGC GCGTCCAGCA ACGACGAGAT ACTGGAATTG |
|
| 301 | GCGCGGATTG CGCCGGACAA GGCGCACAAA ACCATATGCG TGACCCACCT |
|
| 351 | GCAAAGTAAG GGCAGGGGGC GGCAGGGGCG GAAGTGGTCG CACCGTTTGG |
|
| 401 | GCGAGTGCCT GATGTTCAGT TTCGGCTGGG CGTTTGACCG GCCGCAGTAT |
|
| 451 | GAGTTGGGTT CGCTGTCGCC TGTTGCGGCA CTTGCGTGCC GGCGCGCTTT |
|
| 501 | GGGGTGTTTG GGTTTGGAAA CGCAAATCAA GTGGCCAAAC GATTTGGTCG |
|
| 551 | TCGGACGCGA CAAATTGGGC GGCATTCTGA TTGAAACAGT CAGGGCGGGC |
|
| 601 | GGTAAAACGG TTGCCGTGGT CGGTATCGGC ATCAATTTCG TGCTGCCCAA |
|
| 651 | GGAAGTGGAA AACGCCGCTT CCGTGCAGTC GCTGTTTCAG ACGGCATCGC |
|
| 701 | GGCGGGGCAA TGCCGATGCC GCCGTATTGC TGGAAACATT GCTTGCGGAA |
|
| 751 | CTGGGCGCGG TGTTGGAACA ATATGCGGAA GAAGGGTTCG CGCCATTTTT |
|
| 801 | AAATGAGTAT GAAACGGCCA ACCGCGACCA CGGCAAGGCG GTATTGCTGT |
|
| 851 | TGCGCGACGG CGAAACCGTG TGCGAAGGCA CGGTTAAAGG CGTGGACGGA |
|
| 901 | CGAGGCGTTC TGCACTTGGA AACGGCAgaa ggcgaACAGa cggtcgtcag |
|
| 951 | cggcgaaaTC AGcctGCggc ccgacaacaG GTCGGtttcc gtgccgaagc |
|
| 1001 | ggccggatTC GgaacgtTTT tTGCtgttgg aaggcgggaa cagccgGCTC |
|
| 1051 | AAGTGGGCGT GggtggAAAa cggcacgttc gcaaccgtgg gcagcgcgCc |
|
| 1101 | gtaCCGCGAT TTGTCGCCTT TGGGCGCGGA GTGGGCGGAA AAGGCGGATG |
|
| 1151 | GAAATGTCCG CATCGTCGGT TGCGCCGTGT GCGGAGAATC CAAAAAGGCA |
|
| 1201 | CAAGTGAAGG AACAGCTCGC CCGAAAAATC GAGTGGCTGC CGTCTTCCGC |
|
| 1251 | ACAGGCTTTG GGCATACGCA ACCACTACCG CCACCCCGAA GAACACGGTT |
|
| 1301 | CCGACCGTTG GTTCAACGCC TTGGGCAGCC GCCGCTTCAG CCGCAACGCC |
|
| 1351 | TGCGTCGTCG TCAGTTGCGG CACGGCGGTA ACGGTTGACG CGCTCACCGA |
|
| 1401 | TGACGGACAT TATCTCGGCG GAACCATCAT GCCCGGCTTC CACCTGATGA |
|
| 1451 | AAGAATCGCT CGCCGTCCGA ACCGCCAACC TCAACCGCCC CGCCGGCAAA |
|
| 1501 | CGTTACCCTT TCCCGACCAC AACGGGCAAC GCCGTCGCAA GCGGCATGAT |
|
| 1551 | GGACGCGGTT TGCGGCTCGA TAATGATGAT GCACGGCCGT TTGAAAGAAA |
|
| 1601 | AAAACGGCGC GGGCAAGCCT GTCGATGTCA TCATTACCGG CGGCGGCGCG |
|
| 1651 | GCGAAAGTCG CCGAAGCCCT GCCGCCTGCA TTTTTGGCGG AAAATACCGT |
|
| 1701 | GCGCGTGGCG GACAACCTCG TCATCCACGG GCTGCTGAAC CTGATTGCCG |
|
| 1751 | CCGAAGGCGG GGAATCGGAA CACGCTTAA |
[0677]This corresponds to the amino acid sequence <SEQ ID 240; ORF61ng-1>:
[0000]| 1 | MTVLKPSHWR VLAELADGLP QHVSQLAREA DMKPQQLNGF WQQMPAHIRG | |
|
| 51 | LLRQHDGYWR LVRPLAVFDA EGLRDLGERS GFQTALKHEC ASSNDEILEL |
|
| 101 | ARIAPDKAHK TICVTHLQSK GRGRQGRKWS HRLGECLMFS FGWAFDRPQY |
|
| 151 | ELGSLSPVAA LACRRALGCL GLETQIKWPN DLVVGRDKLG GILIETVRAG |
|
| 201 | GKTVAVVGIG INFVLPKEVE NAASVQSLFQ TASRRGNADA AVLLETLLAE |
|
| 251 | LGAVLEQYAE EGFAPFLNEY ETANRDHGKA VLLLRDGETV CEGTVKGVDG |
|
| 301 | RGVLHLETAE GEQTVVSGEI SLRPDNRSVS VPKRPDSERF LLLEGGNSRL |
|
| 351 | KWAWVENGTF ATVGSAPYRD LSPLGAEWAE KADGNVRIVG CAVCGESKKA |
|
| 401 | QVKEQLARKI EWLPSSAQAL GIRNHYRHPE EHGSDRWFNA LGSRRFSRNA |
|
| 451 | CVVVSCGTAV TVDALTDDGH YLGGTIMPGF HLMKESLAVR TANLNRPAGK |
|
| 501 | RYPFPTTTGN AVASGMMDAV CGSIMMMHGR LKEKNGAGKP VDVIITGGGA |
|
| 551 | AKVAEALPPA FLAENTVRVA DNLVIHGLLN LIAAEGGESE HA* |
[0678]ORF61ng-1 and ORF61-1 show 93.9% identity in 591 aa overlap:
[0000]
[0679]Based on this analysis, including the homology with the baf protein of B. pertussis and the presence of a putative prokaryotic membrane lipoprotein lipid attachment site, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 29
[0680]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 241>:
[0000]| 1 | ATGTTTTACC AAATCCTTGC CCTGATTATC TGGAGCAGCT CGTTTATTGC | |
|
| 51 | CGCCAAATAT GTCTATGGCG GCATCGATCC CGCATTGATG GTCGGCGTGC |
|
| 101 | GCCTGCTAAT TGCCGCGCTG CCTGCACTGC CCGCCTGCCG CCGTCATGTC |
|
| 151 | GGCAAGATTC CGCGTGAGGA ATGGAAGCCG TTGCTGATTG TGTCGTTCGT |
|
| 201 | CAACTATGTG CTGACCCTGC TGCTTCAGTT TGTCGGGTTG AAATACACTT |
|
| 251 | CCGCCGCCAG CGCATCGGTC ATTGTCGGAC TCGAGCCGCT GCTGATGGTG |
|
| 301 | TTTGTCGGAC ACTTTTTCTT CAACGACAAA GCGCGTGCCT ACCACTGGAT |
|
| 351 | ATGCGGCGCG GCGGCATTTG CCGGTGTCGC GCTGCTGATG GCGGGCGGTG |
|
| 401 | CGGaAGAGGG CGGCGaAGTC GGCTGGTTCG GCTGCCTGCT GGTGTTGTTG |
|
| 451 | GCGGGCGCGG GCTTTTGTGC CGCTATGCGT CCGACGCAAA GGCTGATTGC |
|
| 501 | ACGCATCGGC GCACCGGCAT TCACATCTGT TTCCATTGCC GCCGCATCGT |
|
| 551 | TGATGTGCCT GCCGTTTTCG CTTGCTTTGG CGCAAAGTTA TACCGTGGAC |
|
| 601 | TGGAGCGTCG GGATGGTATT GTCGCTGCTG TATTTGGGTT TGGGGTGC.. |
[0681]This corresponds to the amino acid sequence <SEQ ID 242; ORF62>:
[0000]| 1 | MFYQILALII WSSSFIAAKY VYGGIDPALM VGVRLLIAAL PALPACRRHV | |
|
| 51 | GKIPREEWKP LLIVSFVNYV LTLLLQFVGL KYTSAASASV IVGLEPLLMV |
|
| 101 | FVGHFFFNDK ARAYHWICGA AAFAGVALLM AGGAEEGGEV GWFGCLLVLL |
|
| 151 | AGAGFCAAMR PTQRLIARIG APAFTSVSIA AASLMCLPFS LALAQSYTVD |
|
| 201 | WSVGMVLSLL YLGLGC.. |
[0682]Further work revealed the complete nucleotide sequence <SEQ ID 243>:
[0000]| 1 | ATGTTTTACC AAATCCTTGC CCTGATTATC TGGAGCAGCT CGTTTATTGC | |
|
| 51 | CGCCAAATAT GTCTATGGCG GCATCGATCC CGCATTGATG GTCGGCGTGC |
|
| 101 | GCCTGCTAAT TGCCGCGCTG CCTGCACTGC CCGCCTGCCG CCGTCATGTC |
|
| 151 | GGCAAGATTC CGCGTGAGGA ATGGAAGCCG TTGCTGATTG TGTCGTTCGT |
|
| 201 | CAACTATGTG CTGACCCTGC TGCTTCAGTT TGTCGGGTTG AAATACACTT |
|
| 251 | CCGCCGCCAG CGCATCGGTC ATTGTCGGAC TCGAGCCGCT GCTGATGGTG |
|
| 301 | TTTGTCGGAC ACTTTTTCTT CAACGACAAA GCGCGTGCCT ACCACTGGAT |
|
| 351 | ATGCGGCGCG GCGGCATTTG CCGGTGTCGC GCTGCTGATG GCGGGCGGTG |
|
| 401 | CGGAAGAGGG CGGCGAAGTC GGCTGGTTCG GCTGCCTGCT GGTGTTGTTG |
|
| 451 | GCGGGCGCGG GCTTTTGTGC CGCTATGCGT CCGACGCAAA GGCTGATTGC |
|
| 501 | ACGCATCGGC GCACCGGCAT TCACATCTGT TTCCATTGCC GCCGCATCGT |
|
| 551 | TGATGTGCCT GCCGTTTTCG CTTGCTTTGG CGCAAAGTTA TACCGTGGAC |
|
| 601 | TGGAGCGTCG GGATGGTATT GTCGCTGCTG TATTTGGGTT TGGGGTGCGG |
|
| 651 | CTGGTACGCC TATTGGCTGT GGAACAAGGG GATGAGCCGT GTTCCTGCCA |
|
| 701 | ATGTTTCGGG ACTGTTGATT TCGCTCGAAC CCGTCGTCGG CGTGCTGCTG |
|
| 751 | GCGGTTTTGA TTTTGGGCGA ACACCTGTCG CCCGTGTCCG CCTTGGGCGT |
|
| 801 | GTTTGTCGTC ATCGCCGCCA CCTTGGTTGC CGGCCGGCTG TCGCATCAAA |
|
| 851 | AATAA |
[0683]This corresponds to the amino acid sequence <SEQ ID 244; ORF62-1>:
[0000]| 1 | MFYQILALII WSSSFIAAKY VYGGIDPALM VGVRLLIAAL PALPACRRHV | |
|
| 51 | GKIPREEWKP LLIVSFVNYV LTLLLQFVGL KYTSAASASV IVGLEPLLMV |
|
| 101 | FVGHFFFNDK ARAYHWICGA AAFAGVALLM AGGAEEGGEV GWFGCLLVLL |
|
| 151 | AGAGFCAAMR PTQRLIARIG APAFTSVSIA AASLMCLPFS LALAQSYTVD |
|
| 201 | WSVGMVLSLL YLGLGCGWYA YWLWNKGMSR VPANVSGLLI SLEPVVGVLL |
|
| 251 | AVLILGEHLS PVSALGVFVV IAATLVAGRL SHQK* |
[0684]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical Transmembrane Protein HI0976 of H. influenzae (Accession Number Q57147)
[0685]ORF62 and HI0976 show 50% aa identity in 114aa overlap:
[0000]| Orf62 | 1 | MFYQILALIIWSSSFIAAKYVYGGIDPALMVGVRXXXXXXXXXXXCRRHVGKIPREEWKP | 60 | |
| | M YQILAL+IWSSS I K Y +DP L+V VR R KI + K | |
| HI0976 | 1 | MYQILALLIWSSSLIVGKLTYSMMDPVLVVQVRLIIAMIIVMPLFLRRWKKIDKPMRKQ | 60 |
|
| Orf62 | 61 | LLIVSFVNYVLTLLLQFVGLKYTSAASASVIVGLEPLLMVFVGHFFFNDKARAY | 114 |
| | L ++F NY LLQF+GLKYTSA+SA ++GLEPLL+VFVGHFFF K + | |
| HI0976 | 61 | LWWLAFFNYTAVFLLQFIGLKYTSASSAVTMIGLEPLLVVFVGHFFFKTKQNGF | 114 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0686]ORF62 shows 99.5% identity over a 216aa overlap with an ORF (ORF62a) from strain A of N. meningitidis.
[0000]
[0687]The complete length ORF62a nucleotide sequence <SEQ ID 245> is:
[0000]| 1 | ATGTTTTACC AAATCCTTGC CCTGATTATC TGGAGCAGCT CGTTTATTGC | |
|
| 51 | CGCCAAATAT GTCTATGGCG GCATCGATCC CGCATTGATG GTCGGCGTGC |
|
| 101 | GCCTGCTGAT TGCTGCGCTG CCTGCACTGC CCGCCTGCCG CCGTCATGTC |
|
| 151 | GGCAAGATTC CGCGTGAGGA ATGGAAGCCG TTGCTGATTG TGTCGTTCGT |
|
| 201 | CAACTATGTG CTGACCCTGC TACTTCAGTT TGTCGGGTTG AAATACACTT |
|
| 251 | CCGCCGCCAG CGCATCGGTC ATTGTCGGAC TCGAGCCACT GCTGATGGTG |
|
| 301 | TTTGTCGGAC ACTTTTTCTT CAACGACAAA GCGCGTGCCT ACCACTGGAT |
|
| 351 | ATGCGGCGCG GCGGCATTTG CCGGTGTCGC GCTGCTGATG GCGGGCGGTG |
|
| 401 | CGGAAGAGGG CGGCGAAGTC GGCTGGTTCG GCTGCCTGCT GGTGTTGTTG |
|
| 451 | GCGGGCGCGG GCTTTTGTGC CGCTATGCGT CCGACGCAAA GGCTGATTGC |
|
| 501 | ACGCATCGGC GCACCGGCAT TCACATCTGT TTCCATTGCC GCCGCATCGT |
|
| 551 | TGATGTGCCT GCCGTTTTCG CTTGCTTTGG CGCAAAGTTA TACCGTGGAC |
|
| 601 | TGGAGCGTCG GAATGGTATT GTCGCTGCTG TATTTGGGCG TGGGGTGCAG |
|
| 651 | CTGGTACGCC TATTGGCTGT GGAACAAGGG GATGAGCCGT GTTCCTGCCA |
|
| 701 | ACGTTTCGGG ACTGTTGATT TCGCTCGAAC CCGTCGTCGG CGTGCTGCTG |
|
| 751 | GCGGTTTTGA TTTTGGGCGA ACACCTGTCG CCCGTGTCCG TCTTGGGCGT |
|
| 801 | GTTTGTCGTC ATCGCCGCCA CCTTGGTTGC CGGCCGGCTG TCGCATCAAA |
|
| 851 | AATAA |
[0688]This encodes a protein having amino acid sequence <SEQ ID 246>:
[0000]| 1 | MFYQILALII WSSSFIAAKY VYGGIDPALM VGVRLLIAAL PALPACRRHV | |
|
| 51 | GKIPREEWKP LLIVSFVNYV LTLLLQFVGL KYTSAASASV IVGLEPLLMV |
|
| 101 | FVGHFFFNDK ARAYHWICGA AAFAGVALLM AGGAEEGGEV GWFGCLLVLL |
|
| 151 | AGAGFCAAMR PTQRLIARIG APAFTSVSIA AASLMCLPFS LALAQSYTVD |
|
| 201 | WSVGMVLSLL YLGVGCSWYA YWLWNKGMSR VPANVSGLLI SLEPVVGVLL |
|
| 251 | AVLILGEHLS PVSVLGVFVV IAATLVAGRL SHQK* |
[0689]ORF62a and ORF62-1 show 98.9% identity in 284 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0690]ORF62 shows 99.5% identity over a 216aa overlap with a predicted ORF (ORF62.ng) from N. gonorrhoeae.
[0000]
[0691]The complete length ORF62ng nucleotide sequence <SEQ ID 247> is:
[0000]| 1 | ATGTTTTACC AAATCCTTGC CCTGATTATC TGGGGCAGCT CGTTTATTGC | |
|
| 51 | CGCCAAATAT GTCTATGGCG GCATCGATCC CGCATTGATG GTCGGCGTGC |
|
| 101 | GCCTGCTGAT TGCCGCGCTG CCTGCACTGC CCGCCTGCCG CCGTCATGTC |
|
| 151 | GGCAAGATTC CGCGTGAGGA ATGGAAGCCG TTGCTGATTG TGTCGTTCGT |
|
| 201 | CAACTATGTG CTGACCCTGC TGCTTCAGTT TGTCGGGTTG AAATACACTT |
|
| 251 | CCGCCGCCAG CGCATCGGTC ATTGTCGGAC TCGAGCCGCT GCTGATGGTG |
|
| 301 | TTTGTCGGAC ACTTTTTCTT CAACGACAAA GCGCGTGCCT ACCACTGGAT |
|
| 351 | ATGCGGCGCG GCGGCATTTG CCGGTGTCGC GCTGCTGATG GCGGGCGGTG |
|
| 401 | CGGAAGAGGG CGGCGAAGTC GGCTGGTTCG GCTGCCTGCT GGTGTTGTTG |
|
| 451 | GCGGGCGCGG GCTTTTGTGC CGCTATGCGT CCGACGCAAA GGCTGATTGC |
|
| 501 | CCGCATCGGC GCACCGGCAT TCACATCTGT TTCCATTGCC GCCGCATCGT |
|
| 551 | TGATGTGCCT GCCGTTTTCG CTTGCTTTGG CGCAAAGTTA TACCGTGGAC |
|
| 601 | TGGAGCGTCG GGATGGTATT GTCGCTGTTG TATTTGGGTT TGGGGTGCGG |
|
| 651 | CTGGTACGCC TATTGGCTGT GGAACAAGGG GATGAGCCGT GTTCCTGCCA |
|
| 701 | ACGCGTCGGG ACTGTTGATT TCGCTCGAAC CCGTCGTCGG CGTGCTGTTG |
|
| 751 | GCGGTTTTGA TTTTGGGCGA ACATTTATCG CCCGTGTCCG CCTTGGGCGT |
|
| 801 | GTTTGTCGTC ATCGCCGCCA CTTTCGCCGC CGGCCGGCTG TCGCGCAGGG |
|
| 851 | ACGCGCAAAA CGGCAATGCC GTCTGA |
[0692]This encodes a protein having amino acid sequence <SEQ ID 248>:
[0000]| 1 | MFYQILALII WGSSFIAAKY VYGGIDPALM VGVRLLIAAL PALPACRRHV | |
|
| 51 | GKIPREEWKP LLIVSFVNYV LTLLLQFVGL KYTSAASASV IVGLEPLLMV |
|
| 101 | FVGHFFFNDK ARAYHWICGA AAFAGVALLM AGGAEEGGEV GWFGCLLVLL |
|
| 151 | AGAGFCAAMR PTQRLIARIG APAFTSVSIA AASLMCLPFS LALAQSYTVD |
|
| 201 | WSVGMVLSLL YLGLGCGWYA YWLWNKGMSR VPANASGLLI SLEPVVGVLL |
|
| 251 | AVLILGEHLS PVSALGVFVV IAATFAAGRL SRRDAQNGNA V* |
[0693]ORF62ng and ORF62-1 show 97.9% identity in 283 aa overlap:
[0000]
[0694]Furthermore, ORF62ng shows significant homology to a hypothetical H. influenzae protein:
[0000]| sp|Q57147|Y976_HAEIN HYPOTHETICAL PROTEIN HI0976 >gi|1074589|pir||B64163 | |
| hypothetical protein HI0976 - Haemophilus influenzae (strain Rd KW20) |
| >gi|1574004 (U32778) hypothetical [Haemophilus influenzae] Length = 128 |
| Score = 106 bits (262), Expect = 2e−22 |
| Identities = 56/114 (49%), Positives = 68/114 (59%) |
|
| Query: | 1 | MFYQILALIIWGSSFIAAKYVYGGIDPALMVGVRXXXXXXXXXXXCRRHVGKIPREEWKP | 60 | |
| | M YQILAL+IW SS I K Y +DP L+V VR R KI + K | |
| Sbjct: | 1 | MLYQILALLIWSSSLIVGKLTYSMMDPVLVVQVRLIIAMIIVMPLFLRRWKKIDKPMRKQ | 60 |
|
| Query: | 61 | LLIVSFVNYVLTLLLQFVGLKYTSAASASVIVGLEPLLMVFVGHFFFNDKARAY | 114 |
| | L ++F NY LLQF+GLKYTSA+SA ++GLEPLL+VFVGHFFF K + | |
| Sbjct: | 61 | LWWLAFFNYTAVFLLQFIGLKYTSASSAVTMIGLEPLLVVFVGHFFFKTKQNGF | 114 |
[0695]Based on this analysis, including the homology with the transmembrane protein of H. influenzae and the putative leader sequence and several transmembrane domains in the gonococcal protein, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 30
[0696]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 249>:
[0000]| 1 | ATGCGCCGTT TTCTACCGAT CGCAGCCATA TGCGCmGwms TCCTGkkGTA | |
|
| 51 | sGGACTGACG GCGGCAACCG GCAGCACCAG TTCGCTGGCG GATTATTTCT |
|
| 101 | GGTGGATTGT TGCGTTCAGC GCAATGCTGC TGCTGGTGTT GTCCGCCGTT |
|
| 151 | TTGGCACGTT ATGTCATATT GCTGTTGAAA GACAGGCGCG ACGGCGTATT |
|
| 201 | CGGTTCGCtA srTyGCCAAA gsGCCTgkks TGGG.ATGTT TACGCTGGTT |
|
| 251 | GCCGkACTGC CCGGCGTGTT TCTGTTCGGC TTTCCCGCAC AGTTCATCAA |
|
| 301 | CGGCACGATT AATTCGTGGT TCGGCAACGA TACCCACGAG GCGCTTGAAC |
|
| 351 | GCAGCCTCAA TTTGAGCAAG TCCGCATTGA ATTTGGCGGC AGACAACGCC |
|
| 401 | CTCGGCAACG CCGTCCCCGT GCAGATAGAC CTCATCGGCG CGGCTTCCCT |
|
| 451 | GCCCGGGGAT ATGGGCAGGG TGCTGGAACA TTACGCCGGC AGCGGTTTTG |
|
| 501 | CCCAGCTTGC CCTGTACAAy ksCGCAAGCG GCAAAATCGA AAAAAGCATC |
|
| 551 | AACCCGCACA AGCTCGATCA GCCGTTTCCA GGTAAGGCGC GTTGGGAaAa |
|
| 601 | AATCCaACGG GCGGGTTCGG TCAGGGATTT GGAAAGCATA GGCGGCGTAT |
|
| 651 | TGTaCGCGCA GGGCTGGCTG TCGGCGGGTA CGCACwACGG GCGCGATTAC |
|
| 701 | GCCTTGTTTT TCCGTCAGCC GGTTCCCAAA GGCGTGGCAG AGGATGCCGT |
|
| 751 | yTTAATCGAA AAGGCAAGGG CGAAATATGC TGAGTTGAGT TACAGCAAAA |
|
| 801 | AAGGTTTGCA GACCTTTTTC CTGGCAACCC TGCTGATTGC CTCGCTGCTG |
|
| 851 | TCGATTTTTC TTGCACTGGT CATGGCACTG TATTTCGCCC GCCGTTTCGT |
|
| 901 | CGAACCCGTC CTATCGCTTG CCGAGGGGGC GAAGGCGGTG GCGCAAGGCG |
|
| 951 | ATTTCAGCCA GACGCGCCCC GTGTTGCGCA ACGACGAGTT CGGACGCTTG |
|
| 1001 | ACCArGTTGT TCAACCACAT GACCGAGCAG CTTTCCATCG CCAAAGATGC |
|
| 1051 | AGACGAGCGC AACCGCCGGC GCGAGGAAGC CGCCAGGCAT TATCTTGAAT |
|
| 1101 | GCGTGTTGGA GGGGCTGACC ACGGGCGTGG TGGTGTTTGA CGAACAAGGC |
|
| 1151 | TGTCTGAAAA CCTTCAACAA AGCGGCGGGT ACC.. |
[0697]This corresponds to the amino acid sequence <SEQ ID 250; ORF64>:
[0000]| 1 | MRRFLPIAAI CAXXLXXGLT AATGSTSSLA DYFWWIVAFS AMLLLVLSAV | |
|
| 51 | LARYVILLLK DRRDGVFGSX XAKXPXXXMF TLVAXLPGVF LFGFPAQFIN |
|
| 101 | GTINSWFGND THEALERSLN LSKSALNLAA DNALGNAVPV QIDLIGAASL |
|
| 151 | PGDMGRVLEH YAGSGFAQLA LYNXASGKIE KSINPHKLDQ PFPGKARWEK |
|
| 201 | IQRAGSVRDL ESIGGVLYAQ GWLSAGTHXG RDYALFFRQP VPKGVAEDAV |
|
| 251 | LIEKARAKYA ELSYSKKGLQ TFFLATLLIA SLLSIFLALV MALYFARRFV |
|
| 301 | EPVLSLAEGA KAVAQGDFSQ TRPVLRNDEF GRLTXLFNHM TEQLSIAKDA |
|
| 351 | DERNRRREEA ARHYLECVLE GLTTGVVVFD EQGCLKTFNK AAGT.. |
[0698]Further work revealed the complete nucleotide sequence <SEQ ID 251>:
[0000]| 1 | ATGCGCCGTT TTCTACCGAT CGCAGCCATA TGCGCCGTCG TCCTGTTGTA | |
|
| 51 | CGGACTGACG GCGGCAACCG GCAGCACCAG TTCGCTGGCG GATTATTTCT |
|
| 101 | GGTGGATTGT TGCGTTCAGC GCAATGCTGC TGCTGGTGTT GTCCGCCGTT |
|
| 151 | TTGGCACGTT ATGTCATATT GCTGTTGAAA GACAGGCGCG ACGGCGTATT |
|
| 201 | CGGTTCGCAG ATTGCCAAAC GCCTTTCTGG GATGTTTACG CTGGTTGCCG |
|
| 251 | TACTGCCCGG CGTGTTTCTG TTCGGCGTTT CCGCACAGTT CATCAACGGC |
|
| 301 | ACGATTAATT CGTGGTTCGG CAACGATACC CACGAGGCGC TTGAACGCAG |
|
| 351 | CCTCAATTTG AGCAAGTCCG CATTGAATTT GGCGGCAGAC AACGCCCTCG |
|
| 401 | GCAACGCCGT CCCCGTGCAG ATAGACCTCA TCGGCGCGGC TTCCCTGCCC |
|
| 451 | GGGGATATGG GCAGGGTGCT GGAACATTAC GCCGGCAGCG GTTTTGCCCA |
|
| 501 | GCTTGCCCTG TACAATGCCG CAAGCGGCAA AATCGAAAAA AGCATCAACC |
|
| 551 | CGCACAAGCT CGATCAGCCG TTTCCAGGTA AGGCGCGTTG GGAAAAAATC |
|
| 601 | CAACGGGCGG GTTCGGTCAG GGATTTGGAA AGCATAGGCG GCGTATTGTA |
|
| 651 | CGCGCAGGGC TGGCTGTCGG CGGGTACGCA CAACGGGCGC GATTACGCCT |
|
| 701 | TGTTTTTCCG TCAGCCGGTT CCCAAAGGCG TGGCAGAGGA TGCCGTCTTA |
|
| 751 | ATCGAAAAGG CAAGGGCGAA ATATGCTGAG TTGAGTTACA GCAAAAAAGG |
|
| 801 | TTTGCAGACC TTTTTCCTGG CAACCCTGCT GATTGCCTCG CTGCTGTCGA |
|
| 851 | TTTTTCTTGC ACTGGTCATG GCACTGTATT TCGCCCGCCG TTTCGTCGAA |
|
| 901 | CCCGTCCTAT CGCTTGCCGA GGGGGCGAAG GCGGTGGCGC AAGGCGATTT |
|
| 951 | CAGCCAGACG CGCCCCGTGT TGCGCAACGA CGAGTTCGGA CGCTTGACCA |
|
| 1001 | AGTTGTTCAA CCACATGACC GAGCAGCTTT CCATCGCCAA AGAAGCAGAC |
|
| 1051 | GAGCGCAACC GCCGGCGCGA GGAAGCCGCC AGGCATTATC TTGAATGCGT |
|
| 1101 | GTTGGAGGGG CTGACCACGG GCGTGGTGGT GTTTGACGAA CAAGGCTGTC |
|
| 1151 | TGAAAACCTT CAACAAAGCG GCGGAACAGA TTTTGGGGAT GCCGCTTACC |
|
| 1201 | CCCCTGTGGG GCAGCAGCCG GCACGGTTGG CACGGCGTTT CGGCGCAGCA |
|
| 1251 | GTCCCTGCTT GCCGAAGTGT TTGCCGCCAT CGGCGCGGCG GCAGGTACGG |
|
| 1301 | ACAAACCGGT CCATGTGAAA TATGCCGCGC CGGACGATGC CAAAATCCTG |
|
| 1351 | CTGGGCAAGG CAACCGTCCT GCCCGAAGAC AACGGCAACG GCGTGGTAAT |
|
| 1401 | GGTGATTGAC GACATCACCG TTTTGATACA CGCGCAAAAA GAAGCCGCGT |
|
| 1451 | GGGGCGAAGT GGCGAAGCGG CTGGCACACG AAATCCGCAA TCCGCTCACG |
|
| 1501 | CCCATCCAGC TTTCCGCCGA ACGGCTGGCG TGGAAATTGG GCGGGAAGCT |
|
| 1551 | GGATGAGCAG GATGCGCAAA TCCTGACGCG TTCGACCGAC ACCATCGTCA |
|
| 1601 | AACAGGTGGC GGCATTGAAG GAAATGGTCG AAGCATTCCG CAATTATGCG |
|
| 1651 | CGTTCCCCTT CGCTCAAATT GGAAAATCAG GATTTGAACG CCTTAATCGG |
|
| 1701 | CGATGTGTTG GCATTGTATG AAGCCGGTCC GTGCCGGTTT GCGGCGGAGC |
|
| 1751 | TTGCCGGCGA ACCGCTGACG GTGGCGGCGG ATACGACCGC CATGCGGCAG |
|
| 1801 | GTGCTGCACA ATATTTTCAA AAATGCCGCC GAAGCGGCGG AAGAAGCCGA |
|
| 1851 | TGTGCCCGAA GTCAGGGTAA AATCGGAAAC AGGGCAGGAC GGTCGGATTG |
|
| 1901 | TCCTGACGGT TTGCGACAAC GGCAAAGGGT TCGGCAGGGA AATGCTGCAC |
|
| 1951 | AACGCCTTCG AGCCGTATGT AACGGACAAA CCGGCGGGAA CGGGATTGGG |
|
| 2001 | TCTGCCTGTG GTGAAAAAAA TCATTGAAGA ACACGGCGGC CGCATCAGCC |
|
| 2051 | TGAGCAATCA GGATGCGGGT GGCGCGTGTG TCAGAATCAT CTTGCCAAAA |
|
| 2101 | ACGGTAAAAA CTTATGCGTA G |
[0699]This corresponds to the amino acid sequence <SEQ ID 252; ORF64-1>:
[0000]| 1 | MRRFLPIAAI CAVVLLYGLT AATGSTSSLA DYFWWIVAFS AMLLLVLSAV | |
|
| 51 | LARYVILLLK DRRDGVFGSQ IAKRLSGMFT LVAVLPGVFL FGVSAQFING |
|
| 101 | TINSWFGNDT HEALERSLNL SKSALNLAAD NALGNAVPVQ IDLIGAASLP |
|
| 151 | GDMGRVLEHY AGSGFAQLAL YNAASGKIEK SINPHKLDQP FPGKARWEKI |
|
| 201 | QRAGSVRDLE SIGGVLYAQG WLSAGTHNGR DYALFFRQPV PKGVAEDAVL |
|
| 251 | IEKARAKYAE LSYSKKGLQT FFLATLLIAS LLSIFLALVM ALYFARRFVE |
|
| 301 | PVLSLAEGAK AVAQGDFSQT RPVLRNDEFG RLTKLFNHMT EQLSIAKEAD |
|
| 351 | ERNRRREEAA RHYLECVLEG LTTGVVVFDE QGCLKTFNKA AEQILGMPLT |
|
| 401 | PLWGSSRHGW HGVSAQQSLL AEVFAAIGAA AGTDKPVHVK YAAPDDAKIL |
|
| 451 | LGKATVLPED NGNGVVMVID DITVLIHAQK EAAWGEVAKR LAHEIRNPLT |
|
| 501 | PIQLSAERLA WKLGGKLDEQ DAQILTRSTD TIVKQVAALK EMVEAFRNYA |
|
| 551 | RSPSLKLENQ DLNALIGDVL ALYEAGPCRF AAELAGEPLT VAADTTAMRQ |
|
| 601 | VLHNIFKNAA EAAEEADVPE VRVKSETGQD GRIVLTVCDN GKGFGREMLH |
|
| 651 | NAFEPYVTDK PAGTGLGLPV VKKIIEEHGG RISLSNQDAG GACVRIILPK |
|
| 701 | TVKTYA* |
|
[0700]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0701]ORF64 shows 92.6% identity over a 392aa overlap with an ORF (ORF64a) from strain A of N. meningitidis:
[0000]
[0702]The complete length ORF64a nucleotide sequence <SEQ ID 253> is:
[0000]| 1 | ATGCGCCGTT TTCTACCGAT CGCAGCCATA TGCGCCGTCG TCCTGTTGTA | |
|
| 51 | CGGACTGACG GCGGCAACCG GCAGCACCAG TTCGCTGGCG GATTATTTCT |
|
| 101 | GGTGGATTGT TGCGTTCAGC GCAATGCTGC TGCTGGTGTT GTCCGCCGTT |
|
| 151 | TTGGCACGTT ATGTCATATT GCTGTTGAAA GACAGGCGCG ACGGCGTATT |
|
| 201 | CGGTTCGCAG ATTGCCAAAC GCCTTTCCGG GATGTTTACG CTGGTTGCCG |
|
| 251 | TACTGCCCGG CGTGTTTCTG TTCGGCGTTT CCGCACAGTT TATCAACGGC |
|
| 301 | ACGATTAATT CGTGGTTCGG CAACGATACC CACGAGGCGC TTGAACGCAG |
|
| 351 | CCTCAATTTG AGCAAGTCCG CATTGAATCT GGCGGCAGAC AACGCCCTTG |
|
| 401 | GCAACGCCAT CCCCGTGCAG ATAGACNTCA TCGGCGCGGC TTCCCTGCCC |
|
| 451 | NGGGATATGG GCAGGGTGCT GGAACATTAC GCCGGCAGCG GTTTTGCCCA |
|
| 501 | GCTTGCCCTG TACAATGCCG CAAGCGGCAA AATCGAAAAA AGCATCAACC |
|
| 551 | CGCACAAGCT CGATCAGCCG TTTCCAGGTA AGGCGCGTTG GGAAAAAATC |
|
| 601 | CAACAGGCGG GTTCGGTCAG GGATNNGGAA AGCATAGGCG GCGTATTGTA |
|
| 651 | CGCGCANGGC TGGCTGTCGG CAGNNACGCA CAACGGGCGC GATTACGCCT |
|
| 701 | TGTTTTTCCG TCAGCCGGTT CCCAAAGGCG TGGCAGAGGA TGCCGTCTTA |
|
| 751 | ATCGAAAAGG CAAGGGCGNA ANANNNTNAG TTGAGTTACA GCAAAAAAGG |
|
| 801 | TTTGCAGACC TTTTTCCTNG CAACCCTGCT GATTGCCTCN CTGCTGTCGA |
|
| 851 | TTTTTCTTGC ACTGGTCATG GCACTGTATT TCGCCCGCCG TTTCGTCGAA |
|
| 901 | CCCGTCCTAT CGCTTGCCGA GGGGGCGAAG GCGGTGGCGC AAGGCGATTT |
|
| 951 | CAGCCAGACG CGCCCCGTGT TGCGCAACGA CGAGTTCGGA CGCTTGACCA |
|
| 1001 | AGTTGTTCAA CCACATGACC GAGCAGCTTT CCATCGCCAA AGAAGCAGAC |
|
| 1051 | GAGCGCAACC GCCGGCGCGA GGAAGCCGCC AGACATTATC TCGAATGCGT |
|
| 1101 | GTTGGAGGGG CTGACCACGG GCGTGGTGGT GTTTGACGAA CAAGGCTGTC |
|
| 1151 | TGAAAACCTT CAACAAAGCG GCGGAACAGA TTTTGGGGAT GCCGCTTACC |
|
| 1201 | CCCCTGTGGG GCAGCAGCCG GCACGGTTGG CACGGCGTTT CGGCGCAGCA |
|
| 1251 | GTCCCTGCTT GCCGAAGTGT TTGCCGCCAT CGGCGCGGCG GCAGGTACGG |
|
| 1301 | ACAAACCGGT CCATGTGAAA TATGCCGCGC CGGACGATGC CAAAATCCTG |
|
| 1351 | CTGGGCAAGG CAACCGTCCT GCCCGAAGAC AACNGCAACG GCGTGGTAAT |
|
| 1401 | GGTGATTGAC GACATCACCG TTTTGATACA CGCGCAAAAA GAAGCCGCGT |
|
| 1451 | GGGGCGAAGT GGCAAAACGG CTGGCACACG AAATCCGCAA TCCGCTCACG |
|
| 1501 | CCCATCCAGC TTTCTGCCGA ACGGCTGGCG TGGAAATTGG GCGGGAAGCT |
|
| 1551 | GGACGAGCAN GACGCGCAAA TCCTGACACG TTCGACCGAC ACCATCATCA |
|
| 1601 | AACAAGTGGC GGCATTAAAA GAAATGGTCG AGGCATTCCG CAATTACNCG |
|
| 1651 | CGTTCCCCTT CGNCTCAATT GGAAAATCAG GATTTGAACG CCTTAATCGG |
|
| 1701 | CGATGTGTTG GCATTGTACG AAGCTGGTCC GTGCCGGTTT GCGGCGGAAC |
|
| 1751 | TTGCCGGCGA ACCGCTGATG ATGGCGGCGG ATACGACCGC CATGCGGCAG |
|
| 1801 | GTGCTGCACA ATATTTTCAA AAATGCCGCC GAAGCGGCGG AAGAAGCCGA |
|
| 1851 | TGTGCCCGAA GTCAGGGTAA AATCGGAAGC GGGGCAGGAC GGACGGATTG |
|
| 1901 | TCCTGACAGT TTGCGACAAC GGCAAGGGGT TCGGCAGGGA AATGCTGCAC |
|
| 1951 | AATGCCTTCG AGCCGTATGT AACGGACAAA CCGGCTGGAA CGGGATTGNG |
|
| 2001 | ACTGCCCGTG GTGAAAAAAA TCATTGAAGA ACACGGCGGC CNCATCAGCC |
|
| 2051 | TGAGCAATCA GGATGCGGGC GGCGCGTNTG TCAGAATCAT CTTGCCAAAA |
|
| 2101 | ACGGTAGAAA CTTATGCGTA G |
[0703]This encodes a protein having amino acid sequence <SEQ ID 254>:
[0000]| 1 | MRRFLPIAAI CAVVLLYGLT AATGSTSSLA DYFWWIVAFS AMLLLVLSAV | |
|
| 51 | LARYVILLLK DRRDGVFGSQ IAKRLSGMFT LVAVLPGVFL FGVSAQFING |
|
| 101 | TINSWFGNDT HEALERSLNL SKSALNLAAD NALGNAIPVQ IDXIGAASLP |
|
| 151 | XDMGRVLEHY AGSGFAQLAL YNAASGKIEK SINPHKLDQP FPGKARWEKI |
|
| 201 | QQAGSVRDXE SIGGVLYAXG WLSAXTHNGR DYALFFRQPV PKGVAEDAVL |
|
| 251 | IEKARAXXXX LSYSKKGLQT FFLATLLIAS LLSIFLALVM ALYFARRFVE |
|
| 301 | PVLSLAEGAK AVAQGDFSQT RPVLRNDEFG RLTKLFNHMT EQLSIAKEAD |
|
| 351 | ERNRRREEAA RHYLECVLEG LTTGVVVFDE QGCLKTFNKA AEQILGMPLT |
|
| 401 | PLWGSSRHGW HGVSAQQSLL AEVFAAIGAA AGTDKPVHVK YAAPDDAKIL |
|
| 451 | LGKATVLPED NXNGVVMVID DITVLIHAQK EAAWGEVAKR LAHEIRNPLT |
|
| 501 | PIQLSAERLA WKLGGKLDEX DAQILTRSTD TIIKQVAALK EMVEAFRNYX |
|
| 551 | RSPSXQLENQ DLNALIGDVL ALYEAGPCRF AAELAGEPLM MAADTTAMRQ |
|
| 601 | VLHNIFKNAA EAAEEADVPE VRVKSEAGQD GRIVLTVCDN GKGFGREMLH |
|
| 651 | NAFEPYVTDK PAGTGLXLPV VKKIIEEHGG XISLSNQDAG GAXVRIILPK |
|
| 701 | TVETYA* |
[0704]ORF64a and ORF64-1 show 96.6% identity in 706 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0705]ORF64 shows 86.6% identity over a 387aa overlap with a predicted ORF (ORF64.ng) from N. gonorrhoeae:
[0000]
[0706]An ORF64ng nucleotide sequence <SEQ ID 255> was predicted to encode a protein having amino acid sequence <SEQ ID 256>:
[0000]| 1 | MRRFLPIAAI CAVVLLYGLT AATGSTSSLA DYFWWIVSFS AMLLLVLSAV | |
|
| 51 | LARYVILLLK DRRNGVFGSQ IAKRLSGMFT LVAVLPGLFL FGISAQFING |
|
| 101 | TINSWFGNDT HEALERSLNL SKSALDLAAD NAVSNAVPVQ IDLIGTASLS |
|
| 151 | GNMGSVLEHY AGSGFAQLAL YNAASGKIEK SINPHQFDQP LPDKEHWEQI |
|
| 201 | QQTGSVRSLE SIGGVLYAQG WLSAGTHNGR DYALFFRQPI PENVAQDAVL |
|
| 251 | IEKARAKYAE LSYSKKGLQT FFLVTLLIAS LLSIFLALVM ALYFARRFVE |
|
| 301 | PILSLAEGAK AVAQGDFSQT RPVLRNDEFG RLTKLFNHMT EQLSIAKEAD |
|
| 351 | ERNRRREEAA RHYLECVLDG LTTGVVVSYP LSCCRTAVFS TCHSSPLSYF* |
[0707]Further work revealed the complete gonococcal DNA sequence <SEQ ID 257>:
[0000]| 1 | ATGCGCCGCT TCCTACCGAT CGCAGCCATA TGCGCCGTCG TCCTGCTGTA | |
|
| 51 | CGGATTGACG GCGGCGACCG GCAGCACCAG TTCGCTGGCG GATTATTTCT |
|
| 101 | GGTGGATAGT CTCGTTCAGC GCAATGCTGC TGCTGGTGTT GTCCGCCGTT |
|
| 151 | TTGGCACGTT ATGTCATATT GCTGTTGAAA GACAGGCGCA ACGGCGTGTT |
|
| 201 | CGGTTCGCAG ATTGCCAAAC GCCTTTCCGG GATGTTCACG CTGGTCGCCG |
|
| 251 | TACTGCCCGG CTTGTTCCTG TTCGGCATTT CCGCGCAGTT TATCAACGGC |
|
| 301 | ACGATTAATT CGTGGTTCGG CAACGACACC CACGAAGCCC TCGAACGCAG |
|
| 351 | CCTTAATTTG AGCAAGTCCG CACTGGATTT GGCGGCAGAC AATGCCGTCA |
|
| 401 | GCAACGCCGT TCCCGTACAG ATAGACCTCA TCGGCACCGC CTCCCTGTCG |
|
| 451 | GGCAATATGG GCAGTGTGCT GGAACACTAC GCCGGCAGCG GTTTTGCCCA |
|
| 501 | GCTTGCCCTG TACAATGCCG CAAGCGGGAA AATCGAAAAA AGCATCAATC |
|
| 551 | CGCACCAATT CGACCAGCCG CTTCCCGACA AAGAACATTG GGAACAGATT |
|
| 601 | CAGCAGACCG GTTCGGTTCG GAGTTTGGAA AGCATAGGCG GCGTATTGTA |
|
| 651 | CGCGCAGGGA TGGTTGTCGG CAGGTACGCA CAACGGGCGC GATTACGCGC |
|
| 701 | TGTTCTTCCG CCAGCCGATT CCCGAAAATG TCGCACAGGA TGCCGTTCTG |
|
| 751 | ATTGAAAAGG CGCGGGCGAA ATATGCCGAA TTGAGTTACA GCAAAAAAGG |
|
| 801 | TTTGCAGACC TTTTTTCTGG TAACCCTGCT GATTGCCTCG CTGCTGTCGA |
|
| 851 | TTTTTCTTGC GCTGGTAATG GCACTGTATT TTGCCCGCCG TTTCGTCGAA |
|
| 901 | CCCATTCTGT CGCTTGCCGA GGGCGCAAAG GCGGTGGCGC AGGGTGATTT |
|
| 951 | CAGCCAGACG CGCCCCGTAT TGCGCAACGA CGAGTTCGGA CGTTTGACCA |
|
| 1001 | AGCTGTTCAA CCATATGACC GAGCAGCTTT CCATCGCCAA AGAAGCAGAC |
|
| 1051 | GAACGCAACC GCCGGCGCGA GGAAGCCGCC CGTCACTACC TCGAGTGCGT |
|
| 1101 | GTTGGATGGG TTGACTACCG GTGTGGTGGT GTTTGACGAA AAAGGCCGTT |
|
| 1151 | TGAAAACCTT CAACAAGGCG GCGGAACAGA TTTTGGGGAT GCCGCTCGCC |
|
| 1201 | CCCCTGTGGG GCAGCAGCCG GCACGGTTGG CACGGCGTTT CGGCGCAGCA |
|
| 1251 | GTCCCTGCTT GCCGAAGTGT TtgccgccAT CGGTGCGGCG GCAGGTACGG |
|
| 1301 | ACAAACCGGT CCAGGTGGAA TATGCCGCGC CGGACGATGC CAAAATCCTG |
|
| 1351 | CTGGGCAAGG CGACGGTATT GCCCGAAGAC AACGGCAACG GCGTGGTGAT |
|
| 1401 | GGTGATTGAC GACATCACCG TGCTGATACG CGCGCAAAAA GAAGCCGCGT |
|
| 1451 | GGGGTGAAGT GGCGAAGCGG CTGGCACACG AAATCCGCAA TCCGCTCACG |
|
| 1501 | CCCATCCAGC TTTCCGCCGA ACGGCTGGCG TGGAAATTGG GCGGGAAGCT |
|
| 1551 | GGACGATCAG GACGCGCAAA TCCTGACGCG TtcgACCGAC ACCATCATCA |
|
| 1601 | AACAGgtggc gGCGTTAAAA GAAATGGTCG AGGCATTCCG CAATTACGCG |
|
| 1651 | CGCGCCCCTT CGCTCAAACT GGAAAATCAG GATTTGAACG CCTTAATCGG |
|
| 1701 | CGATGTTTTG GCCCTGTACG AAGCCGGCCC GTGCCGGTTT GAGGCGGAAC |
|
| 1751 | TTGCCGGCGA ACCGCTGATG ATGGCGGCGG ATACGACCGC CATGCGGCAG |
|
| 1801 | GTGCTGCACA ATATTTTCAA AAATGCCGCC GAAGCGGCGG AAGAAGCCGA |
|
| 1851 | TATGCCCGAA GTCAGGGTAA AATCGGAAAC GGGGCAGGAC GGACGGATTG |
|
| 1901 | TCCTGACGGT TTGCGACAAC GGCAAGGGAT TCGGCAAGGA AATGCTGCAC |
|
| 1951 | AATGCTTTCG AGCCGTATGT GACGGATAAG CCGGCGGGAA CGGGACTGGG |
|
| 2001 | TCTGCCTGTA GTGAAAAAAA TCATTGGAGA ACACGGCGGC CGCATCAGCC |
|
| 2051 | TGAGCAATCA GGATGCGGGT GGGGCGTGTG TCAGAATCAT CTTGCCAAAA |
|
| 2101 | ACGGTAGAAA CTTATGCGTA G |
[0708]This corresponds to the amino acid sequence <SEQ ID 258; ORF64ng-1>:
[0000]| 1 | MRRFLPIAAI CAVVLLYGLT AATGSTSSLA DYFWWIVSFS AMLLLVLSAV | |
|
| 51 | LARYVILLLK DRRNGVFGSQ IAKRLSGMFT LVAVLPGLFL FGISAQFING |
|
| 101 | TINSWFGNDT HEALERSLNL SKSALDLAAD NAVSNAVPVQ IDLIGTASLS |
|
| 151 | GNMGSVLEHY AGSGFAQLAL YNAASGKIEK SINPHQFDQP LPDKEHWEQI |
|
| 201 | QQTGSVRSLE SIGGVLYAQG WLSAGTHNGR DYALFFRQPI PENVAQDAVL |
|
| 251 | IEKARAKYAE LSYSKKGLQT FFLVTLLIAS LLSIFLALVM ALYFARRFVE |
|
| 301 | PILSLAEGAK AVAQGDFSQT RPVLRNDEFG RLTKLFNHMT EQLSIAKEAD |
|
| 351 | ERNRRREEAA RHYLECVLDG LTTGVVVFDE KGRLKTFNKA AEQILGMPLA |
|
| 401 | PLWGSSRHGW HGVSAQQSLL AEVFAAIGAA AGTDKPVQVE YAAPDDAKIL |
|
| 451 | LGKATVLPED NGNGVVMVID DITVLIRAQK EAAWGEVAKR LAHEIRNPLT |
|
| 501 | PIQLSAERLA WKLGGKLDDQ DAQILTRSTD TIIKQVAALK EMVEAFRNYA |
|
| 551 | RAPSLKLENQ DLNALIGDVL ALYEAGPCRF EAELAGEPLM MAADTTAMRQ |
|
| 601 | VLHNIFKNAA EAAEEADMPE VRVKSETGQD GRIVLTVCDN GKGFGKEMLH |
|
| 651 | NAFEPYVTDK PAGTGLGLPV VKKIIGEHGG RISLSNQDAG GACVRIILPK |
|
| 701 | TVETYA* |
[0709]ORF64ng-1 and ORF64-1 show 93.8% identity in 706 aa overlap:
[0000]
[0710]Furthermore, ORF64ng-1 shows significant homology to a protein from A. caulinodans:
[0000]| sp|Q04850|NTRY_AZOCA NITROGEN REGULATION PROTEIN NTRY >gi|77479|pir||S18624 ntrY | |
| protein —Azorhizobium caulinodans >gi|38737 (X63841) NtrY gene product |
| [Azorhizobium caulinodans] Length = 771 |
| Score = 218 bits (550), Expect = 7e−56 |
| Identities = 195/720 (27%), Positives = 320/720 (44%), Gaps = 58/720 (8%) |
|
| Query: | 7 | IAAICAVVLLYGLTAATGSTSSLADYFWWIXXXXXXXXXXXXXXXXRYVILLLKDRRNGV | 66 | |
| | I+A+ ++L GLT + + + R + + K R G |
| Sbjct: | 35 | ISALATFLILMGLTPVVPTHQVVIS----VLLVNAAAVLILSAMVGREIWRIAKARARGR | 90 |
|
| Query: | 67 | FGSQIAKRLSGMFTLVAVLPGLFLFGISAQFINGTINSWFGNDTHEALERSLNLSKSALD | 126 |
| | +++ R+ G+F +V+V+P + + +++ ++ ++ WF T E + S++++++ + |
| Sbjct: | 91 | AAARLHIRIVGLFAVVSVVPAILVAVVASLTLDRGLDRWFSMRTQEIVASSVSVAQTYVR | 150 |
|
| Query: | 127 | LAADNAVSNAVPVQIDLIGTASLSGNMGSVLEHYAG--SGFAQLALYNAASGKIEKSINP | 184 |
| | A N + + + DL S+ Y G S F Q+ AA + ++ |
| Sbjct: | 151 | EHALNIRGDILAMSADLTRLKSV----------YEGDRSRFNQILTAQAALRNLPGAMLI | 200 |
|
| Query: | 185 | HQFDQPLPDKEHWEQIQQTGSVRSLESIGGVLYAQGWLSAGTHNGRDYA----------- | 233 |
| | + D + ++ + I + V + +IG Q + N DY |
| Sbjct: | 201 | RR-DLSVVERAN-VNIGREFIVPANLAIGDATPDQPVIYLP--NDADYVAAVVPLKDYDD | 256 |
|
| Query: | 234 | --LFFRQPIPENVAQDAVLIEKARAKYAELSYSKKGLQTFFLVTXXXXXXXXXXXXXVMA | 291 |
| | L+ + I V ++ A Y L + G+Q F + + |
| Sbjct: | 257 | LYLYVARLIDPRVIGYLKTTQETLADYRSLEERRFGVQVAFALMYAVITLIVLLSAVWLG | 316 |
|
| Query: | 292 | LYFARRFVEPILSLAEGAKAVAQGDFSQTRPVLRND-EFGRLTKLFNHMTEQLSIXXXXX | 350 |
| | L F++ V PI L A VA+G+ P+ R + + L + FN MT +L |
| Sbjct: | 317 | LNFSKWLVAPIRRLMSAADHVAEGNLDVRVPIYRAEGDLASLAETFNKMTHELRSQREAI | 376 |
|
| Query: | 351 | XXXXXXXXXXXHYLECVLDGLTTGVVVFDEKGRLKTFNKAAEQILGMPLAPLWGSSRHGW | 410 |
| | + E VL G+ GV+ D + R+ N++AE++LG L+ + RH |
| Sbjct: | 377 | LTARDQIDSRRRFTEAVLSGVGAGVIGLDSQERITILNRSAERLLG--LSEVEALHRHLA | 434 |
|
| Query: | 411 | HGVSAQQSLLAEVFXXXXXXXXTDKPVQVEYAAPDDAKILLGKATVLPEDNG---NGVVM | 467 |
| | V LL E + VQ D + + V E + +G V+ |
| Sbjct: | 435 | EVVPETAGLLEEA------EHARQRSVQGNITLTRDGRERVFAVRVTTEQSPEAEHGWVV | 488 |
|
| Query: | 468 | VIDDITVLIRAQKEAAWGEVAKRLAHEIRNPLTPIQLSAERLAWKLGGKLDDQDAQILTR | 527 |
| | +DDIT LI AQ+ +AW +VA+R+AHEI+NPLTPIQLSAERL K G + QD +I + |
| Sbjct: | 489 | TLDDITELISAQRTSAWADVARRIAHEIKNPLTPIQLSAERLKRKFGRHV-TQDREIFDQ | 547 |
|
| Query: | 528 | STDTIIKQVAALKEMVEAFRNYARAPSLKLENQDLNALIGDVLALYEAGPCRFEAELAGE | 587 |
| | TDTII+QV + MV+ F ++AR P +++QD++ +I + L G + |
| Sbjct: | 548 | CTDTIIRQVGDIGRMVDEFSSFARMPKPVVDSQDMSEIIRQTVFLMRVGHPEVVFDSEVP | 607 |
|
| Query: | 588 | PLMMAA-DTTAMRQVLHNIFKNXXXXXXXXDMPEVRVK------- SETGQDGRIVLTVCD | 639 |
| | P M A D + Q L NI KN P+VR + + G+D +V+ + D |
| Sbjct: | 608 | PAMPARFDRRLVSQALTNILKNAAEAIEAVP-PDVRGQGRIRVSANRVGED--LVIDIID | 664 |
|
| Query: | 640 | NGKGFGKEMLHNAFEPYVTDKPAGTGLGLPVVKKIIGEHGGRISLSNQDAG-GACVRIIL | 698 |
| | NG G +E + EPYVT + GTGLGL +V KI+ EHGG I L++ G GA +R+ L |
| Sbjct: | 665 | NGTGLPQESRNRLLEPYVTTREKGTGLGLAIVGKIMEEHGGGIELNDAPEGRGAWIRLTL | 724 |
[0711]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 31
[0712]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 259>:
[0000]| 1 | ATGTACGCAT TTACCGCCGC ACAGCAACAG AAGGCACTCT TCCGGCTGGT | |
|
| 51 | GCTTTTTCAT ATCCTCATCA TCGCCGCCAG CAACTATCTG GTGCAGTTCC |
|
| 101 | CTTTCCAAAT TTTCGGCATC CACACCACTT GGGGCGCATT TTCCTTTCCC |
|
| 151 | TTCATCTTCC TTGCCACCGA CCTGACCGTC CGCATTTTCG GTTCTCACTT |
|
| 201 | GGCACGGCGG ATTATCTTTT GGGTGATGTT CCCCGCCCTT TTGCTTTCCT |
|
| 251 | ACGTCTTTTC CGTTTTGTTC CACAACGGCA GTTGGACAGG CTTGGGCGCG |
|
| 301 | CTGTCCGAAT TCAACACCTT TGTCGGACGC ATCGCCTTAG CCAGCTTTGC |
|
| 351 | CGCCTACGCG ATCGGACAAA TCCTTGATAT TTTTGTATTC AACAAATTAC |
|
| 401 | GCCGTCTGAA AGCGTGGTGG ATTGCACCGA ACGCATCAAC CGTCATCGGG |
|
| 451 | CACGCGTTGG ATACG... |
[0713]This corresponds to the amino acid sequence <SEQ ID 260; ORF66>:
[0000]| 1 | MYAFTAAQQQ KALFRLVLFH ILIIAASNYL VQFPFQIFGI HTTWGAFSFP | |
|
| 51 | FIFLATDLTV RIFGSHLARR IIFWVMFPAL LLSYVFSVLF HNGSWTGLGA |
|
| 101 | LSEFNTFVGR IALASFAAYA IGQILDIFVF NKLRRLKAWW IAPNASTVIG |
|
| 151 | HALDT... |
[0714]Further work revealed the complete nucleotide sequence <SEQ ID 261>:
[0000]| 1 | ATGTACGCAT TTACCGCCGC ACAGCAACAG AAGGCACTCT TCCGGCTGGT | |
|
| 51 | GCTTTTTCAT ATCCTCATCA TCGCCGCCAG CAACTATCTG GTGCAGTTCC |
|
| 101 | CTTTCCAAAT TTTCGGCATC CACACCACTT GGGGCGCATT TTCCTTTCCC |
|
| 151 | TTCATCTTCC TTGCCACCGA CCTGACCGTC CGCATTTTCG GTTCTCACTT |
|
| 201 | GGCACGGCGG ATTATCTTTT GGGTGATGTT CCCCGCCCTT TTGCTTTCCT |
|
| 251 | ACGTCTTTTC CGTTTTGTTC CACAACGGCA GTTGGACAGG CTTGGGCGCG |
|
| 301 | CTGTCCGAAT TCAACACCTT TGTCGGACGC ATCGCCTTAG CCAGCTTTGC |
|
| 351 | CGCCTACGCG ATCGGACAAA TCCTTGATAT TTTTGTATTC AACAAATTAC |
|
| 401 | GCCGTCTGAA AGCGTGGTGG ATTGCACCGA CCGCATCAAC CGTCATCGGC |
|
| 451 | AACGCCTTGG ATACGCTGGT ATTTTTCGCC GTTGCCTTCT ACGCAAGCAG |
|
| 501 | CGATGGATTT ATGGCGGCAA ACTGGCAGGG CATCGCTTTT GTCGATTACC |
|
| 551 | TGTTCAAACT TACCGTCTGC ACCCTCTTCT TCCTGCCCGC CTACGGCGTG |
|
| 601 | ATACTGAATC TGCTGACGAA AAAACTGACA ACCCTGCAAA CCAAACAGGC |
|
| 651 | GCAAGACCGC CCCGCGCCCT CGCTGCAAAA TCCGTAA |
[0715]This corresponds to the amino acid sequence <SEQ ID 262; ORF66-1>:
[0000]| 1 | MYAFTAAQQQ KALFRLVLFH ILIIAASNYL VQFPFQIFGI HTTWGAFSFP | |
|
| 51 | FIFLATDLTV RIFGSHLARR IIFWVMFPAL LLSYVFSVLF HNGSWTGLGA |
|
| 101 | LSEFNTFVGR IALASFAAYA IGQILDIFVF NKLRRLKAWW IAPTASTVIG |
|
| 151 | NALDTLVFFA VAFYASSDGF MAANWQGIAF VDYLFKLTVC TLFFLPAYGV |
|
| 201 | ILNLLTKKLT TLQTKQAQDR PAPSLQNP* |
[0716]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the Hypothetical Protein o221 of E. Coli (Accession Number P37619)
[0717]ORF66 and o221 protein show 67% aa identity in 155aa overlap:
[0000]| orf66 | 1 | MYAFTAAQQQKALFRLVLFHILIIAASNYLVQFPFQIFGIHTTWGAFSFPFIFLATDLTV | 60 | |
| | M F+ Q+ KALF L LFH+L+I +SNYLVQ P I G HTTWGAFSFPFIFLATDLTV |
| o221 | 1 | MNVFSQTQRYKALFWLSLFHLLVITSSNYLVQLPVSILGFHTTWGAFSFPFIFLATDLTV | 60 |
|
| orf66 | 61 | RIFGSHLARRIIFWVMFPALLLSYVFSVLFHNGSWTGLGALSEFNTFVGRIALASFAAYA | 120 |
| | RIFG+ LARRIIF VM PALL+SYV S LF+ GSW G GAL+ FN FV RIA ASF AYA |
| o221 | 61 | RIFGAPLARRIIFAVMIPALLISYVISSLFYMGSWQGFGALAHFNLFVARIATASFMAYA | 120 |
|
| orf66 | 121 | IGQILDIFVFNKLRRLKAWWIAPNASTVIGHALDT | 155 |
| | +GQILD+ VFN+LR+ + WW+AP AST+ G+ DT |
| o221 | 121 | LGQILDVHVFNRLRQSRRWWLAPTASTLFGNVSDT | 155 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0718]ORF66 shows 96.1% identity over a 155aa overlap with an ORF (ORF66a) from strain A of N. meningitidis:
[0000]
[0719]The complete length ORF66a nucleotide sequence <SEQ ID 263> is:
[0000]| 1 | ATGTACGCAT TTACCGCCGC ACAGCAACAG AAGGCACTCT TCTGGCTGGT | |
|
| 51 | GCTTTTTCAT ATCCTCATCA TCGCCGCCAG CAACTATCTG GTGCAGTTCC |
|
| 101 | CCTTCCAAAT TTCCGGCATC CACACCACTT GGGGCGCGTT TTCCTTTCCC |
|
| 151 | TTCATCTTCC TCGCCACCGA CCTGACCGTC CGCATTTTCG GTTCGCACTT |
|
| 201 | GGCACGGCGG ATTATCTTTT GGGTCATGTT CCCCGCCCTT TTGCTTTCCT |
|
| 251 | ACGTCTTTTC CGTTTTGTTC CACAACGGCA GTTGGACGGG CTTGGGCGCG |
|
| 301 | CTGTCCGAAT TCAACACCTT TGTCGGACGC ATCGCGCTGG CAAGTTTTGC |
|
| 351 | CGCCTACGCG CTCGGACAAA TCCTTGATAT TTTTGTGTTC AACAAATTAC |
|
| 401 | GCCGTCTGAA AGCGTGGTGG GTTGCCCCGA CTGCATCAAC CGTCATCGGC |
|
| 451 | AACGCCTTAG ATACGTTGGT ATTTTTCGCC GTTGCCTTCT ACGCAAGCAG |
|
| 501 | CGATGGATTT ATGGCGGCAA ACTGGCAGGG CATCGCTTTT GTCGATTACC |
|
| 551 | TGTTCAAACT CACCGTCTGC GGTCTGTTTT TCCTGCCCGC CTACGGCGTG |
|
| 601 | ATTCTGAATC TGCTGACGAA AAAACTGACG ACCCTGCAAA CCAAACAGGC |
|
| 651 | GCAAGACCGC CCCGCGCCCT CGCTGCAAAA TCCGTAA |
[0720]This encodes a protein having amino acid sequence <SEQ ID 264>:
[0000]| 1 | MYAFTAAQQQ KALFWLVLFH ILIIAASNYL VQFPFQISGI HTTWGAFSFP | |
|
| 51 | FIFLATDLTV RIFGSHLARR IIFWVMFPAL LLSYVFSVLF HNGSWTGLGA |
|
| 101 | LSEFNTFVGR IALASFAAYA LGQILDIFVF NKLRRLKAWW VAPTASTVIG |
|
| 151 | NALDTLVFFA VAFYASSDGF MAANWQGIAF VDYLFKLTVC GLFFLPAYGV |
|
| 201 | ILNLLTKKLT TLQTKQAQDR PAPSLQNP* |
[0721]ORF66a and ORF66-1 show 97.8% identity in 228 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0722]ORF66 shows 94.2% identity over a 155aa overlap with a predicted ORF (ORF66.ng) from N. gonorrhoeae.
[0000]
[0723]The complete length ORF66ng nucleotide sequence <SEQ ID 265> is:
[0000]| 1 | ATGTACGCAT TGACCGCCGC ACAGCAACAG AAGGCACTCT TCCGGCTGGT | |
|
| 51 | GCTTTTCCAT ATCCTCATCA TCGCCGCCAG CAACTATCTG GTGCAGTTCC |
|
| 101 | CCTTCCGGAT TTTCGGCATC CACACCACTT GGGGCGCGTT TTCCTTTCCC |
|
| 151 | TTCATCTTCC TCGCCACCGA CCTGACCGTC CGCATTTTCG GTTCGCACTT |
|
| 201 | GGCGCGGCGG ATTATCTTTT GGGTGATGTT CCCCGCCCTT ttgCTTTcat |
|
| 251 | aCGTCTTTTC CGTTTTGTTC CACAACGGCA GTTGGACGGG CTTGGGCGCG |
|
| 301 | ctgTCCCAAT TCAACACCTT TGTCGGACGC ATCGCGCTGG CAAGTTTTGC |
|
| 351 | CGCCTACGCG CTCGGACAAA TCCTTGATAT TTTCGTATTC GACAAATTAC |
|
| 401 | GCCGTCTGAA AGCGTGGTGG ATTGCCCCGG CCGCATCAAC CGTCATCGGC |
|
| 451 | AATGCACTGG ACACGTTAGT ATTTTTTGCC GTTGCCTTTT ACGCAAGCAG |
|
| 501 | CGATGAATTT ATGGCGGCAA ACTGGCAGGG CATCGCTTTT GTCGATTACC |
|
| 551 | TGTTCAAACT TACCGTCTGC ACCCTCTTCT TCCTGCCCGC CTACGGCGTG |
|
| 601 | ATACTGAATC TGCTGACGAA AAAACTGACG GCCCTGCAAA CCAAACAGGC |
|
| 651 | GCAAGACCGC CCCGTGCCCT CGCTGCAAAA TCCGTAA |
[0724]This encodes a protein having amino acid sequence <SEQ ID 266>:
[0000]| 1 | MYALTAAQQQ KALFRLVLFH ILIIAASNYL VQFPFRIFGI HTTWGAFSFP | |
|
| 51 | FIFLATDLTV RIFGSHLARR IIFWVMFPAL SLSYVFSVLF HNGSWTGLGA |
|
| 101 | PSQFNTFVGR IALASFAAYA LGQILDIFVF DKLRRLKAWW IAPAASTVIG |
|
| 151 | NALDTLVFFA VAFYASSDEF MAANWQGIAF VDYLFKLTVC TLFFLPAYGV |
|
| 201 | ILNLLTKKLT ALQTKQAQDR PVPSLQNP* |
[0725]An alternative annotated sequence is:
[0000]| 1 | MYALTAAQQQ KALFRLVLFH ILIIAASNYL VQFPFRIFGI HTTWGAFSFP | |
|
| 51 | FIFLATDLTV RIFGSHLARR IIFWVMFPAL LLSYVFSVLF HNGSWTGLGA |
|
| 101 | LSQFNTFVGR IALASFAAYA LGQILDIFVF DKLRRLKAWW IAPAASTVIG |
|
| 151 | NALDTLVFFA VAFYASSDEF MAANWQGIAF VDYLFKLTVC TLFFLPAYGV |
|
| 201 | ILNLLTKKLT ALQTKQAQDR PVPSLQNP* |
[0726]ORF66ng and ORF66-1 show 96.1% identity in 228 aa overlap:
[0000]
[0727]Furthermore, ORF66ng shows significant homology with an E. coli ORF:
[0000]| sp|P37619|YHHQ_ECOLI HYPOTHETICAL 25.3 KD PROTEIN IN FTSY-NIKA INTERGENIC | |
| REGION (O221) |
| >gi|1073495|pir||S47690 hypothetical protein o221 - |
| Escherichia coli >gi|466607 (U00039) No definition line found |
| [Escherichia coli] >gi|1789882 (AE000423) hypothetical 25.3 kD protein in |
| ftsY-nikA intergenic region [Escherichia coli] |
| Length = 221 |
| Score = 273 bits (692), Expect = 5e−73 |
| Identities = 132/203 (65%), Positives = 155/203 (76%) |
|
| Query: | 1 | MYALTAAQQQKALFRLVLFHILIIAASNYLVQFPFRIFGIHTTWGAFSFPFIFLATDLTV | 60 | |
| | M + Q+ KALF L LFH+L+I +SNYLVQ P I G HTTWGAFSFPFIFLATDLTV |
| Sbjct: | 1 | MNVFSQTQRYKALFWLSLFHLLVITSSNYLVQLPVSILGFHTTWGAFSFPFIFLATDLTV | 60 |
|
| Query: | 61 | RIFGSHLARRIIFWVMFPALLLSYVFSVLFHNGSWTGLGALSQFNTFVGRIALASFAAYA | 120 |
| | RIFG+ LARRIIF VM PALL+SYV S LF+ GSW G GAL+ FN FV RIA ASF AYA |
| Sbjct: | 61 | RIFGAPLARRIIFAVMIPALLISYVISSLFYMGSWQGFGALAHFNLFVARIATASFMAYA | 120 |
|
| Query: | 121 | LGQILDIFVFDKLRRLKAWWIAPAASTVIGNALDTLVFFAVAFYASSDEFMAANWQGIAF | 180 |
| | LGQILD+ VF++LR+ + WW+AP AST+ GN DTL FF +AF+ S D FMA +W IA |
| Sbjct: | 121 | LGQILDVHVFNRLRQSRRWWLAPTASTLFGNVSDTLAFFFIAFWRSPDAFMAEHWMEIAL | 180 |
|
| Query: | 181 | VDYLFKLTVCTLFFLPAYGVILN | 203 |
| | VDY FK+ + +FFLP YGV+LN |
| Sbjct: | 181 | VDYCFKVLISIVFFLPMYGVLLN | 203 |
[0728]Based on this analysis, including the homology with the E. coli protein and the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 32
[0729]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 267>:
[0000]| 1 | ATGGTCATAA AATATACAAA TTTGAATTTT GCGAAATTGT CGATAATTGC | |
|
| 51 | AATTTTGATG ATGTATTCGT TTGAAGCGAA TGCAAAyGCA GTmwrAATAT |
|
| 101 | CTGAAACTGT TTCAGTTGAT ACCGGACAAG GTGCGAAAAT TCATAAGTTT |
|
| 151 | GTACCTAAAA ATAGTAAAAC TTATTCATCT GATTTAATAA AAACGGTAGA |
|
| 201 | TTTAACACAC AyyCCTACGG GCGCAAAAGC CCGAATCAAC GCCAAAATAA |
|
| 251 | CCGCCAGCGT ATCCCGCGCC GGCGTATTGG CGGGGGTCGG CAAACTTGCC |
|
| 301 | CGCTTAGgCG CGAAATTCAG CACAAGGGCG GTtCCCTATG TCGGAACAGC |
|
| 351 | CcTTTTAGCC CACGACGTAT ACGAAAcTTT CAAAGAAGAC ATACAGGCAC |
|
| 401 | GAGGCTACCA ATACGACCCC GAAACCGACA AATTTGTAAA AGGCTACGAA |
|
| 451 | TATAGTAATT GCCTTTGGTA CGAAGACAAA AGACGTATTA ATAGAACCTA |
|
| 501 | TGGCTGCTAC GGCGTTGAT.. |
[0730]This corresponds to the amino acid sequence <SEQ ID 268; ORF72>:
[0000]| 1 | MVIKYTNLNF AKLSIIAILM MYSFEANANA VXISETVSVD TGQGAKIHKF | |
|
| 51 | VPKNSKTYSS DLIKTVDLTH XPTGAKARIN AKITASVSRA GVLAGVGKLA |
|
| 101 | RLGAKFSTRA VPYVGTALLA HDVYETFKED IQARGYQYDP ETDKFVKGYE |
|
| 151 | YSNCLWYEDK RRINRTYGCY GVD.. |
[0731]Further work revealed the complete nucleotide sequence <SEQ ID 269>:
[0000]| 1 | ATGGTCATAA AATATACAAA TTTGAATTTT GCGAAATTGT CGATAATTGC | |
|
| 51 | AATTTTGATG ATGTATTCGT TTGAAGCGAA TGCAAATGCA GTAAAAATAT |
|
| 101 | CTGAAACTGT TTCAGTTGAT ACCGGACAAG GTGCGAAAAT TCATAAGTTT |
|
| 151 | GTACCTAAAA ATAGTAAAAC TTATTCATCT GATTTAATAA AAACGGTAGA |
|
| 201 | TTTAACACAC ATCCCTACGG GCGCAAAAGC CCGAATCAAC GCCAAAATAA |
|
| 251 | CCGCCAGCGT ATCCCGCGCC GGCGTATTGG CGGGGGTCGG CAAACTTGCC |
|
| 301 | CGCTTAGGCG CGAAATTCAG CACAAGGGCG GTTCCCTATG TCGGAACAGC |
|
| 351 | CCTTTTAGCC CACGACGTAT ACGAAACTTT CAAAGAAGAC ATACAGGCAC |
|
| 401 | GAGGCTACCA ATACGACCCC GAAACCGACA AATTTGCAAA GGTCTCAGGC |
|
| 451 | TAA |
[0732]This corresponds to the amino acid sequence <SEQ ID 270; ORF72-1>:
[0000]| 1 | MVIKYTNLNF AKLSIIAILM MYSFEANANA VKISETVSVD TGQGAKIHKF | |
|
| 51 | VPKNSKTYSS DLIKTVDLTH IPTGAKARIN AKITASVSRA GVLAGVGKLA |
|
| 101 | RLGAKFSTRA VPYVGTALLA HDVYETFKED IQARGYQYDP ETDKFAKVSG |
|
| 151 | * |
[0733]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0734]ORF72 shows 98.0% identity over a 147aa overlap with an ORF (ORF72a) from strain A of N. meningitidis.
[0000]
[0735]The complete length ORF72a nucleotide sequence <SEQ ID 271> is:
[0000]| 1 | ATGGTCATAA AATATACAAA TTTGAATTTT GCGAAATTGT CGATAATTGC | |
|
| 51 | AATTTTGATG ATGTATTCGT TTGAAGCGAA TGCAAATGCA GTAAAAATAT |
|
| 101 | CTGAAACTGT TTCAGTTGAT ACCGGACAAG GTGCGAAAAT TCATAAGTTT |
|
| 151 | GTACCTAAAA ATAGTAAAAC TTATTCATCT GATTTAATAA AAACGGTAGA |
|
| 201 | TTTAACACAC ATCCCTACGG GCGCAAAAGC CCGAATCAAC GCCAAAATAA |
|
| 251 | CCGCCAGCGT ATCCCGCGCC GGCGTATTGG CGGGGGTCGG CAAACTTGCC |
|
| 301 | CGCTTAGGCG CGAAATTCAG CACAAGGGCG GTTCCCTATG TCGGAACAGC |
|
| 351 | CCTTTTAGCC CACGACGTAT ACGAAACTTT CAAAGAAGAC ATACAGGCAC |
|
| 401 | GAGGCTACCA ATACGACCCC GAAACCGACA AATTTGCAAA GGTCTCAGGC |
|
| 451 | TAA |
[0736]This encodes a protein having amino acid sequence <SEQ ID 272>:
[0000]| 1 | MVIKYTNLNF AKLSIIAILM MYSFEANANA VKISETVSVD TGQGAKIHKF | |
|
| 51 | VPKNSKTYSS DLIKTVDLTH IPTGAKARIN AKITASVSRA GVLAGVGKLA |
|
| 101 | RLGAKFSTRA VPYVGTALLA HDVYETFKED IQARGYQYDP ETDKFAKVSG |
|
| 151 | * |
[0737]ORF72a and ORF72-1 show 100.0% identity in 150 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0738]ORF72 shows 89% identity over a 173aa overlap with a predicted ORF (ORF72.ng) from N. gonorrhoeae:
[0000]
[0739]An ORF72ng nucleotide sequence <SEQ ID 273> was predicted to encode a protein having amino acid sequence <SEQ ID 274>:
[0000]| 1 | MVTKHTNLNF AKLSIIAILM MYSFEANANA VKISETLSVD TGQGAKVHKF | |
|
| 51 | VPKSSNIYSS DLTKAVDLTH IPTGAKARIN AKITASVSRA GVLSGVGKLV |
|
| 101 | RQGAKFGTRA VPYVGTALLA HDVYETFKED IQARGCRYDP ETDKFVKGYE |
|
| 151 | YANCLWYEDE RRINRTYGCY GVDSSIMRLM PDRSRFPEVK QLMESQMYRL |
|
| 201 | ARPFWNWRKE ELNKLSSLDW NNFVLNRCTF DWNGGGCAVN KGDDFRAGAS |
|
| 251 | FSLGRNPKYK EEMDAKKPEE ILSLKVDADP DKYIEATGYP GYSEKVEVAP |
|
| 301 | GTKVNMGPVT DRNGNPVQVA ATFGRDAQGN TTADVQVIPR PDLTPASAEA |
|
| 351 | PHAQPLPEVS PAENPANNPD PDENPGTRPN PEPDPDLNPD ANPDTDGQPG |
|
| 401 | TSPDSPAVPD RPNGRHRKER KEGEDGGLSC DYFPEILACQ EMGKPSDRMF |
|
| 451 | HDISIPQVTD DKTWSSHNFL PSNGVCPQPK TFHVFGRQYR ASYEPLCVFA |
|
| 501 | EKIRFAVLLA FIIMSAFVVF GSLGGE* |
[0740]After further analysis, the following gonococcal DNA sequence <SEQ ID 275> was identified:
[0000]| 1 | ATGGTCACAA AACATACAAA TTTGAATTTT GCGAAATTGT CGATAATTGC | |
|
| 51 | AATTTTGATG ATGTATTCGT TTGAAGCGAA TGCAAATGCA GTAAAAATAT |
|
| 101 | CTGAAACTCT TTCGGTTGAT ACCGGACAAG GCGCGAAAGT TCATAAGTTC |
|
| 151 | GTTCCTAAAT CAAGTAATAT TTATTCATCT GATTTAACAA AAGCGGTAGA |
|
| 201 | TTTAACGCAT ATCCCCACGG GCGCAAAAGC CCGAATCAAC GCCAAAATAA |
|
| 251 | CCGCCAGCGT ATCCCGCGCC GGCGTATTGT CGGGGGTCGG CAAACTTGTC |
|
| 301 | CGCCAAGGCG CGAAATTCGG CACAAGGGCG GTTCCCTATG TCGGAACAGC |
|
| 351 | CCTTTTAGCC CACGACGTAT ACGAAACTTT CAAAGAAGAC ATACAGGCAC |
|
| 401 | GAGGCTGCCG ATACGATCCC GAAACCGACA AATTT |
[0741]This corresponds to the amino acid sequence <SEQ ID 276; ORF72ng-1>:
[0000]| 1 | MVTKHTNLNF AKLSIIAILM MYSFEANANA VKISETLSVD TGQGAKVHKF | |
|
| 51 | VPKSSNIYSS DLTKAVDLTH IPTGAKARIN AKITASVSRA GVLSGVGKLV |
|
| 101 | RQGAKFGTRA VPYVGTALLA HDVYETFKED IQARGCRYDP ETDKF |
[0742]ORF72ng-1 and ORF721-1 show 89.7% identity in 145 aa overlap:
[0000]
[0743]Based on this analysis, including the presence of a putative leader sequence and transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 33
[0744]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 277>:
[0000]| 1 | ATGAGATTTT TCGGTATCGG TTTTTTGGTG CTGCTGTTTT TGGAGATTAT | |
|
| 51 | GTCGATTGTG TGGGTTGCCG ATTGGCTGGG CGGCGGCTGG ACGTTGTTTT |
|
| 101 | TGATGGCGGC AGGTTTTGCC GCCGGCGTGC TGATGCTCAG GCAAACCGGG |
|
| 151 | GCTGACCGGT CTTTTATTGG CGGGCGCGGC AATGAGAAGC GGCGGGAAGG |
|
| 201 | TATCCGTTTA TCAGATGTTG TGGCCTATC.. |
[0745]This corresponds to the amino acid sequence <SEQ ID 278; ORF73>:
[0000]| 1 | MRFFGIGFLV LLFLEIMSIV WVADWLGGGW TLFLMAAGFA AGVLMLRQTG | |
|
| 51 | LTGLLLAGAA MRSGGKVSVY QMLWPI.. |
[0746]Further work revealed the complete nucleotide sequence <SEQ ID 279>:
[0000]| 1 | ATGAGATTTT TCGGTATCGG TTTTTTGGTG CTGCTGTTTT TGGAGATTAT | |
|
| 51 | GTCGATTGTG TGGGTTGCCG ATTGGCTGGG CGGCGGCTGG ACGTTGTTTT |
|
| 101 | TGATGGCGGC AGGTTTTGCC GCCGGCGTGC TGATGCTCAG GCATACGGGG |
|
| 151 | CTGTCCGGTC TTTTATTGGC GGGCGCGGCA ATGAGAAGCG GCGGGAGGGT |
|
| 201 | ATCCGTTTAT CAGATGTTGT GGCCTATCCG TTATACGGTG GCGGCTGTGT |
|
| 251 | GTCTGATGAG TCCGGGATTC GTATCCTCGG TGTTGGCGGT ATTGCTGCTG |
|
| 301 | CTGCCGTTTA AGGGAGGGGC AGTGTTGCAG GCAGGAGGTG CGGAAAATTT |
|
| 351 | TTTCAACATG AACCAATCGG GCAGAAAAGA GGGCTTTTCC CGCGATGACG |
|
| 401 | ATATTATCGA GGGAGAATAT ACGGTTGAAG AGCCTTACGG CGGCAATCGT |
|
| 451 | TCCCGAAACG CCATCGAACA CAAAAAAGAC GAATAA |
[0747]This corresponds to the amino acid sequence <SEQ ID 280; ORF73-1>:
[0000]| 1 | MRFFGIGFLV LLFLEIMSIV WVADWLGGGW TLFLMAAGFA AGVLMLRHTG | |
|
| 51 | LSGLLLAGAA MRSGGRVSVY QMLWPIRYTV AAVCLMSPGF VSSVLAVLLL |
|
| 101 | LPFKGGAVLQ AGGAENFFNM NQSGRKEGFS RDDDIIEGEY TVEEPYGGNR |
|
| 151 | SRNAIEHKKD E* |
[0748]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0749]ORF73 shows 90.8% identity over a 76aa overlap with an ORF (ORF73a) from strain A of N. meningitidis.
[0000]
[0750]The complete length ORF73a nucleotide sequence <SEQ ID 281> is:
[0000]| 1 | ATGAGATTTT TCGGTATCGG TTTTTTGGTG CTGCTGTTTT TGGAGATTAT | |
|
| 51 | GTCGATTGTG TGGGTTGCCG ATTGGTTGGG CGGCGGTTGG ACGCTGTTTC |
|
| 101 | TAATGGCGGC AACCTTTGCC GCCGGCGTGG TGATGCTCAG GCATACGGGG |
|
| 151 | CTGTCCGGTC TTTTATTGGC GGGCGCGGCA ATGAGAAGCG GCGGGAGGGT |
|
| 201 | ATCCGTTTAT CANATGTTGT GGCNTATCCG TTATACGGTG GCGGCGGTGT |
|
| 251 | GTCNGATGAG TCCGGGATTC GTATCCTCGG TGTNGGCGGT ATTGCTGNTG |
|
| 301 | CTNCCGTTTA AGGGAGGTGC AGTGTTGCAG GCAGGAGGTG CGGAAAATTT |
|
| 351 | TTTCAACATG AACCANTCGG GCAGAAAAGA NGGCNTTTCC CGCGATGACG |
|
| 401 | ATATTATCGA GGGGGAATAT ACGGTTGAAG ANCCTTACGG CGGCANTCGT |
|
| 451 | TTCCGAAACG CCNTNGAACA CAAAAAAGAC GAATAA |
[0751]This encodes a protein having amino acid sequence <SEQ ID 282>:
[0000]| 1 | MRFFGIGFLV LLFLEIMSIV WVADWLGGGW TLFLMAATFA AGVVMLRHTG | |
|
| 51 | LSGLLLAGAA MRSGGRVSVY XMLWXIRYTV AAVCXMSPGF VSSVXAVLLX |
|
| 101 | LPFKGGAVLQ AGGAENFFNM NXSGRKXGXS RDDDIIEGEY TVEXPYGGXR |
|
| 151 | FRNAXEHKKD E* |
[0752]ORF73a and ORF73-1 show 91.3% identity in 161 aa overlap
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0753]ORF73 shows 92.1% identity over a 76aa overlap with a predicted ORF (ORF73.ng) from N. gonorrhoeae:
[0000]
[0754]The complete length ORF73ng nucleotide sequence <SEQ ID 283> is:
[0000]| 1 | ATGAGATTTT TCGGTATCGG TTTTTTGGTG CTGCTGTTTT TGGAAATTAT | |
|
| 51 | GTCGATTGTG TGGGTTGCCG ATTGGCTGGG CGGCGGTTGG AcgcTGTTTC |
|
| 101 | TAATGGCGGC AACCTTTGCC GCCGGTGTGC TGATGCTCAG GCATAcggGG |
|
| 151 | CTGTCCGGTC TTTTATTGGC TGGCGCGGCG GTAAAAagta gtgGGAAGGT |
|
| 201 | ATCTGTTTAT CagatgtTGT GGCCTATCCG TTATAcggtg gcggcggtgT |
|
| 251 | GTCTGatgag tCcggGATTC GTATCCTccg tgttggCGGT ATTGCTGCTG |
|
| 301 | CTGCcgttta aggGaggGgc agtgttgcag gcaggaggtg cggaaaATTT |
|
| 351 | TTTCAACATg aaCcaatcgg gcagaaAaga gggatttttc cacgatgacg |
|
| 401 | atattatcga gggagaatat acggttgaaa aacctgacgg cggcaatcgt |
|
| 451 | tcccgaAAcg ccatcgaaca cgaaaAagac gaataA |
[0755]This encodes a protein having amino acid sequence <SEQ ID 284>:
[0000]| 1 | MRFFGIGFLV LLFLEIMSIV WVADWLGGGW TLFLMAATFA AGVLMLRHTG | |
|
| 51 | LSGLLLAGAA VKSSGKVSVY QMLWPIRYTV AAVCLMSPGF VSSVLAVLLL |
|
| 101 | LPFKGGAVLQ AGGAENFFNM NQSGRKEGFF HDDDIIEGEY TVEKPDGGNR |
|
| 151 | SRNAIEHEKD E* |
[0756]ORF73ng and ORG73-1 show 93.8% identity in 161 aa overlap
[0000]
[0757]Based on this analysis, including the presence of a putative leader sequence and putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 34
[0758]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 285>:
[0000]| 1 | ATGTTTGTTT TTCAGACGGC ATTCTT.ATG TTTCAGAAAC ATTTGCAGAA | |
|
| 51 | AGCCTCCGAC AGCGTCGTCG GAGGGACATT ATACGTGGTT GCCACGCCCA |
|
| 101 | TCGGCAATTT GGCGGACATT ACCCTGCGCG CTTTGGCGGT ATTGCAAAAG |
|
| 151 | GCG....... .....GCCGA AGACACGCGC GTTACCGCAC AGCTTTTGAG |
|
| 201 | CGCGTACGGC ATTCAGGGCA AACTCGTCAG TGTGCGCGAA CACAACGAAC |
|
| 251 | GGCAGATGGC GGACAAGATT GTCGGCTATC TTTCAGACGG CATGGTTGTG |
|
| 301 | GCACAGGTTT CCGATGCGGG TACGCCGGCC GTGTGCGACC CGGGCGCGAA |
|
| 351 | ACTCGCCCGC CGCGTGCGTG AGGCCGGGTT TAAAGTCGTT CCCGTCGTGG |
|
| 401 | GCGCAAC.GC GGTGATGGCG GCTTTGAGCG TGGCCGGTGT GGAAGGATCC |
|
| 451 | GATTTTTATT TCAACGGTTT TGTACCGCCG AAATCGGGAG AACGCAGGAA |
|
| 501 | ACTGTTTGCC AAATGGGTGC GGGCGGCGTT TCCTATCGTC ATGTTTGAAA |
|
| 551 | CGCCGCACCG CATCGGTGCA GCGCTTGCCG ATATGGCGGA ACTGTTCCCC |
|
| 601 | GAACGCCGAT TAATGCTGGC GCGCGAAATT ACGAAAACGT TTGAAACGTT |
|
| 651 | CTTAAGCGGC ACGGTTGGGG AAATTCAGAC GGCATTGTCT GCCGACGGCG |
|
| 701 | ACCAATCGCG CGGCGAGATG GTGTTGGTGC TTTATCCGGC GCAGGATGAA |
|
| 751 | AAACACGAAG GCTTGTCCGA GTCCGCGCAA AACATCATGA AAATCCTCAC |
|
| 801 | AGCCGAGCTG CCGACCAAAC AGGCGGCGGA GCTTGCTGCC AAAATCACGG |
|
| 851 | GCGAGGGAAA GAAAGCTTTG TACGAT.. |
[0759]This corresponds to the amino acid sequence <SEQ ID 286; ORF75>:
[0000]| 1 | MFVFQTAFXM FQKHLQKASD SVVGGTLYVV ATPIGNLADI TLRALAVLQK | |
|
| 51 | A....AEDTR VTAQLLSAYG IQGKLVSVRE HNERQMADKI VGYLSDGMVV |
|
| 101 | AQVSDAGTPA VCDPGAKLAR RVREAGFKVV PVVGAXAVMA ALSVAGVEGS |
|
| 151 | DFYFNGFVPP KSGERRKLFA KWVRAAFPIV MFETPHRIGA ALADMAELFP |
|
| 201 | ERRLMLAREI TKTFETFLSG TVGEIQTALS ADGDQSRGEM VLVLYPAQDE |
|
| 251 | KHEGLSESAQ NIMKILTAEL PTKQAAELAA KITGEGKKAL YD.. |
[0760]Further work revealed the complete nucleotide sequence <SEQ ID 287>:
[0000]| 1 | ATGTTTCAGA AACATTTGCA GAAAGCCTCC GACAGCGTCG TCGGAGGGAC | |
|
| 51 | ATTATACGTG GTTGCCACGC CCATCGGCAA TTTGGCGGAC ATTACCCTGC |
|
| 101 | GCGCTTTGGC GGTATTGCAA AAGGCGGACA TCATCTGTGC CGAAGACACG |
|
| 151 | CGCGTTACCG CACAGCTTTT GAGCGCGTAC GGCATTCAGG GCAAACTCGT |
|
| 201 | CAGTGTGCGC GAACACAACG AACGGCAGAT GGCGGACAAG ATTGTCGGCT |
|
| 251 | ATCTTTCAGA CGGCATGGTT GTGGCACAGG TTTCCGATGC GGGTACGCCG |
|
| 301 | GCCGTGTGCG ACCCGGGCGC GAAACTCGCC CGCCGCGTGC GTGAGGCCGG |
|
| 351 | GTTTAAAGTC GTTCCCGTCG TGGGCGCAAG CGCGGTGATG GCGGCTTTGA |
|
| 401 | GCGTGGCCGG TGTGGAAGGA TCCGATTTTT ATTTCAACGG TTTTGTACCG |
|
| 451 | CCGAAATCGG GAGAACGCAG GAAACTGTTT GCCAAATGGG TGCGGGCGGC |
|
| 501 | GTTTCCTATC GTCATGTTTG AAACGCCGCA CCGCATCGGT GCGACGCTTG |
|
| 551 | CCGATATGGC GGAACTGTTC CCCGAACGCC GATTAATGCT GGCGCGCGAA |
|
| 601 | ATTACGAAAA CGTTTGAAAC GTTCTTAAGC GGCACGGTTG GGGAAATTCA |
|
| 651 | GACGGCATTG TCTGCCGACG GCAACCAATC GCGCGGCGAG ATGGTGTTGG |
|
| 701 | TGCTTTATCC GGCGCAGGAT GAAAAACACG AAGGCTTGTC CGAGTCCGCG |
|
| 751 | CAAAACATCA TGAAAATCCT CACAGCCGAG CTGCCGACCA AACAGGCGGC |
|
| 801 | GGAGCTTGCT GCCAAAATCA CGGGCGAGGG AAAGAAAGCT TTGTACGATC |
|
| 851 | TGGCTCTGTC TTGGAAAAAC AAATAG |
[0761]This corresponds to the amino acid sequence <SEQ ID 288; ORF75-1>:
[0000]| 1 | MFQKHLQKAS DSVVGGTLYV VATPIGNLAD ITLRALAVLQ KADIICAEDT | |
|
| 51 | RVTAQLLSAY GIQGKLVSVR EHNERQMADK IVGYLSDGMV VAQVSDAGTP |
|
| 101 | AVCDPGAKLA RRVREAGFKV VPVVGASAVM AALSVAGVEG SDFYFNGFVP |
|
| 151 | PKSGERRKLF AKWVRAAFPI VMFETPHRIG ATLADMAELF PERRLMLARE |
|
| 201 | ITKTFETFLS GTVGEIQTAL SADGNQSRGE MVLVLYPAQD EKHEGLSESA |
|
| 251 | QNIMKILTAE LPTKQAAELA AKITGEGKKA LYDLALSWKN K* |
[0762]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0763]ORF75 shows 95.8% identity over a 283aa overlap with an ORF (ORF75a) from strain A of N. meningitidis.
[0000]
[0764]The complete length ORF75a nucleotide sequence <SEQ ID 289> is:
[0000]| 1 | ATGTTTCAGA AACATTTGCA GAAAGCCTCC GACAGCGTCG TCGGAGGGAC | |
|
| 51 | ATTATACGTG GTTGCCACGC CCATCGGCAA TTTGGCGGAC ATTACCCTGC |
|
| 101 | GCGCTTTGGC GGTATTGCAA AAGGCGGACA TCATCTGTGC CGAAGACACG |
|
| 151 | CGCGTTACCG CGCAGCTTTT GAGCGCGTAC GGCATTCAGG GCAAACTCGT |
|
| 201 | CAGCGTGCGC GAACACAACG AACGGCAGAT GGCGGACAAG ATTGTCGGCT |
|
| 251 | ATCTTTCAGA CGGCATGGTT GTGGCACAGG TTTCCGATGC GGGTACGCCG |
|
| 301 | GCCGTGTGCG ACCCGGGCGC GAAACTCGCC CGCCGCGTGC GTGAGGTCGG |
|
| 351 | GTTTAAAGTT GTCCCTGTTG TCGGCGCAAG CGCGGTGATG GCGGCTTTGA |
|
| 401 | GTGTGGCTGG TGTGGCGGGA TCCGATTTTT ATTTCAACGG TTTTGTACCG |
|
| 451 | CCGAAATCGG GCGAACGTAG GAAATTGTTT GCCAAATGGG TGCGGGTGGC |
|
| 501 | GTTTCCCGTC GTGATGTTTG AAACGCCGCA CCGCATCGGG GCGACGCTTG |
|
| 551 | CCGATATGGC GGAACTGTTC CCCGAACGCC GATTAATGCT GGCGCGCGAA |
|
| 601 | ATCACGAAAA CGTTTGAAAC GTTCTTAAGC GGCACGGTTG GGGAAATTCA |
|
| 651 | GACGGCATTG GCGGCGGACG GCAACCAATC GCGCGGCGAG ATGGTGTTGG |
|
| 701 | TGCTTTATCC GGCGCAGGAT GAAAAACACG AAGGCTTGTC CGAGTCCGCG |
|
| 751 | CAAAACATCA TGAAAATCCT CACAGCCGAG CTGCCGACCA AACAGGCGGC |
|
| 801 | GGAGCTTGCC GCCAAAATCA CGGGCGAGGG AAAAAAAGCT TTGTACGATC |
|
| 851 | TGGCACTGTC TTGGAAAAAC AAATGA |
[0765]This encodes a protein having amino acid sequence <SEQ ID 290>:
[0000]| 1 | MFQKHLQKAS DSVVGGTLYV VATPIGNLAD ITLRALAVLQ KADIICAEDT | |
|
| 51 | RVTAQLLSAY GIQGKLVSVR EHNERQMADK IVGYLSDGMV VAQVSDAGTP |
|
| 101 | AVCDPGAKLA RRVREVGFKV VPVVGASAVM AALSVAGVAG SDFYFNGFVP |
|
| 151 | PKSGERRKLF AKWVRVAFPV VMFETPHRIG ATLADMAELF PERRLMLARE |
|
| 201 | ITKTFETFLS GTVGEIQTAL AADGNQSRGE MVLVLYPAQD EKHEGLSESA |
|
| 251 | QNIMKILTAE LPTKQAAELA AKITGEGKKA LYDLALSWKN K* |
[0766]ORF75a and ORF75-1 show 98.3% identity in 291 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0767]ORF75 shows 93.2% identity over a 292aa overlap with a predicted ORF (ORF75.ng) from N. gonorrhoeae:
[0000]
[0768]An ORF75ng nucleotide sequence <SEQ ID 291> was predicted to encode a protein having amino acid sequence <SEQ ID 292>:
[0000]| 1 | MSVFQTAFFM FQKHLQKASD SVVGGTLYVV ATPIGNLADI TLRALAVLQK | |
|
| 51 | ADIICAEDTR VTAQLLSAYG IQGRLVSVRE HNERQMADKV IGFLSDGLVV |
|
| 101 | AQVSDAGTPA VCDPGAKLAR RVREAGFKVV PVVGASAVMA ALSVAGVAES |
|
| 151 | DFYFNGFVPP KSGERRKLFA KWVRAAFPVV MFETPHRIGA TLADMAELFP |
|
| 201 | ERRLMLAREI TKTFETFLSG TVGEIQTALA ADGNQSRGEM VLVLYPAQDE |
|
| 251 | KHEGLSESAQ NAMKILAAEL PTKQAAELAA KITGEGKKAL YDLALSWKNK |
|
| 301 | * |
[0769]After further analysis, the following gonococcal DNA sequence <SEQ ID 293> was identified:
[0000]| 1 | ATGTTTCAGA AACACTTGCA GAAAGCCTCC GACAGCGTCG TCGGAGGGAC | |
|
| 51 | ATTATACGTG GTTGCCACGC CCATCGGCAA TTTGGCAGAC ATTACCCTGC |
|
| 101 | GCGCTTTGGC GGTATTGCAA AAGGCGGACA TCATTTGTGC CGAAGACACG |
|
| 151 | CGCGTTACTG CGCAGCTTTT GAGCGCGTAC GGCATTCAGG GCAGGTTGGT |
|
| 201 | CAGTGTGCGC GAACACAACG AGCGGCAGAT GGCGGACAAG GTAATCGGTT |
|
| 251 | TCCTTTCAGA CGGCCTGGTT GTGGCGCAGG TTTCCGATGC GGGTACGCCG |
|
| 301 | GCCGTGTGCG ACCCGGGCGC GAAACTCGCC CGCCGCGTGC GCGAAGCAGG |
|
| 351 | GTTCAAAGTC GTTCCCGTCG TGGGCGCAAG CGCGGTAATG GCGGCGTTGA |
|
| 401 | GTGTGGCCGG TGTGGCGGAA TCCGATTTTT ATTTCAACGG TTTTGTACCG |
|
| 451 | CCGAAATCGG GCGAACGTAG GAAATTGTTT GCCAAATGGG TGCGGGCGGC |
|
| 501 | ATTTCCTGTC GTCATGTTTG AAACGCCGCA CCGAATCGGG GCAACGCTTG |
|
| 551 | CCGATATGGC GGAATTGTTC CCCGAACGCC GTCTGATGCT GGCGCGCGAA |
|
| 601 | ATCACGAAAA CGTTTGAAAC GTTCTTAAGC GGCACGGTTG GGGAAATTCA |
|
| 651 | GACGGCATTG GCGGCGGACG GCAACCAATC GCGCGGCGAG ATGGTGTTGG |
|
| 701 | TGCTTTATCC GGCGCAGGAT GAAAAACACG AAGGCTTGTC CGAGTCTGCG |
|
| 751 | CAAAATGCGA TGAAAATCCT TGCGGCCGAG CTGCCGACCA AGCAGGCGGC |
|
| 801 | GGAGCTTGCC GCCAAGATTA CAGGTGAGGG CAAAAAGGCT TTGTACGATT |
|
| 851 | TGGCACTGTC GTGGAAAAAC AAATGA |
[0770]This corresponds to the amino acid sequence <SEQ ID 294; ORF75ng-1>:
[0000]| 1 | MFQKHLQKAS DSVVGGTLYV VATPIGNLAD ITLRALAVLQ KADIICAEDT | |
|
| 51 | RVTAQLLSAY GIQGRLVSVR EHNERQMADK VIGFLSDGLV VAQVSDAGTP |
|
| 101 | AVCDPGAKLA RRVREAGFKV VPVVGASAVM AALSVAGVAE SDFYFNGFVP |
|
| 151 | PKSGERRKLF AKWVRAAFPV VMFETPHRIG ATLADMAELF PERRLMLARE |
|
| 201 | ITKTFETFLS GTVGEIQTAL AADGNQSRGE MVLVLYPAQD EKHEGLSESA |
|
| 251 | QNAMKILAAE LPTKQAAELA AKITGEGKKA LYDLALSWKN K* |
[0771]ORF75ng-1 and ORF75-1 show 96.2% identity in 291 aa overlap:
[0000]
[0772]Furthermore, ORG75ng-1 shows significant homology to a hypothetical E. coli protein:
[0000]| sp|P45528|YRAL_ECOLI HYPOTHETICAL 31.3 KD PROTEIN IN | |
| AGAI-MTR INTERGENIC REGION (F286) |
| >gi|606086 (U18997) ORF_f286 [Escherichia coli] |
| >gi|1789535 (AE000395) hypothetical 31.3 kD protein in agai-mtr intergenic |
| region [Escherichia coli] Length = 286 |
| Score = 218 bits (550), Expect = 3e−56 |
| Identities = 128/284 (45%), Positives = 171/284 (60%), Gaps = 4/284 (1%) |
|
| Query: | 4 | KHLQKASDSVVGGTLYVVATPIGNLADITLRALAVLQKADIICAEDTRVTAQLLSAYGIQ | 63 | |
| | K Q A +S G LY+V TPIGNLADIT RAL VLQ D+I AEDTR T LL +GI |
| Sbjct: | 2 | KQHQSADNSQ--GQLYIVPTPIGNLADITQRALEVLQAVDLIAAEDTRHTGLLLQHFGIN | 59 |
|
| Query: | 64 | GRLVSVREHNERQMADKVIGFLSDGLVVAQVSDAGTPAVCDPGAKLARRVREAGFKVVPV | 123 |
| | RL ++ +HNE+Q A+ ++ L +G +A VSDAGTP + DPG L R REAG +VVP+ |
| Sbjct: | 60 | ARLFALHDHNEQQKAETLLAKLQEGQNIALVSDAGTPLINDPGYHLVRTCREAGIRVVPL | 119 |
|
| Query: | 124 | VGASAVMAALSVAGVAESDFYFNGFVPPKSGERRKLFAKWVRAAFPVVMFETPHRIGATL | 183 |
| | G A + ALS AG+ F + GF+P KS RR ++ +E+ HR+ +L |
| Sbjct: | 120 | PGPCAAITALSAAGLPSDRFCYEGFLPAKSKGRRDALKAIEAEPRTLIFYESTHRLLDSL | 179 |
|
| Query: | 184 | ADMAELFPERR-LMLAREITKTFETFLSGTVGEIQTALAADGNQSRGEMVLVLYPAQDEK | 242 |
| | D+ + E R ++LARE+TKT+ET VGE+ + D N+ +GEMVL++ + |
| Sbjct: | 180 | EDIVAVLGESRYVVLARELTKTWETIHGAPVGELLAWVKEDENRRKGEMVLIV-EGHKAQ | 238 |
|
| Query: | 243 | HEGLSESAQNAMKILAAELPTKQAAELAAKITGEGKKALYDLAL | 286 |
| | E L A + +L AELP K+AA LAA+I G K ALY AL |
| Sbjct: | 239 | EEDLPADALRTLALLQAELPLKKAAALAAEIHGVKKNALYKYAL | 282 |
[0773]Based on this analysis, including the presence of a putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 35
[0774]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 295>:
[0000]
[0775]This corresponds to the amino acid sequence <SEQ ID 296; ORF76>:
[0000]
[0776]Further work revealed the complete nucleotide sequence <SEQ ID 297>:
[0000]| 1 | ATGAAACAGA AAAAAACCGC TGCCGCAGTT ATTGCTGCAA TGTTGGCAGG | |
|
| 51 | TTTTGCGGCA GCCAAAGCAC CCGAAATCGA CCCGGCTTTG GTGGATACGC |
|
| 101 | TGGTGGCGCA GATCATGCAG CAGGCAGACC GGCATGCGGA GCAGTCCCAA |
|
| 151 | AAACCGGACG GGCAGGCAAT CCGAAACGAT GCCGTCCGCC GGCTACAAAC |
|
| 201 | TTTGGAAGTT TTGAAAAACA GGGCATTGAA GGAAGGTTTG GATAAGGATA |
|
| 251 | AGGATGTCCA AAACCGCTTT AAAATCGCCG AAGCGTCTTT TTATGCCGAG |
|
| 301 | GAGTACGTCC GTTTTCTGGA ACGTTCGGAA ACGGTTTCCG AAGACGAGCT |
|
| 351 | GCACAAGTTT TACGAACAGC AAATCCGCAT GATCAAATTG CAGCAGGTCA |
|
| 401 | GCTTCGCAAC CGAAGAGGAG GCGCGTCAGG CGCAGCAGCT CCTGCTCAAA |
|
| 451 | GGGCTGTCTT TTGAAGGGCT GATGAAGCGT TATCCGAACG ACGAGCAGGC |
|
| 501 | TTTTGACGGT TTCATTATGG CGCAGCAGCT TCCCGAGCCG CTGGCTTCGC |
|
| 551 | AGTTTGCCGC GATGAATCGG GGCGACGTTA CCCGCGATCC GGTCAAATTG |
|
| 601 | GGCGAACGCT ATTATCTGTT CAAACTCAGC GAGGTCGGGA AAAACCCCGA |
|
| 651 | CGCGCAGCCT TTCGAGTTGG TCAGAAACCA GTTGGAGCAG GGTTTGAGAC |
|
| 701 | AGGAAAAAGC CCGCTTGAAA ATCGATGCCC TTTTGGAAGA AAACGGTGTC |
|
| 751 | AAACCGTAA |
[0777]This corresponds to the amino acid sequence <SEQ ID 298; ORF76-1>:
[0000]| 1 | MKQKKTAAAV IAAMLAGFAA AKAPEIDPAL VDTLVAQIMQ QADRHAEQSQ | |
|
| 51 | KPDGQAIRND AVRRLQTLEV LKNRALKEGL DKDKDVQNRF KIAEASFYAE |
|
| 101 | EYVRFLERSE TVSEDELHKF YEQQIRMIKL QQVSFATEEE ARQAQQLLLK |
|
| 151 | GLSFEGLMKR YPNDEQAFDG FIMAQQLPEP LASQFAAMNR GDVTRDPVKL |
|
| 201 | GERYYLFKLS EVGKNPDAQP FELVRNQLEQ GLRQEKARLK IDALLEENGV |
|
| 251 | KP* |
[0778]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0779]ORF76 shows 96.7% identity over a 30aa overlap and 96.8% identity over a 31 aa overlap with an ORF (ORF76a) from strain A of N. meningitidis.
[0000]
[0780]The complete length ORF76a nucleotide sequence <SEQ ID 299> is:
[0000]| 1 | ATGAAACAGA AAAAAACCGC TGCCGCAGTT ATTGCTGCAA TGTTGGCAGG | |
|
| 51 | TTTTGCGGCA GCCAAAGCAC CCGAAATCGA CCCGGCTTTG GTGGATACGC |
|
| 101 | TGGTGGCGCA GATCATGCAG CAGGCAGACC GGCATGCGGA GCAGTCCCAA |
|
| 151 | AAACCGGACG GGCAGGCAAT CCGAAACGAT GCCGTCCGTC GGCTGCAAAC |
|
| 201 | TTTGGAAGTT TTGAAAAACA GGGCATTGAA GGAAGGTTTG GATAAGGATA |
|
| 251 | AGGATGTCCA AAACCGCTTT AAAATCGCCG AAGCGTCTTT TTATGCCGAG |
|
| 301 | GAGTACGTCC GTTTTCTGGA ACGTTCGGAA ACGGTTTCCG AAAGCGCACT |
|
| 351 | GCGTCAGTTT TATGAGCGGC AAATCCGCAT GATCAAATTG CAGCAGGTCA |
|
| 401 | GCTTCGCAAC CGAAGAGGAG GCGCGTCAGG CGCAGCAGCT CCTGCTCAAA |
|
| 451 | GGGCTGTCTT TTGAAGGGCT GATGAAGCGT TATCCGAACG ACGAGCAGGC |
|
| 501 | TTTTGACGGT TTCATTATGG CGCAGCAGCT TCCCGAGCCG CTGGCTTCGC |
|
| 551 | AGTTTGCAGC GATGAATCGG GGCGACGTTA CCCGCGATCC GGTCAAATTG |
|
| 601 | GGCGAACGCT ATTATCTGTT CAAACTCAGC GAGGTCGGGA AAAACCCCGA |
|
| 651 | CGCGCAGCCT TTCGAGTTGG TCAGAAACCA GTTGGAACAA GGTTTGAGAC |
|
| 701 | AGGAAAAAGC CCGCTTGAAA ATCGATGCCA TTTTGGAAGA AAACGGTGTC |
|
| 751 | AAACCGTAA |
[0781]This encodes a protein having amino acid sequence <SEQ ID 300>:
[0000]| 1 | MKQKKTAAAV IAAMLAGFAA AKAPEIDPAL VDTLVAQIMQ QADRHAEQSQ | |
|
| 51 | KPDGQAIRND AVRRLQTLEV LKNRALKEGL DKDKDVQNRF KIAEASFYAE |
|
| 101 | EYVRFLERSE TVSESALRQF YERQIRMIKL QQVSFATEEE ARQAQQLLLK |
|
| 151 | GLSFEGLMKR YPNDEQAFDG FIMAQQLPEP LASQFAAMNR GDVTRDPVKL |
|
| 201 | GERYYLFKLS EVGKNPDAQP FELVRNQLEQ GLRQEKARLK IDAILEENGV |
|
| 251 | KP* |
[0782]ORF76a and ORF76-1 show 97.6% identity in 252 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0783]The aligned aa sequences of ORF76 and a predicted ORF (ORF76.ng) from N. gonorrhoeae of the N- and C-termini show 96.7% and 100% identity in 30 and 31 overlap, respectively:
[0000]
[0784]The complete length ORF76ng nucleotide sequence <SEQ ID 301> is:
[0000]| 1 | ATGAAACAGA AAAAGACCGC TGCCGCAGTT ATTGCTGCAA TGTTGGCAGG | |
|
| 51 | TTTTGCGGCA GCCAAAGCAC CCGAAATCGA CCCGGCTTTG GTGGATACGC |
|
| 101 | TGGTGGCGCA GATCATGCAG CAGGCAGACC GGCATGCGGA GCAGTCCCAA |
|
| 151 | AGACCGGACG GGCAGGCAAT CCGAAACGAT GCCGTCCGCC GGCTGCAAAC |
|
| 201 | TTTGGAAGTT TTGAAAAACA GGGCATTGAA GGAAGGTTTG GATAAGGATA |
|
| 251 | AGGATGTCCA AAACCGCTTT AAAATCGCCG AAGCGTCTTT TTATGCCGAG |
|
| 301 | GAGTACGTCC GTTTTCTGGA ACGTTCGGAA ACGGTTTCCG AAAGCGCACT |
|
| 351 | GCGTCAGTTT TATGAGCGGC AAATCCGCAT GATCAAATTG CAGCAGGTCA |
|
| 401 | GCTTCGCAAC CGAAGAGGAG GCGCGTCAGG CGCAGCAGCT CCTGCTCAAA |
|
| 451 | GGGCTGTCTT TTGAAGGGCT GATGAAGCGT TATCCGAACG ACGAGCAGGC |
|
| 501 | GTTCGACGGT TTCATTATGG CGCAGCAGCT TCCCGAGCCG CTGGCTTcgc |
|
| 551 | agtttgCCGG TATGAACCGT GGCGACGTTA CCCGCAATCC GGTCAAATTG |
|
| 601 | GGCGAACGCT ATTACCTGTT CAAACTCGGC GCGGTCGGGA AAAACCCCGA |
|
| 651 | CGCGCAGCCT TTCGAGTTGG TCAGAAACCA GTTGGAACAA GGTTTGAGGC |
|
| 701 | AGGAAAAAGC CCGCTTGAAA ATCGATGCCC TTTTGGAaga Aaacggtgtc |
|
| 751 | AaacCGTAA |
[0785]This encodes a protein having amino acid sequence <SEQ ID 302>:
[0000]| 1 | MKQKKTAAAV IAAMLAGFAA AKAPEIDPAL VDTLVAQIMQ QADRHAEQSQ | |
|
| 51 | RPDGQAIRND AVRRLQTLEV LKNRALKEGL DKDKDVQNRF KIAEASFYAE |
|
| 101 | EYVRFLERSE TVSESALRQF YERQIRMIKL QQVSFATEEE ARQAQQLLLK |
|
| 151 | GLSFEGLMKR YPNDEQAFDG FIMAQQLPEP LASQFAGMNR GDVTRNPVKL |
|
| 201 | GERYYLFKLG AVGKNPDAQP FELVRNQLEQ GLRQEKARLK IDALLEENGV |
|
| 251 | KP* |
[0786]ORF76ng and ORF76-1 show 96.0% identity in 252 aa overlap
[0000]
[0787]Furthermore, ORF76ng shows significant homology to a B. subtilis export protein precursor:
[0000]| sp|P24327|PRSA_BACSU PROTEIN EXPORT PROTEIN PRSA | |
| PRECURSOR >gi|98227|pir||S15269 |
| 33K lipoprotein - Bacillus subtilis >gi|39782 (X57271) 33 kDa lipoprotein |
| [Bacillus subtilis] |
| >gi|2226124|gnl|PID|e325181 (Y14077) 33 kDa lipoprotein [Bacillus subtilis] |
| >gi|2633331|gnl|PID|e1182997 (Z99109) molecular chaperonin |
| [Bacillus subtilis] |
| Length = 292 |
| Score = 50.4 bits (118), Expect = 1e−05 |
| Identities = 48/199 (24%), Positives = 82/199 (41%), Gaps = 32/199 (16%) |
|
| Query: | 70 | VLKNRALKEGLDK-----DKDVQNRFKIAEASF----------YAEEYVRFLERSETVSE | 114 | |
| | VL ++ LDK DK++ N+ K + Y ++Y++ + E +++ |
|
| Sbjct: | 53 | VLTQLVQEKVLDKKYKVSDKEIDNKLKEYKTQLGDQYTALEKQYGKDYLKEQVKYELLTQ | 112 |
|
| Query: | 115 | SA-----------LRQFYERQIRMIKLQQVSFATEEEARQAQQLLLKGLSFEGLMKRYPN | 163 |
| | A +++++E I+ + A ++ A + ++ L KG FE L K Y |
|
| Sbjct: | 113 | KAAKDNIKVTDADIKEYWEGLKGKIRASHILVADKKTAEEVEKKLKKGEKFEDLAKEYST | 172 |
|
| Query: | 164 | DEQAFDG-----FIMAQQLPEPLASQFAAMNRGDVTRDPVKLGERYYLFKLSEVGKNPDA | 218 |
| | D A G F Q+ E + + G+V+ DPVK Y++ K +E D |
|
| Sbjct: | 173 | DSSASKGGDLGWFAKEGQMDETFSKAAFKLKTGEVS-DPVKTQYGYHIIKKTEERGKYDD | 231 |
|
| Query: | 219 | QPFELVRNQLEQGLRQEKA | 237 |
| | EL LEQ L A |
|
| Sbjct: | 232 | MKKELKSEVLEQKLNDNAA | 250 |
[0788]Based on this analysis, including the presence of a putative leader sequence and a RGD motif in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0789]ORF76-1 (27.8 kDa) was cloned in the pET vector and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 10A shows the results of affinity purification of the His-fusion protein, Purified His-fusion protein was used to immunise mice, whose sera were used for Western blot (FIG. 10B), ELISA (positive result), and FACS analysis (FIG. 10C). These experiments confirm that ORF76-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 36
[0790]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 303>:
[0000]
[0791]This corresponds to the amino acid sequence <SEQ ID 304; ORF81>:
[0000]
[0792]Further work revealed the complete nucleotide sequence <SEQ ID 305>:
[0000]| 1 | ATGAAAAAAT CTTTCCTTAC GCTTGTTCTG TATTCGTCTT TACTTACCGC | |
|
| 51 | CAGCGAAATT GCCTATCGCT TTGTATTTGG GATTGAAACC TTACCGGCGG |
|
| 101 | CAAAAATTGC GGAAACGTTT GCGCTGACAT TTGTGATTGC TGCGCTGTAT |
|
| 151 | CTGTTTGCGC GTTATAAGGT GACGCGTTTG TTGATTGCGG TGTTTTTTGC |
|
| 201 | GTTCAGCATT ATTGCCAACA ATGTGCATTA CGCGGTTTAT CAAAGCTGGA |
|
| 251 | TGACGGGCAT CAATTATTGG CTGATGCTGA AAGAGGTTAC CGAAGTCGGC |
|
| 301 | AGCGCGGGTG CGTCGATGTT GGATAAGTTG TGGCTGCCTG TGTTGTGGGG |
|
| 351 | CGTGTTGGAA GTCATGTTGT TTTGCAGCCT TGCCAAGTTC CGCCGTAAGA |
|
| 401 | CGCATTTTTC TGCCGATATA CTGTTTGCCT TCCTAATGCT GATGATTTTC |
|
| 451 | GTGCGTTCGT TCGACACGAA ACAAGAGCAC GGTATTTCGC CCAAACCGAC |
|
| 501 | ATACAGCCGC ATCAAAGCCA ATTATTTCAG CTTCGGTTAT TTTGTCGGAC |
|
| 551 | GCGTGTTGCC GTATCAGTTG TTTGATTTAA GCAGGATTCC CGCCTTTAAG |
|
| 601 | CAGCCTGCTC CAAGCAAAAT CGGGCAGGGC AGTGTTCAAA ATATCGTCCT |
|
| 651 | GATTATGGGC GAAAGCGAAA GCGCGGCGCA TTTGAAGCTG TTTGGCTACG |
|
| 701 | GACGCGAAAC TTCGCCGTTT TTAACCCGGC TGTCGCAAGC CGATTTTAAG |
|
| 751 | CCGATTGTGA AACAAAGTTA TTCCGCAGGC TTTATGACTG CAGTGTCCCT |
|
| 801 | GCCCAGTTTT TTCAATGCGA TACCGCACGC CAACGGCTTG GAACAAATCA |
|
| 851 | GCGGCGGCGA TACCAATATG TTCCGCCTCG CCAAAGAGCA GGGCTATGAA |
|
| 901 | ACGTATTTTT ACAGCGCGCA GGCGGAAAAC GAGATGGCGA TTTTGAACTT |
|
| 951 | AATCGGTAAG AAATGGATAG ACCATCTGAT TCAGCCGACG CAACTTGGCT |
|
| 1001 | ACGGCAACGG CGACAATATG CCCGATGAGA AGCTGCTGCC GTTGTTCGAC |
|
| 1051 | AAAATCAATT TGCAGCAGGG CAAGCATTTT ATCGTGTTGC ACCAACGCGG |
|
| 1101 | TTCGCACGCC CCATACGGCG CATTGTTGCA GCCTCAAGAT AAAGTATTCG |
|
| 1151 | GCGAAGCCGA TATTGTGGAT AAGTACGACA ACACCATCCA CAAAACCGAC |
|
| 1201 | CAAATGATTC AAACCGTATT CGAGCAGCTG CAAAAGCAGC CTGACGGCAA |
|
| 1251 | CTGGCTGTTT GCCTATACCT CCGATCATGG CCAGTATGTT CGCCAAGATA |
|
| 1301 | TCTACAATCA AGGCACGGTG CAGCCCGACA GCTATCTCGT GCCGCTAGTG |
|
| 1351 | TTGTACAGCC CGGATAAGGC CGTGCAACAG GCTGCCAACC AGGCTTTTGC |
|
| 1401 | GCCTTGCGAG ATTGCCTTCC ATCAGCAGCT TTCAACGTTC CTGATTCACA |
|
| 1451 | CGTTGGGCTA CGATATGCCG GTTTCAGGTT GTCGCGAAGG CTCGGTAACG |
|
| 1501 | GGCAACCTGA TTACGGGTGA TGCAGGCAGC TTGAACATTC GCGACGGCAA |
|
| 1551 | GGCGGAATAT GTTTATCCGC AATGA |
[0793]This corresponds to the amino acid sequence <SEQ ID 306; ORF81-1>:
[0000]| 1 | MKKSFLTLVL YSSLLTASEI AYRFVFGIET LPAAKIAETF ALTFVIAALY | |
|
| 51 | LFARYKVTRL LIAVFFAFSI IANNVHYAVY QSWMTGINYW LMLKEVTEVG |
|
| 101 | SAGASMLDKL WLPVLWGVLE VMLFCSLAKF RRKTHFSADI LFAFLMLMIF |
|
| 151 | VRSFDTKQEH GISPKPTYSR IKANYFSFGY FVGRVLPYQL FDLSRIPAFK |
|
| 201 | QPAPSKIGQG SVQNIVLIMG ESESAAHLKL FGYGRETSPF LTRLSQADFK |
|
| 251 | PIVKQSYSAG FMTAVSLPSF FNAIPHANGL EQISGGDTNM FRLAKEQGYE |
|
| 301 | TYFYSAQAEN EMAILNLIGK KWIDHLIQPT QLGYGNGDNM PDEKLLPLFD |
|
| 351 | KINLQQGKHF IVLHQRGSHA PYGALLQPQD KVFGEADIVD KYDNTIHKTD |
|
| 401 | QMIQTVFEQL QKQPDGNWLF AYTSDHGQYV RQDIYNQGTV QPDSYLVPLV |
|
| 451 | LYSPDKAVQQ AANQAFAPCE IAFHQQLSTF LIHTLGYDMP VSGCREGSVT |
|
| 501 | GNLITGDAGS LNIRDGKAEY VYPQ* |
[0794]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0795]ORF81 shows 84.7% identity over a 85aa overlap and 99.2% identity over a 121aa overlap with an ORF (ORF81a) from strain A of N. meningitidis:
[0000]
[0796]The complete length ORF81a nucleotide sequence <SEQ ID 307> is:
[0000]| 1 | ATGAAAAAAT CCCTTTTCGT TCTCTTTCTG TATTCGTCCC TACTTACTGC | |
|
| 51 | CAGCGAAATT GCTTATCGCT TTGTATTCGG AATTGAAACC TTACCGGCTG |
|
| 101 | CAAAAATGGC AGAAACGTTT GCGCTGACAT TTGTGATTGC TGCGCTGTAT |
|
| 151 | CTGTTTGCGC GTTATAAGGC AACGCGTTTG TTGATTGCGG TGTTTTTCGC |
|
| 201 | GTTCAGCATT ATTGCCAACA ATGTGCATTA CGCGGTTTAT CAAAGCTGGA |
|
| 251 | TAACGGGCAT TAATTATTGG CTGATGCTGA AAGAGATTAC CGAAGTTGGC |
|
| 301 | GGCGCAGGGG CGTCGATGTT GGATAAGTTG TGGCTGCCTG CGTTGTGGGG |
|
| 351 | CGTGTTGGAA GTCATGTTGT TTTGCAGCCT TGCCAAGTTC CGCCGTAAGA |
|
| 401 | CGCATTTTTC TGCCGATATA CTGTTTGCCT TCCTAATGCT GATGATTTTC |
|
| 451 | GTGCGTTCGT TCGACACGAA ACAAGAACAC GGTATTTCGC CCAAACCGAC |
|
| 501 | ATACAGCCGC ATCAAAGCCA ATTATTTCAG CTTCGGTTAT TTTGTCGGAC |
|
| 551 | GCGTGTTGCC GTATCAGTTG TTTGATTTAA GCAAGATTCC TGTGTTCAAA |
|
| 601 | CAGCCTGCTC CAAGCAGAAT CGGGCAAGGC AGTATTCAAA ATATCGTCCT |
|
| 651 | GATTATGGGC GAAAGCGAAA GCGCGGCGCA TTTGAAATTG TTTGGCTACG |
|
| 701 | GGCGCGAAAC TTCGCCGTTT TTGACCCAGC TTTCGCAAGC CGATTTTAAG |
|
| 751 | CCGATTGTGA AACAAAGTTA TTCCGCAGGC TTTATGACGG CAGTATCCCT |
|
| 801 | GCCCAGTTTC TTTAACGTCA TACCGCATGC CAACGGCTTG GAACAAATCA |
|
| 851 | GCGGCGGCGA TATTGTGGAT AAGTACGACA ACACCATCCA CAAAACCGAC |
|
| 901 | CAAATGATTC AAACCGTATT CGAGCAGCTG CAAAAGCAGC CTGACGGCAA |
|
| 951 | CTGGCTGTTT GCCTATACCT CCGATCATGG CCAGTATGTT CGCCAAGATA |
|
| 1001 | TCTACAATCA AGGCACGGTG CAGCCCGACA GCTATCTCGT GCCGCTGGTG |
|
| 1051 | TTGTACAGCC CGGATAAGGC CGTGCAACAG GCTGCCAACC AGGCTTTTGC |
|
| 1101 | GCCTTGCGAG ATTGCCTTCC ATCAGCAGCT TTCAACGTTC CTGATTCACA |
|
| 1151 | CGTTGGGCTA CGATATGCCG GTTTCAGGTT GTCGCGAAGG CTCGGTAACG |
|
| 1201 | GGCAACCTGA TTACGGGTGA TGCAGGCAGC TTGAACATTC GCGACGGCAA |
|
| 1251 | GGCGGAATAT GTTTATCCGC AATGA |
[0797]This encodes a protein having amino acid sequence <SEQ ID 308>:
[0000]| 1 | MKKSLFVLFL YSSLLTASEI AYRFVFGIET LPAAKMAETF ALTFVIAALY | |
|
| 51 | LFARYKATRL LIAVFFAFSI IANNVHYAVY QSWITGINYW LMLKEITEVG |
|
| 101 | GAGASMLDKL WLPALWGVLE VMLFCSLAKF RRKTHFSADI LFAFLMLMIF |
|
| 151 | VRSFDTKQEH GISPKPTYSR IKANYFSFGY FVGRVLPYQL FDLSKIPVFK |
|
| 201 | QPAPSRIGQG SIQNIVLIMG ESESAAHLKL FGYGRETSPF LTQLSQADFK |
|
| 251 | PIVKQSYSAG FMTAVSLPSF FNVIPHANGL EQISGGDIVD KYDNTIHKTD |
|
| 301 | QMIQTVFEQL QKQPDGNWLF AYTSDHGQYV RQDIYNQGTV QPDSYLVPLV |
|
| 351 | LYSPDKAVQQ AANQAFAPCE IAFHQQLSTF LIHTLGYDMP VSGCREGSVT |
|
| 401 | GNLITGDAGS LNIRDGKAEY VYPQ* |
[0798]ORF81a and ORF81-1 show 77.9% identity in 524 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0799]The aligned aa sequences of ORF81 and a predicted ORF (ORF81.ng) from N. gonorrhoeae of the N- and C-termini show 82.4% and 97.5% identity in 85 and 121 overlap, respectively:
[0000]
[0800]The complete length ORF81ng nucleotide sequence <SEQ ID 309> is:
[0000]| 1 | ATGAAAAAAT CCCTTTTCGT TCTCTTTCTG TATTCATCCC TACTTACCGC | |
|
| 51 | CAGCGAAATC GCCTATCGCT TTGTATTCGG AATTGAAACC TTACCGGCTG |
|
| 101 | CAAAAATGGC GGAAACGTTT GCGCTGACAT TTATGATTGC TGCGCTGTAT |
|
| 151 | CTGTTTGCGC GTTATAAGGC TTCGCGGCTG CTGATTGCGG TGTTTTTCGC |
|
| 201 | GTTCAGCATG ATTGCCAACA ATGTGCATTA CGCGGTTTAT CAAAGCTGGA |
|
| 251 | TGACGGGTAT TAACTATTGG CTGATGCTGA AAGAGGTTAC CGAAGTCGGC |
|
| 301 | AGCGCGGGCG CGTCGATGTT GGATAAGTTG TGGCTGCCTG CTTTGTGGGG |
|
| 351 | CGTGGCGGAA GTCATGTTGT TTTGCAGCCT TGCCAAGTTC CGCCGTAAGA |
|
| 401 | CGCATTTTTC TGCCGATATA CTGTTTGCCT TCCTAATGCT GATGATTTTC |
|
| 451 | GTGCGTTCGT TCGACACGAA ACAAGAGCAC GGTATTTCGC CCAAACCGAC |
|
| 501 | ATACAGCCGC ATCAAAGCCA ATTATTTCAG CTTCGGTTAT TTTGTCGGGC |
|
| 551 | GCGTGTTGCC GTATCAGTTG TTTGATTTAA GCAAGATCCC TGTGTTCAAA |
|
| 601 | CAGCCTGCTC CAAGCAAAAT CGGGCAAGGC AGTATTCAAA ATATCGTCCT |
|
| 651 | GATTATGGGC GAAAGCGAAA GCGCGGCGCA TTTGAAATTG TTTGGTTACG |
|
| 701 | GGCGCGAAAC TTCGCCGTTT TTAACCCGGC TGTCGCAAGC CGATTTTAAG |
|
| 751 | CCGATTGTGA AACAAAGTTA TTCCGCAGGC TTTATGACGG CAGTATCCCT |
|
| 801 | GCCCAGTTTC TTTAACGTCA TACCGCACGC CAACGGCTTG GAACAAATCA |
|
| 851 | GCGGCGGCGA TACCAATATG TTCCGCCTCG CCAAAGAGCA GGGCTATGAA |
|
| 901 | ACGTATTTTT ACAGTGCCCA GGCTGAAAAC CAAATGGCAA TTTTGAACTT |
|
| 951 | AATCGGTAAG AAATGGATAG ACCATCTGAT TCAGCCGACG CAACTTGGCT |
|
| 1001 | ACGGCAACGG CGACAATATG CCCGATGAGA AGCTGCTGCC GTTGTTCGAC |
|
| 1051 | AAAATCAATT TGCAGCAGGG CAGGCATTTT ATCGTGTTGC ACCAACGCGG |
|
| 1101 | TTCGCACGCC CCATACGGCG CATTGTTGCA GCCTCAAGAT AAAGTATTCG |
|
| 1151 | GCGAAGCCGA TATTGTGGAT AAGTACGACA ACACCATCCA CAAAACCGAC |
|
| 1201 | CAAATGATTC AAACCGTATT CGAGCAGCTG CAAAAGCAGC CTGACGGCAA |
|
| 1251 | CTGGCTGTTT GCCTATACCT CCGATCATGG CCAGTATGTG CGCCAAGATA |
|
| 1301 | TCTACAATCA AGGCACGGTG CAGCCCGACA GCTATATTGT GCCTCTGGTT |
|
| 1351 | TTGTACAGCC CGGATAAGGC CGTGCAACAG GCTGCCAACC AGGCTTTTGC |
|
| 1401 | GCCTTGCGAG ATTGCCTTCC ATCAGCAGCT TTCAACGTTC CTGATTCACA |
|
| 1451 | CGTTGGGCTA CGATATGCCG GTTTCAGGTT GTCGCGAAGG CTCGGTAACA |
|
| 1501 | GGCAACCTGA TTACGGGCGA TGCAGGCAGC TTGAACATTC GCAACGGCAA |
|
| 1551 | GGCGGAATAT GTTTATCCGC AATAA |
[0801]This encodes a protein having amino acid sequence <SEQ ID 310>:
[0000]| 1 | MKKSLFVLFL YSSLLTASEI AYRFVFGIET LPAAKMAETF ALTFMIAALY | |
|
| 51 | LFARYKASRL LIAVFFAFSM IANNVHYAVY QSWMTGINYW LMLKEVTEVG |
|
| 101 | SAGASMLDKL WLPALWGVAE VMLFCSLAKF RRKTHFSADI LFAFLMLMIF |
|
| 151 | VRSFDTKQEH GISPKPTYSR IKANYFSFGY FVGRVLPYQL FDLSKIPVFK |
|
| 201 | QPAPSKIGQG SIQNIVLIMG ESESAAHLKL FGYGRETSPF LTRLSQADFK |
|
| 251 | PIVKQSYSAG FMTAVSLPSF FNVIPHANGL EQISGGDTNM FRLAKEQGYE |
|
| 301 | TYFYSAQAEN QMAILNLIGK KWIDHLIQPT QLGYGNGDNM PDEKLLPLFD |
|
| 351 | KINLQQGRHF IVLHQRGSHA PYGALLQPQD KVFGEADIVD KYDNTIHKTD |
|
| 401 | QMIQTVFEQL QKQPDGNWLF AYTSDHGQYV RQDIYNQGTV QPDSYIVPLV |
|
| 451 | LYSPDKAVQQ AANQAFAPCE IAFHQQLSTF LIHTLGYDMP VSGCREGSVT |
|
| 501 | GNLITGDAGS LNIRNGKAEY VYPQ* |
[0802]ORF81ng and ORF81-1 show 96.4% identity in 524 aa overlap:
[0000]
[0803]Furthermore, ORF81ng shows significant homology to an E. coli OMP:
[0000]| gi|1256380 (050906) outer membrane adherence protein-associated | |
| protein [E. coli] Length = 547 |
| Score = 87.4 bits (213), Expect = 2e−16 |
| Identities = 122/468 (26%), Positives = 198/468 (42%), |
| Gaps = 70/468 (14%) |
|
| Query: | 25 | VFGIETLPAAKMAETFA-LTFMIAALYLFARYKAS--RLLIAVFFAFSMIANNVHYAVYQ | 81 | |
| | VFGI L A+ A L F + + + R + RLL+A F + A ++ ++Y |
| Sbjct: | 29 | VFGITNLVASSGAHMVQRLLFFVLTILVVKRISSLPLRLLVAAPFVL-LTAADMSISLY- | 86 |
|
| Query: | 82 | SWMT-------GINYWLMLKEVTEVGSAGASMLDKLWLPALWGVAEVMLFCSLAKFRRKT | 134 |
| | SW T G ++ + EV A ML ++ P L A + L + |
| Sbjct: | 87 | SWCTFGTTFNDGFAISVLQSDPDEV----AKMLG-MYSPYLCAFAFLSLLFLAVIIKYDV | 141 |
|
| Query: | 135 | HFSADILFAFLMLMIFVRSF DTKQEHGISPKPTYSRIKAN--YFSFGYFVG | 183 |
| | + L+L++ S D K ++ SP SR +F+ YF |
| Sbjct: | 142 | SLPTKKVTGILLLIVISGSLFSACQFAYKDAKNKNAFSPYILASRFATYTPFFNLNYFAL | 201 |
|
| Query: | 184 | RVLPYQ--LFDLSKIPVFKQPAPSKIGQGSIQNIVLIMGESESAAHLKLFGYGRETSPFL | 241 |
| | +Q L + +P F+ + I VLI+GES ++ L+GY R T+P + |
| Sbjct: | 202 | AAKEHQRLLSIANTVPYFQL----SVRDTGIDTYVLIVGESVRVDNMSLYGYTRSTTPQV | 257 |
|
| Query: | 242 | TRLSQADFKPIVKQSYSAGFMTAVSLP---SFFNVIPHANGLEQISGGDTNMFRLAKEQG | 298 |
| | +Q + Q+ S TA+S+P + +V+ H I N+ +A + G |
| Sbjct: | 258 | E--AQRKQIKLFNQAISGAPYTALSVPLSLTADSVLSH-----DIHNYPDNIINMANQAG | 310 |
|
| Query: | 299 | YETYFYSAQA---ENQMAILNLIGKKWIDHLIQPTQLGYGNGDNMPDEKLLPLFDKINLQ | 355 |
| | ++T++ S+Q+ +N A+ ++ ++ + Y G DE LLP + Q |
| Sbjct: | 311 | FQTFWLSSQSAFRQNGTAVTSI--------AMRAMETVYVRGF---DELLLPHLSQALQQ | 359 |
|
| Query: | 356 | --QGRHFIVLHQRGSHAPYGALLQPQDKVFGEADIVDK-YDNTIHKTDQMIQTVFEQLQK | 412 |
| | Q + IVLH GSH P + VF D D YDN+IH TD ++ VFE L+ |
| Sbjct: | 360 | NTQQKKLIVLHLNGSHEPACSAYPQSSAVFQPQDDQDACYDNSIHYTDSLLGQVFELLK- | 418 |
|
| Query: | 413 | QPDGNWLFAYTSDHG---QYVRQDIYNQG--TVQPDSYIVPL-VLYSP | 454 |
| | D Y +DHG ++++Y G +Y VP+ + YSP |
| Sbjct: | 419 | --DRRASVMYFADHGLERDPTKKNVYFHGGREASQQAYHVPMFIWYSP | 464 |
[0804]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 37
[0805]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 311>:
[0000]| 1 | ...ACCCTGCTCC TCTTCATCCC CCTCGTCCTC ACAC.GTGCG GCACACTGAC | |
|
| 51 | CGGCATACTC GCCCaCGGCG GCGGCAAACG CTTTGCCGTC GAACAAGAAC |
|
| 101 | TCGTCGCCGC ATCGTCCCGC GCCGCCGTCA AAGAAATGGA TTTGTCCGCC |
|
| 151 | yTAAAAGGAC GCAAAGCCGC CyTTTACGTC TCCGTTATGG GCGACCAAGG |
|
| 201 | TTCGGGCAAC ATAAGCGGCG GACGCTACTC TATCGACGCA CTGATACGCG |
|
| 251 | GCGGCTACCA CAACAACCCC GAAAGTGCCA CCCAATACAG CTACCCCGCC |
|
| 301 | TACGACACTA CCGCCACCAC CAAATCCGAC GCGCTCTCCA GCGTAACCAC |
|
| 351 | TTCCACATCG CTTTTGAACG CCCCCGCCGC CGyCyTGACG AAAAACAGCG |
|
| 401 | GACGCAAAGG CGAACGcTCC GCCGGACTGT CCGTCAACGG CACGGGCGAC |
|
| 451 | TACCGCAACG AAACCCTGCT CGCCAACCCC CGCGACGTTT CCTTCCTGAC |
|
| 501 | CAACCTCATC CAAACCGTCT TCTACCTGCG CGGCATCGAA GTCgTACCGC |
|
| 551 | CCGrATACGC CGACACCGAC GTATTCGTAA CCGTCGACGT A... |
[0806]This corresponds to the amino acid sequence <SEQ ID 312; ORF83>:
[0000]| 1 | ..TLLLFIPLVL TXCGTLTGIL AHGGGKRFAV EQELVAASSR AAVKEMDLSA | |
|
| 51 | LKGRKAAXYV SVMGDQGSGN ISGGRYSIDA LIRGGYHNNP ESATQYSYPA |
|
| 101 | YDTTATTKSD ALSSVTTSTS LLNAPAAXLT KNSGRKGERS AGLSVNGTGD |
|
| 151 | YRNETLLANP RDVSFLTNLI QTVFYLRGIE VVPPXYADTD VFVTVDV.. |
[0807]Further work revealed the complete nucleotide sequence <SEQ ID 313>:
[0000]| 1 | ATGAAAACCC TGCTCCTCCT CATCCCCCTC GTCCTCACAG CCTGCGGCAC | |
|
| 51 | ACTGACCGGC ATACCCGCCC ACGGCGGCGG CAAACGCTTT GCCGTCGAAC |
|
| 101 | AAGAACTCGT CGCCGCATCG TCCCGCGCCG CCGTCAAAGA AATGGATTTG |
|
| 151 | TCCGCCCTAA AAGGACGCAA AGCCGCCCTT TACGTCTCCG TTATGGGCGA |
|
| 201 | CCAAGGTTCG GGCAACATAA GCGGCGGACG CTACTCTATC GACGCACTGA |
|
| 251 | TACGCGGCGG CTACCACAAC AACCCCGAAA GTGCCACCCA ATACAGCTAC |
|
| 301 | CCCGCCTACG ACACTACCGC CACCACCAAA TCCGACGCGC TCTCCAGCGT |
|
| 351 | AACCACTTCC ACATCGCTTT TGAACGCCCC CGCCGCCGCC CTGACGAAAA |
|
| 401 | ACAGCGGACG CAAAGGCGAA CGCTCCGCCG GACTGTCCGT CAACGGCACG |
|
| 451 | GGCGACTACC GCAACGAAAC CCTGCTCGCC AACCCCCGCG ACGTTTCCTT |
|
| 501 | CCTGACCAAC CTCATCCAAA CCGTCTTCTA CCTGCGCGGC ATCGAAGTCG |
|
| 551 | TACCGCCCGA ATACGCCGAC ACCGACGTAT TCGTAACCGT CGACGTATTC |
|
| 601 | GGCACCGTCC GCAGCCGTAC CGAACTGCAC CTCTACAACG CCGAAACCCT |
|
| 651 | TAAAGCCCAA ACCAAGCTCG AATATTTCGC CGTTGACCGC GACAGCCGGA |
|
| 701 | AACTGCTGAT TACCCCTAAA ACCGCCGCCT ACGAATCCCA ATACCAAGAA |
|
| 751 | CAATACGCCC TTTGGACCGG CCCTTACAAA GTCAGCAAAA CCGTCAAAGC |
|
| 801 | CTCAGACCGC CTGATGGTCG ATTTCTCCGA CATTACCCCC TACGGCGACA |
|
| 851 | CAACCGCCCA AAACCGTCCC GACTTCAAAC AAAACAACGG TAAAAAACCC |
|
| 901 | GATGTCGGCA ACGAAGTCAT CCGCCGCCGC AAAGGAGGAT AA |
[0808]This corresponds to the amino acid sequence <SEQ ID 314; ORF83-1>:
[0000]| 1 | MKTLLLLIPL VLTACGTLTG IPAHGGGKRF AVEQELVAAS SRAAVKEMDL | |
|
| 51 | SALKGRKAAL YVSVMGDQGS GNISGGRYSI DALIRGGYHN NPESATQYSY |
|
| 101 | PAYDTTATTK SDALSSVTTS TSLLNAPAAA LTKNSGRKGE RSAGLSVNGT |
|
| 151 | GDYRNETLLA NPRDVSFLTN LIQTVFYLRG IEVVPPEYAD TDVFVTVDVF |
|
| 201 | GTVRSRTELH LYNAETLKAQ TKLEYFAVDR DSRKLLITPK TAAYESQYQE |
|
| 251 | QYALWTGPYK VSKTVKASDR LMVDFSDITP YGDTTAQNRP DFKQNNGKKP |
|
| 301 | DVGNEVIRRR KGG* |
[0809]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0810]ORF83 shows 96.4% identity over a 197aa overlap with an ORF (ORF83a) from strain A of N. meningitidis:
[0000]
[0811]The complete length ORF83a nucleotide sequence <SEQ ID 315> is:
[0000]| 1 | ATGAAAACCC TGCTCNTCCT CATCCCCCTC GTCCTCACAG CCTGCGGCAC | |
|
| 51 | ACTGACCGGC ATACCCGCCC ACGGCGGCGG CAAACGCTTT GCCGTCGAAC |
|
| 101 | AAGAACTCGT CGCCGCATCG TCCCGCGCCG CCGTCAAAGA AATGGACTTG |
|
| 151 | TCCGCCCTGA AAGGACGCAA AGCCGCCCTT TACGTCTCCG TTATGGGCGA |
|
| 201 | CCAAGGTTCG GGCAACATAA GCGGCGGACG CTACTCTATC GACGCACTGA |
|
| 251 | TACGCGGCGG CTACCACAAC AACCCCGAAA GTGCCACCCA ATACAGCTAC |
|
| 301 | CCCGCCTACG ACACTACCGC CACCACCAAA TCCGACGCGC TCTCCAGCGT |
|
| 351 | AACCACTTCC ACATCGCTTT TGAACGCCCC CGCCGCCGCC CTGACGAAAA |
|
| 401 | ACAGCGGACG CAAAGGCGAA CGCTCCGCCG GACTGTCCGT CAACGGCACG |
|
| 451 | GGCGACTACC GCAACGAAAC CCTGCTCGCC AACCCCCGCG ACGTTTCCTT |
|
| 501 | CCTGACCAAC CTCATCCAAA CCGTCTTCTA CCTGCGCGGC ATCGAAGTCG |
|
| 551 | TACCGCCCGA ATACGCCGAC ACCGACGTAT TCGTAACCGT CGACGTATTC |
|
| 601 | GGCACCGTCC GCAGCCGCAC CGAACTGCAC CTCTACAACG CCGAAACCCT |
|
| 651 | TAAAGCCCAA ACCAAGCTCG AATATTTCGC CGTTGACCGC GACAGCCGGA |
|
| 701 | AACTGCTGAT TGCCCCTAAA ACCGCCGCCT ACGAATCCCA ATACCAAGAA |
|
| 751 | CAATACGCCC TCTGGATGGG ACCTTACAGC GTCGGCAAAA CCGTCAAAGC |
|
| 801 | CTCAGACCGC CTGATGGTCG ATTTCTCCGA CATCACCCCC TACGGCGACA |
|
| 851 | CAACCGCCCA AAACCGTCCC GACTTCAAAC AAAACAACGG TAAAAAACCC |
|
| 901 | GATGTCGGCA ACGAAGTCAT CCGCCGCCGC AAAGGAGGAT AA |
[0812]This encodes a protein having amino acid sequence <SEQ ID 316>:
[0000]| 1 | MKTLLXLIPL VLTACGTLTG IPAHGGGKRF AVEQELVAAS SRAAVKEMDL | |
|
| 51 | SALKGRKAAL YVSVMGDQGS GNISGGRYSI DALIRGGYHN NPESATQYSY |
|
| 101 | PAYDTTATTK SDALSSVTTS TSLLNAPAAA LTKNSGRKGE RSAGLSVNGT |
|
| 151 | GDYRNETLLA NPRDVSFLTN LIQTVFYLRG IEVVPPEYAD TDVFVTVDVF |
|
| 201 | GTVRSRTELH LYNAETLKAQ TKLEYFAVDR DSRKLLIAPK TAAYESQYQE |
|
| 251 | QYALWMGPYS VGKTVKASDR LMVDFSDITP YGDTTAQNRP DFKQNNGKKP |
|
| 301 | DVGNEVIRRR KGG* |
[0813]ORF83a and ORF83-1 show 98.4% identity in 313 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0814]ORF83 shows 94.9% identity over a 197aa overlap with a predicted ORF (ORF83.ng) from N. gonorrhoeae:
[0000]
[0815]The complete length ORF83ng nucleotide sequence <SEQ ID 317> is:
[0000]| 1 | ATGAAAACCC TGCTCCTCCT CATCCCCCTC GTACTCACCG CCTGCGGCAC | |
|
| 51 | ACTGACCGGC ATACCCGCCC ACGGCGGCGG CAAACGCTTT GCCGTCGAAC |
|
| 101 | AGGAACTCGT CGCCGCATCG TCCCGCGCCG CCGTCAAAGA AATGGACTTG |
|
| 151 | TCCGCCCTGA AAGGACGCAA AGCCGCCCTT TACGTCTCCG TTATGGGCGA |
|
| 201 | CCAAGGTTCG GGCAACATAA GCGGCGGACG CTACTCCATC GACGCACTGA |
|
| 251 | TACGCGGCGG CTACCACAAC AACCCCGACA GCGCCACCCG ATACAGCTAC |
|
| 301 | CCCGCCTATG ACACTACCGC CACCACCAAA TCCGACGCGC TCTCCGGCGT |
|
| 351 | AACCACTTCC ACATCGCTTT TGAACGCCCC CGCCGCCGCC CTGACGAAAA |
|
| 401 | ACAACGGACG CAAAGGCGAA CGCTCCGCCG GACTGTCCGT CAACGGCACG |
|
| 451 | GGCGACTACC GCAACGAAAC CCTGCTCGCC AACCCCCGCG ACGTTTCCTT |
|
| 501 | CCTGACCAAC CTCATCCAAA CCGTCTTCTA CCTGCGCGGC ATCGAAGTCG |
|
| 551 | TACCGCCCGA ATACGCCGAC ACCGACGTAT TCGTAACCGT CGACGTATTC |
|
| 601 | GGCACCGTCC GCAGCCGTAC CGAACTGCAC CTCTACAACG CCGAAACCCT |
|
| 651 | TAAAGCCCAA ACCAAGCTCG AATATTTCGC CGTCGACCGC GACAGCCGGA |
|
| 701 | AACTGCTGAT TGCCCCTAAA ACCGCCGCCT ACGAATCCCA ATACCAAGAA |
|
| 751 | CAATACGCCC TCTGGATGGG ACCTTACAGC GTCGGCAAAA CCGTCAAAGC |
|
| 801 | CTCAGACCGC CTGATGGTCG ATTTCTCCGA CATCACCCCC TACGGCGACA |
|
| 851 | CAACCGCCCA AAACCGTCCC GACTTCAAAC AAAACAACGG TAAAAACCCC |
|
| 901 | GATGTCGGCA ACGAAGTCAT CCGCCGCCGC AAAGGAGGAT AA |
[0816]This encodes a protein having amino acid sequence <SEQ ID 318>:
[0000]| 1 | MKTLLLLIPL VLTACGTLTG IPAHGGGKRF AVEQELVAAS SRAAVKEMDL | |
|
| 51 | SALKGRKAAL YVSVMGDQGS GNISGGRYSI DALIRGGYHN NPDSATRYSY |
|
| 101 | PAYDTTATTK SDALSGVTTS TSLLNAPAAA LTKNNGRKGE RSAGLSVNGT |
|
| 151 | GDYRNETLLA NPRDVSFLTN LIQTVFYLRG IEVVPPEYAD TDVFVTVDVF |
|
| 201 | GTVRSRTELH LYNAETLKAQ TKLEYFAVDR DSRKLLIAPK TAAYESQYQE |
|
| 251 | QYALWMGPYS VGKTVKASDR LMVDFSDITP YGDTTAQNRP DFKQNNGKNP |
|
| 301 | DVGNEVIRRR KGG* |
[0817]ORF83ng and ORF83-1 show 97.1% identity in 313 aa overlap
[0000]
[0818]Based on this analysis, including the presence of a putative ATP/GTP-binding site motif A (P-loop) in the gonococcal protein (double-underlined) and a putative prokaryotic membrane lipoprotein lipid attachment site (single-underlined), it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 38
[0819]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 319>:
[0000]| 1 | ATGGCAGAGA TCTGTTTGAT AACCGGCACG CCCGGTTCAG GGAAAACATT | |
|
| 51 | AAAAATGGTT TCCATGATGG CGAATGATGA AATGTTTAAG CCTGATGAAA |
|
| 101 | AAGCCATACG CCGTAAAGTA TTTACGAACA TAAAAGGCTT GAAAATACCG |
|
| 151 | CACACCTACA TAGAAACGGA CGCAAAAAAG CTGCCGAAAT CGACAGATGA |
|
| 201 | GCAGCTTTCG GCGCATGATA TGTACGAATG GATAAAGAAG CCCGAAAATA |
|
| 251 | TCGGGTCTAT TGTCATTGTA GATGAAGCTC AAGACGTATG GCCGGCACGC |
|
| 301 | TCGGCAGGTT CAAAAATCCC TGAAAATGTC CAATGGCTGA ATACGCACAG |
|
| 351 | ACATCAGGGC ATTGATATAT TTGTTTTGAC TCAAGGTCCT AAGCTTCTAG |
|
| 401 | ATCAAAATCT TAGAACGCTT GTACGGAAAC ATTACCACAT CGCTTCAAAC |
|
| 451 | AAGATGGGTA TGCGTACGCT TTTAGAATGG AAAATATGCG CGGACGATCC |
|
| 501 | CGTAAAAATG GCATCAAGCG CATTCTCCAG TATCTATACA CTGGATAAAA |
|
| 551 | AAGTTTATGA CTTGTAysrr TmmGCGGAAG TTCATACCGT AAATAAGGTC |
|
| 601 | AAGCGGTCAA AGTGGTTTTA CACTCTGCCa GTAATAGTAT TGCTGATTCC |
|
| 651 | CGTGTTTGTC GGCCTGTCCT ATAAAATGTT GagCaGTTAC GGAAAAAAAC |
|
| 701 | aGGAAGAACC CGCAGCACAA GAATCGGCGG CAACAGAACA GCAGGCAGTA |
|
| 751 | CTTCCGGATA AAACAGAAGG CGAGCCGGTA AATAACGGCA ACCTTACCGC |
|
| 801 | AGATATGTTT GTTCCGACAT TGTCCGAaAA ACCCGrAAGC AAGCcgaTTT |
|
| 851 | ATAACGGTGT AAGGCAGGTA AGAACCTTTG AATATATAGC AGGCTGTATA |
|
| 901 | GAAGGCGGAA GAACCGGATG CGCCTGCTAT TCGCaTCAAG GGACGGCATt |
|
| 951 | gaAAGAAGTG ACGGaGTTGA TGTGccaAgG aCTATGTaAA AAacGGCTTG |
|
| 1001 | CCGTTTAACC CaTACAAAGA AGAAAGCCAA GGGCAGGAAG TTCAGCAAAG |
|
| 1051 | CGCGCAgCAA CATTCGGACA GGGCGcCAAG TTGCCACATT GGGCGGAAAA |
|
| 1101 | CCGTAGCAGA ACCTAATGTA CGATAATTGG GAAGAACGCG GGAAACCGTT |
|
| 1151 | TGAAGGAATC GGaCGGGGGC GTGGTCGGAT CGGCAAACTG A |
[0820]This corresponds to the amino acid sequence <SEQ ID 320; ORF84>:
[0000]| 1 | MAEICLITGT PGSGKTLKMV SMMANDEMFK PDEKAIRRKV FTNIKGLKIP | |
|
| 51 | HTYIETDAKK LPKSTDEQLS AHDMYEWIKK PENIGSIVIV DEAQDVWPAR |
|
| 101 | SAGSKIPENV QWLNTHRHQG IDIFVLTQGP KLLDQNLRTL VRKHYHIASN |
|
| 151 | KMGMRTLLEW KICADDPVKM ASSAFSSIYT LDKKVYDLYX XAEVHTVNKV |
|
| 201 | KRSKWFYTLP VIVLLIPVFV GLSYKMLSSY GKKQEEPAAQ ESAATEQQAV |
|
| 251 | LPDKTEGEPV NNGNLTADMF VPTLSEKPXS KPIYNGVRQV RTFEYIAGCI |
|
| 301 | EGGRTGCACY SHQGTALKEV TELMCKDYVK NGLPFNPYKE ESQGQEVQQS |
|
| 351 | AQQHSDRAQV ATLGGKPXQN LMYDNWEERG KPFEGIGGGV VGSAN* |
[0821]Further work revealed the complete nucleotide sequence <SEQ ID 321>:
[0000]| 1 | ATGGCAGAGA TCTGTTTGAT AACCGGCACG CCCGGTTCAG GGAAAACATT | |
|
| 51 | AAAAATGGTT TCCATGATGG CGAATGATGA AATGTTTAAG CCTGATGAAA |
|
| 101 | ACGGCATACG CCGTAAAGTA TTTACGAACA TAAAAGGCTT GAAAATACCG |
|
| 151 | CACACCTACA TAGAAACGGA CGCAAAAAAG CTGCCGAAAT CGACAGATGA |
|
| 201 | GCAGCTTTCG GCGCATGATA TGTACGAATG GATAAAGAAG CCCGAAAATA |
|
| 251 | TCGGGTCTAT TGTCATTGTA GATGAAGCTC AAGACGTATG GCCGGCACGC |
|
| 301 | TCGGCAGGTT CAAAAATCCC TGAAAATGTC CAATGGCTGA ATACGCACAG |
|
| 351 | ACATCAGGGC ATTGATATAT TTGTTTTGAC TCAAGGTCCT AAGCTTCTAG |
|
| 401 | ATCAAAATCT TAGAACGCTT GTACGGAAAC ATTACCACAT CGCTTCAAAC |
|
| 451 | AAGATGGGTA TGCGTACGCT TTTAGAATGG AAAATATGCG CGGACGATCC |
|
| 501 | CGTAAAAATG GCATCAAGCG CATTCTCCAG TATCTATACA CTGGATAAAA |
|
| 551 | AAGTTTATGA CTTGTACGAA TCAGCGGAAG TTCATACCGT AAATAAGGTC |
|
| 601 | AAGCGGTCAA AGTGGTTTTA CACTCTGCCA GTAATAGTAT TGCTGATTCC |
|
| 651 | CGTGTTTGTC GGCCTGTCCT ATAAAATGTT GAGCAGTTAC GGAAAAAAAC |
|
| 701 | AGGAAGAACC CGCAGCACAA GAATCGGCGG CAACAGAACA GCAGGCAGTA |
|
| 751 | CTTCCGGATA AAACAGAAGG CGAGCCGGTA AATAACGGCA ACCTTACCGC |
|
| 801 | AGATATGTTT GTTCCGACAT TGTCCGAAAA ACCCGAAAGC AAGCCGATTT |
|
| 851 | ATAACGGTGT AAGGCAGGTA AGAACCTTTG AATATATAGC AGGCTGTATA |
|
| 901 | GAAGGCGGAA GAACCGGATG CGCCTGCTAT TCGCATCAAG GGACGGCATT |
|
| 951 | GAAAGAAGTG ACGGAGTTGA TGTGCAAGGA CTATGTAAAA AACGGCTTGC |
|
| 1001 | CGTTTAACCC ATACAAAGAA GAAAGCCAAG GGCAGGAAGT TCAGCAAAGC |
|
| 1051 | GCGCAGCAAC ATTCGGACAG GGCGCAAGTT GCCACATTGG GCGGAAAACC |
|
| 1101 | GTAGCAGAAC CTAATGTACG ATAATTGGGA AGAACGCGGG AAACCGTTTG |
|
| 1151 | AAGGAATCGG CGGGGGCGTG GTCGGATCGG CAAACTGA |
[0822]This corresponds to the amino acid sequence <SEQ ID 322; ORF84-1>:
[0000]| 1 | MAEICLITGT PGSGKTLKMV SMMANDEMFK PDENGIRRKV FTNIKGLKIP | |
|
| 51 | HTYIETDAKK LPKSTDEQLS AHDMYEWIKK PENIGSIVIV DEAQDVWPAR |
|
| 101 | SAGSKIPENV QWLNTHRHQG IDIFVLTQGP KLLDQNLRTL VRKHYHIASN |
|
| 151 | KMGMRTLLEW KICADDPVKM ASSAFSSIYT LDKKVYDLYE SAEVHTVNKV |
|
| 201 | KRSKWFYTLP VIVLLIPVFV GLSYKMLSSY GKKQEEPAAQ ESAATEQQAV |
|
| 251 | LPDKTEGEPV NNGNLTADMF VPTLSEKPES KPIYNGVRQV RTFEYIAGCI |
|
| 301 | EGGRTGCACY SHQGTALKEV TELMCKDYVK NGLPFNPYKE ESQGQEVQQS |
|
| 351 | AQQHSDRAQV ATLGGKP*QN LMYDNWEERG KPFEGIGGGV VGSAN* |
[0823]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0824]ORF84 shows 93.9% identity over a 395aa overlap with an ORF (ORF84a) from strain A of N. meningitidis:
[0000]
[0825]The complete length ORF84a nucleotide sequence <SEQ ID 323> is:
[0000]| 1 | ATGGCAGAGA TCTGTTTGAT AACCGGCACG CCCGGTTCAG GGAAAACATT | |
|
| 51 | AAAAATGGTT TCCATGATGG CAAACGATGA AATGTTTAAG CCGGATGAAA |
|
| 101 | ACGGCATACG CCGTAAAGTA TTTACGAACA TCAAAGGCTT GAAGATACCG |
|
| 151 | CACACCTACA TAGAAACGGA CGCGAAAAAG CTGCCGAAAT CGACAGATGA |
|
| 201 | GCAGCTTTCG GCGCATGATA TGTACGAATG GATAAAGAAG CCCGAAAATA |
|
| 251 | TCGGGTCTAT TGTCATTGTA GATGAAGCTC AAGACGTATG GCCGGCACGC |
|
| 301 | TCGGCAGGTT CAAAAATCCC TGAAAATGTC CAATGGCTGA ATACGCACAG |
|
| 351 | ACATCAGGGC ATTGATATAT TTGTTTTGAC TCAAGGCTCT AAGCTTCTAG |
|
| 401 | ATCAAAATCT TAGAACGCTT GTACGGAAAC ATTACCACAT CGCTTCAAAC |
|
| 451 | AAGATGGGTA TGCGTACGCT TTTAGAATGG AAAATATGCG CGGACGATCC |
|
| 501 | CGTAAAAATG GCATCAAGCG CATTCTCCAG TATCTATACA CTGGATAAAA |
|
| 551 | AAGTTTATGA CTTGTACGAA TCAGCGGAAG TTCATACCGT AAATAAGGTC |
|
| 601 | AAGCGGTCAA AATGGTTTTA TACTCTGCCA GTAATAATAT TGCTGATTCC |
|
| 651 | CGTTTTTGTC GGCCTGTCCT ATAAAATGTT AAGTAGTTAT GGAAAAAAAC |
|
| 701 | AGGAAGAACC CGCAGCACAA GAATCGGCGG CAACAGAACA TCAGGCAGTA |
|
| 751 | TTTCAGGATA AAACAGAAGG CGAGCCGGTA AACAACGGTA ACCTTACCGC |
|
| 801 | AGATATGTTT GTTCCGACAT TGTCCGAAAA ACCCGAAAGC AAGCCGATTT |
|
| 851 | ATAACGGTGT AAGGCAGGTA AGAACCTTTG AATATATAGC AGGCTGTGTA |
|
| 901 | GAAGGCGGAA GAACCGGATG CACATGCTAT TCGCATCAAG GGACGGCATT |
|
| 951 | GAAAGAAATT ACAAAGGAAA TGTGCAAGGA TTACGCAAGA AACGGATTGC |
|
| 1001 | CGTTTAACCC ATATAAAGAA GAAAGCCAAG GGCGGGATGT CCAGCAAAGT |
|
| 1051 | GAGCAGCACC ATTCGGACAG ACCGCAAGTT GCCACGTTGG GCGGAAAGCC |
|
| 1101 | GTGGCAAAAT CTTATGTATG ATAATTGGCA GGAGCGCGGA AAACCGTTTG |
|
| 1151 | AAGGAATCGG CGGGGGCGTG GTCGGATCGG CAAACTGA |
[0826]This encodes a protein having amino acid sequence <SEQ ID 324>:
[0000]| 1 | MAEICLITGT PGSGKTLKMV SMMANDEMFK PDENGIRRKV FTNIKGLKIP | |
|
| 51 | HTYIETDAKK LPKSTDEQLS AHDMYEWIKK PENIGSIVIV DEAQDVWPAR |
|
| 101 | SAGSKIPENV QWLNTHRHQG IDIFVLTQGS KLLDQNLRTL VRKHYHIASN |
|
| 151 | KMGMRTLLEW KICADDPVKM ASSAFSSIYT LDKKVYDLYE SAEVHTVNKV |
|
| 201 | KRSKWFYTLP VIILLIPVFV GLSYKMLSSY GKKQEEPAAQ ESAATEHQAV |
|
| 251 | FQDKTEGEPV NNGNLTADMF VPTLSEKPES KPIYNGVRQV RTFEYIAGCV |
|
| 301 | EGGRTGCTCY SHQGTALKEI TKEMCKDYAR NGLPFNPYKE ESQGRDVQQS |
|
| 351 | EQHHSDRPQV ATLGGKPWQN LMYDNWQERG KPFEGIGGGV VGSAN* |
[0827]ORF84a and ORF84-1 show 95.2% identity in 395 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0828]ORF84 shows 94.2% identity over a 395aa overlap with a predicted ORF (ORF84.ng) from N. gonorrhoeae.
[0000]
[0829]The complete length ORF84ng nucleotide sequence <SEQ ID 325> is:
[0000]| 1 | ATGGCAGAAA TCTGTTTGAT AACCGGCACG CCCGGTTCAG GGAAAACATT | |
|
| 51 | AAAAATGGTT TCCATGATGG CAAACGATGA AATGTTTAAG CCAGATGAAA |
|
| 101 | ACGGCGTACG CCGTAAAGTA TTTACGAACA TCAAAGGTTT GAAGATACCG |
|
| 151 | CACACCCACA TAGAAACAGA CGCAAAGAAG CTGCCGAAAT CAACCGATGA |
|
| 201 | ACAGCTTTCG GCGCATGATA TGTATGAATG GATCAAGAAG CCTGAAAacg |
|
| 251 | tcggcgCAAT CGTTATTGTC GATGAGGCGC AAGACGTATG GCCCGCACGC |
|
| 301 | TccgCAGGTT CGAAAATCCC CGAAAACGTC CAATGGCTGA ACACACACAG |
|
| 351 | GCATCAGGGC ATAGATATAT TTGTATTGAC ACAAGGTCCT AAACTCTTAG |
|
| 401 | ATCAGAACTT GCGAACATTG GTTAAAAGAC ATTACCACAT TGCGGCCAAC |
|
| 451 | AAAATGGGTT TGCGTACCCT GCTTGAATGG AAAGTATGCG CGGATGACCC |
|
| 501 | GGTAAAAATG GCATCAAGTG CATTTTCCAG TATCTACACA CTGGATAAAA |
|
| 551 | AAGTTTATGA CTTGTACGAA TCCGCAGAAA TTCACACGGT AAACAAAGTC |
|
| 601 | AAGCGTTCAA AATGGTTTTA TGCATTGCCC GTCATCATAT TATTGATTCC |
|
| 651 | GCTATTTGTC GGTTTGTCTT ACAAAATGTT GGGCAGTTAC GGAAAAAAAC |
|
| 701 | AGGAAGAACC CGCAGCACAA GAATCGGCGG CAACAGAACA GCAGGCAGTA |
|
| 751 | CTTCCGGATA AAACAGAAGG AGAATCGGTG AATAACGGAA ACCTTACGGC |
|
| 801 | AGATATGTTT GTTCCGACAT TGCCCGAAAA ACCCGAAAGC AAGCCGATTT |
|
| 851 | ATAACGGTGT AAGGCAGGTA AGGACCTTTG AATATATAGC AGGCTGTATA |
|
| 901 | GAAGGCGGAA GAACCGGATG CACCTGCTAT TCGCATCAAG GGACGGCATT |
|
| 951 | GAAAGAAGTG ACGGAGTTGA TGTGCAAGGA CTATGTAAAA AACGGCTTGC |
|
| 1001 | CGTTTAACCC ATACAAAGAA GAAAGCCAAG GGCAGGAAGT TCAGCAAAGC |
|
| 1051 | GCGCAGCAAC ATTCGGACAG GGCGCAAGTT GCCACCTTGG GCGGAAAACC |
|
| 1101 | GCAGCAGAAC CTAATGTACG ACAATTGGGA AGAACGCGGG AAACCGTTTG |
|
| 1151 | AAGGAATCGG CGGGGGCGTG GTCGGATCGG CAAACTGA |
[0830]This encodes a protein having amino acid sequence <SEQ ID 326>:
[0000]| 1 | MAEICLITGT PGSGKTLKMV SMMANDEMFK PDENGVRRKV FTNIKGLKIP | |
|
| 51 | HTHIETDAKK LPKSTDEQLS AHDMYEWIKK PENVGAIVIV DEAQDVWPAR |
|
| 101 | SAGSKIPENV QWLNTHRHQG IDIFVLTQGP KLLDQNLRTL VKRHYHIAAN |
|
| 151 | KMGLRTLLEW KVCADDPVKM ASSAFSSIYT LDKKVYDLYE SAEIHTVNKV |
|
| 201 | KRSKWFYALP VIILLIPLFV GLSYKMLGSY GKKQEEPAAQ ESAATEQQAV |
|
| 251 | LPDKTEGESV NNGNLTADMF VPTLPEKPES KPIYNGVRQV RTFEYIAGCI |
|
| 301 | EGGRTGCTCY SHQGTALKEV TELMCKDYVK NGLPFNPYKE ESQGQEVQQS |
|
| 351 | AQQHSDRAQV ATLGGKPQQN LMYDNWEERG KPFEGIGGGV VGSAN* |
[0831]ORF84ng and ORF84-1 show 95.4% identity in 395 aa overlap:
[0000]
[0832]Based on this analysis, including the presence of a putative transmembrane domain (single-underlined) in the gonococcal protein, and a putative ATP/GTP-binding site motif A (P-loop, double-underlined), it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 39
[0833]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 327>:
[0000]| 1 | GTGGTTTTCC TGAATGCCGA CAACGGGATA TTGGTTCAGG ACTTGCCTTT | |
|
| 51 | TGAAGTCAAA CTGAAAAAAT TCCATATCGA TTTTTACAAT ACGGGTATGC |
|
| 101 | CGCGTGATTT CGCCAGCGAT ATTGAAGTGA CGGACAAGGC AACCGGTGAG |
|
| 151 | AAACTCGAGC GCACCATCCG CGTGAACCAT CCTTTGACCT TGCACGGCAT |
|
| 201 | CACGATTTAT CAGGCGAGTT TTGCCGACGG CGGTTCGGAT TTGACATTCA |
|
| 251 | AGGCGTGGAA TTTGGGTGAT GCTTCGCGCG AGCCTGTCGT GTTGAAGGCA |
|
| 301 | ACATCCATAC ACCAGTTTCC GTTGGAAATT GGCAAACACA AATATCGTCT |
|
| 351 | TGAGTTCGAT CAGTTCACTT CTATGAATGT GGAGGACATG AGCGAGGGCG |
|
| 401 | CGGAACGGGA AAAAAGCCTG AAATCCACGC TGCCCGATGT CCGCGCCGTT |
|
| 451 | ACTCAGGAAG GTCACAAATA CACCAAT... .......... .....TACCG |
|
| 501 | TATCCGTGAT GCGCCAGGCC AGGCGGTCGA ATATAAAAAC TATATGCTGC |
|
| 551 | CGGTTTTGCA GGAACAGGAT TATTTTTGGA TTACCGGCAC GCGCAGCGC. |
|
| 601 | TTGCAGCAGC AATACCGCTG GCTGCGTATC CCCTTGGACA AGCAGTTGAA |
|
| 651 | AGCGGACACC TTTATGGCAT TGCGTGAGTT TTTGAAAGAT GGGGAAGGGC |
|
| 701 | GCAAACGTCT .GTTGCCGAC GCAACCAAAG GCGCACCTGC CGAAATCCGC |
|
| 751 | GAACAATTCA TGCTGGCTGC GGAAAACACG CTGAACATCT TTGCACAAAA |
|
| 801 | AGGCTATTTG GGATTGGACG AATTTATTAC GTCCAATATC CCGAAAGAGC |
|
| 851 | AGCAGGATAA GATGCAGGGC TATTTCTACG AAATGCTTTA CGGCGTGATG |
|
| 901 | AACGCTGCTT TGGATGAAAC CAT.ACCCGG TACGGCTTGC CCGAATGGCA |
|
| 951 | GCAGGATGAA GCGCGGAATC GTTTCCTGCT GCACAGTATG GATGCGTACA |
|
| 1001 | CGGGTTTGAC CGAATATCCC GCGCCTATGC TGCTGCAACT TGATGGGTTT |
|
| 1051 | TCCGAGGTGC GTTCGTCGGG TTTGCAGATG ACCCGTTCCC C.GGTCCGCT |
|
| 1101 | TTTGGTCTAT CTC... |
[0834]This corresponds to the amino acid sequence <SEQ ID 328; ORF88>:
[0000]| 1 | MVFLNADNGI LVQDLPFEVK LKKFHIDFYN TGMPRDFASD IEVTDKATGE | |
|
| 51 | KLERTIRVNH PLTLHGITIY QASFADGGSD LTFKAWNLGD ASREPVVLKA |
|
| 101 | TSIHQFPLEI GKHKYRLEFD QFTSMNVEDM SEGAEREKSL KSTLPDVRAV |
|
| 151 | TQEGHKYTNX XXXXXYRIRD APGQAVEYKN YMLPVLQEQD YFWITGTRSX |
|
| 201 | LQQQYRWLRI PLDKQLKADT FMALREFLKD GEGRKRXVAD ATKGAPAEIR |
|
| 251 | EQFMLAAENT LNIFAQKGYL GLDEFITSNI PKEQQDKMQG YFYEMLYGVM |
|
| 301 | NAALDETXTR YGLPEWQQDE ARNRFLLHSM DAYTGLTEYP APMLLQLDGF |
|
| 351 | SEVRSSGLQM TRSXGPLLVY L... |
[0835]Further work revealed the complete nucleotide sequence <SEQ ID 329>:
[0000]| 1 | ATGAGTAAAT CCCGTAGATC TCCCCCACTT CTTTCCCGTC CGTGGTTCGC | |
|
| 51 | TTTTTTCAGC TCCATGCGCT TTGCAGTCGC TTTGCTCAGT CTGCTGGGTA |
|
| 101 | TTGCATCGGT TATCGGTACG GTGTTGCAGC AAAACCAGCC GCAGACGGAT |
|
| 151 | TATTTGGTCA AATTCGGATC GTTTTGGGCG CAGATTTTTG GTTTTCTGGG |
|
| 201 | ACTGTATGAC GTCTATGCTT CGGCATGGTT TGTCGTTATC ATGATGTTTT |
|
| 251 | TGGTGGTTTC TACCAGTTTG TGCCTGATTC GCAATGTGCC GCCGTTCTGG |
|
| 301 | CGCGAAATGA AGTCTTTTCG GGAAAAGGTT AAAGAAAAAT CTCTGGCGGC |
|
| 351 | GATGCGCCAT TCTTCGCTGT TGGATGTAAA AATTGCGCCC GAGGTTGCCA |
|
| 401 | AACGTTATCT GGAAGTACAA GGTTTTCAGG GAAAAACCAT TAACCGTGAA |
|
| 451 | GACGGGTCGG TTCTGATTGC CGCCAAAAAA GGCACAATGA ACAAATGGGG |
|
| 501 | CTATATCTTT GCCCATGTTG CTTTGATTGT CATTTGCCTG GGCGGGTTGA |
|
| 551 | TAGACAGTAA CCTGCTGTTG AAACTGGGTA TGCTGACCGG TCGGATTGTT |
|
| 601 | CCGGACAATC AGGCGGTTTA TGCCAAGGAT TTCAAGCCCG AAAGTATTTT |
|
| 651 | GGGTGCGTCC AATCTCTCAT TTAGGGGCAA CGTCAATATT TCCGAGGGGC |
|
| 701 | AGAGTGCGGA TGTGGTTTTC CTGAATGCCG ACAACGGGAT ATTGGTTCAG |
|
| 751 | GACTTGCCTT TTGAAGTCAA ACTGAAAAAA TTCCATATCG ATTTTTACAA |
|
| 801 | TACGGGTATG CCGCGTGATT TCGCCAGCGA TATTGAAGTG ACGGACAAGG |
|
| 851 | CAACCGGTGA GAAACTCGAG CGCACCATCC GCGTGAACCA TCCTTTGACC |
|
| 901 | TTGCACGGCA TCACGATTTA TCAGGCGAGT TTTGCCGACG GCGGTTCGGA |
|
| 951 | TTTGACATTC AAGGCGTGGA ATTTGGGTGA TGCTTCGCGC GAGCCTGTCG |
|
| 1001 | TGTTGAAGGC AACATCCATA CACCAGTTTC CGTTGGAAAT TGGCAAACAC |
|
| 1051 | AAATATCGTC TTGAGTTCGA TCAGTTCACT TCTATGAATG TGGAGGACAT |
|
| 1101 | GAGCGAGGGC GCGGAACGGG AAAAAAGCCT GAAATCCACG CTGAACGATG |
|
| 1151 | TCCGCGCCGT TACTCAGGAA GGTAAAAAAT ACACCAATAT CGGCCCTTCC |
|
| 1201 | ATTGTTTACC GTATCCGTGA TGCGGCAGGG CAGGCGGTCG AATATAAAAA |
|
| 1251 | CTATATGCTG CCGGTTTTGC AGGAACAGGA TTATTTTTGG ATTACCGGCA |
|
| 1301 | CGCGCAGCGG CTTGCAGCAG CAATACCGCT GGCTGCGTAT CCCCTTGGAC |
|
| 1351 | AAGCAGTTGA AAGCGGACAC CTTTATGGCA TTGCGTGAGT TTTTGAAAGA |
|
| 1401 | TGGGGAAGGG CGCAAACGTC TGGTTGCCGA CGCAACCAAA GGCGCACCTG |
|
| 1451 | CCGAAATCCG CGAACAATTC ATGCTGGCTG CGGAAAACAC GCTGAACATC |
|
| 1501 | TTTGCACAAA AAGGCTATTT GGGATTGGAC GAATTTATTA CGTCCAATAT |
|
| 1551 | CCCGAAAGAG CAGCAGGATA AGATGCAGGG CTATTTCTAC GAAATGCTTT |
|
| 1601 | ACGGCGTGAT GAACGCTGCT TTGGATGAAA CCATACGCCG GTACGGCTTG |
|
| 1651 | CCCGAATGGC AGCAGGATGA AGCGCGGAAT CGTTTCCTGC TGCACAGTAT |
|
| 1701 | GGATGCGTAC ACGGGTTTGA CCGAATATCC CGCGCCTATG CTGCTGCAAC |
|
| 1751 | TTGATGGGTT TTCCGAGGTG CGTTCGTCGG GTTTGCAGAT GACCCGTTCC |
|
| 1801 | CCGGGTGCGC TTTTGGTCTA TCTCGGCTCG GTGCTGTTGG TATTGGGTAC |
|
| 1851 | GGTATTGATG TTTTATGTGC GCGAAAAACG GGCGTGGGTA TTGTTTTCAG |
|
| 1901 | ACGGCAAAAT CCGTTTTGCC ATGTCTTCGG CCCGCAGCGA ACGGGATTTG |
|
| 1951 | CAGAAGGAAT TTCCAAAACA CGTCGAGAGT CTGCAACGGC TCGGCAAGGA |
|
| 2001 | CTTGAATCAT GACTGA |
[0836]This corresponds to the amino acid sequence <SEQ ID 330; ORF88-1>:
[0000]| 1 | MSKSRRSPPL LSRPWFAFFS SMRFAVALLS LLGIASVIGT VLQQNQPQTD | |
|
| 51 | YLVKFGSFWA QIFGFLGLYD VYASAWFVVI MMFLVVSTSL CLIRNVPPFW |
|
| 101 | REMKSFREKV KEKSLAAMRH SSLLDVKIAP EVAKRYLEVQ GFQGKTINRE |
|
| 151 | DGSVLIAAKK GTMNKWGYIF AHVALIVICL GGLIDSNLLL KLGMLTGRIV |
|
| 201 | PDNQAVYAKD FKPESILGAS NLSFRGNVNI SEGQSADVVF LNADNGILVQ |
|
| 251 | DLPFEVKLKK FHIDFYNTGM PRDFASDIEV TDKATGEKLE RTIRVNHPLT |
|
| 301 | LHGITIYQAS FADGGSDLTF KAWNLGDASR EPVVLKATSI HQFPLEIGKH |
|
| 351 | KYRLEFDQFT SMNVEDMSEG AEREKSLKST LNDVRAVTQE GKKYTNIGPS |
|
| 401 | IVYRIRDAAG QAVEYKNYML PVLQEQDYFW ITGTRSGLQQ QYRWLRIPLD |
|
| 451 | KQLKADTFMA LREFLKDGEG RKRLVADATK GAPAEIREQF MLAAENTLNI |
|
| 501 | FAQKGYLGLD EFITSNIPKE QQDKMQGYFY EMLYGVMNAA LDETIRRYGL |
|
| 551 | PEWQQDEARN RFLLHSMDAY TGLTEYPAPM LLQLDGFSEV RSSGLQMTRS |
|
| 601 | PGALLVYLGS VLLVLGTVLM FYVREKRAWV LFSDGKIRFA MSSARSERDL |
|
| 651 | QKEFPKHVES LQRLGKDLNH D* |
[0837]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0838]ORF88 shows 95.7% identity over a 371aa overlap with an ORF (ORF88a) from strain A of N. meningitidis.
[0000]
[0839]The complete length ORF88a nucleotide sequence <SEQ ID 331> is:
[0000]| 1 | ATGAGTAAAT CCCGTAGATC TCCCCCACTT CTTTCCCGTC CGTGGTTCGC | |
|
| 51 | TTTTTTCAGC TCCATGCGCT TTGCGGTCGC TTTGCTCAGT CTGCTGGGTA |
|
| 101 | TTGCATCGGT TATCGGTACG GTGTTGCAGC AAAACCAGCC GCAGACGGAT |
|
| 151 | TATTTGGTCA AATTCGGATC GTTTTGGGCG CAGATTTTTG GTTTTCTGGG |
|
| 201 | ACTGTATGAC GTCTATGCTT CGGCATGGTT TGTCGTTATC ATGATGTTTT |
|
| 251 | TGGTGGTTTC TACCAGTTTG TGCCTGATTC GCAATGTGCC GCCGTTCTGG |
|
| 301 | CGCGAAATGA AGTCTTTTCG GGAAAAGGTT AAAGAAAAAT CTCTGGCGGC |
|
| 351 | GATGCGCCAT TCTTCGCTGT TGGATGTAAA AATTGCGCCC GAGGTTGCCA |
|
| 401 | AACGTTATCT GGAAGTACAA GGTTTTCAGG GAAAAACCAT TAACCGTGAA |
|
| 451 | GACGGGTCGG TTCTGATTGC CGCCAAAAAA GGCACAATGA ACAAATGGGG |
|
| 501 | CTATATCTTT GCCCATGTTG CTTTGATTGT CATTTGCCTG GGCGGGTTGA |
|
| 551 | TAGACAGTAA CCTGCTGTTG AAACTGGGTA TGCTGACCGG TCGGATTGTT |
|
| 601 | CCGGACAATC AGGCGGTTTA TGCCAAGGAT TTCAAGCCCG AAAGTATTTT |
|
| 651 | GGGTGCGTCC AATCTCTCAT TTAGGGGCAA CGTCAATATT TCCGAGGGGC |
|
| 701 | AGAGTGCGGA TGTGGTTTTC CTGAATGCCG ACAACGGGAT ATTGGTTCAG |
|
| 751 | GACTTGCCTT TTGAAGTCAA ACTGAAAAAA TTCCATATCG ATTTTTACAA |
|
| 801 | TACGGGTATG CCGCGCGATT TTGCCAGTGA TATTGAAGTA ACGGATAAGG |
|
| 851 | CAACCGGTGA GAAACTCGAG CGCACCATCC GCGTGAACCA TCCTTTGACC |
|
| 901 | TTGCACGGCA TCACGATTTA TCAGGCGAGT TTTGCCGACG GCGGTTCGGA |
|
| 951 | TTTGACATTC AAGGCGTGGA ATTTGGGTGA TGCTTCGCGC GAGCCTGTCG |
|
| 1001 | TGTTGAAGGC AACATCCATA CACCAGTTTC CGTTGGAAAT TGGCAAACAC |
|
| 1051 | AAATATCGTC TTGAGTTCGA TCAGTTTACT TCTATGAATG TGGAGGACAT |
|
| 1101 | GAGCGAGGGC GCGGAACGGG AAAAAAGCCT GAAATCCACG CTGAACGATG |
|
| 1151 | TCCGCGCCGT TACTCAGGAA GGTAAAAAAT ACACCAATAT CGGCCCTTCC |
|
| 1201 | ATTGTTTACC GTATCCGTGA TGCGGCAGGG CAGGCGGTCG AATATAAAAA |
|
| 1251 | CTATATGCTG CCGGTTTTGC AGGAACAGGA TTATTTTTGG ATTACCGGCA |
|
| 1301 | CGCGCAGCGG CTTGCAGCAG CAATACCGCT GGCTGCGTAT CCCCTTGGAC |
|
| 1351 | AAGCAGTTGA AAGCGGACAC CTTTATGGCA TTGCGTGAGT TTTTGAAAGA |
|
| 1401 | TGGGGAAGGG CGCAAACGTC TGGTTGCCGA CGCAACCAAA GGCGCACCTG |
|
| 1451 | CCGAAATCCG CGAACAATTC ATGCTGGCTG CGGAAAACAC GCTGAACATC |
|
| 1501 | TTTGCACAAA AAGGCTATTT GGGATTGGAC GAATTTATTA CGTCCAATAT |
|
| 1551 | CCCGAAAGAG CAGCAGGATA AGATGCAGGG CTATTTCTAC GAAATGCTTT |
|
| 1601 | ACGGCGTGAT GAACGCTGCT TTGGATGAAA CCATACGCCG GTACGGCTTG |
|
| 1651 | CCCGAATGGC AGCAGGATGA AGCGCGGAAT CGTTTCCTGC TGCACAGTAT |
|
| 1701 | GGATGCGTAC ACGGGTTTGA CCGAATATCC CGCGCCTATG CTGCTGCAAC |
|
| 1751 | TTGATGGGTT TTCCGAGGTG CGTTCGTCGG GTTTGCAGAT GACCCGTTCC |
|
| 1801 | CCGGGTGCGC TTTTGGTCTA TCTCGGCTCG GTGCTGTTGG TATTGGGTAC |
|
| 1851 | GGTATTGATG TTTTATGTGC GCGAAAAACG GGCGTGGGTA TTGTTTTCAG |
|
| 1901 | ACGGCAAAAT CCGTTTTGCC ATGTCTTCGG CCCGCAGCGA ACGGGATTTG |
|
| 1951 | CAGAAGGAAT TTCCAAAACA CGTCGAGAGT CTGCAACGGC TCGGCAAGGA |
|
| 2001 | CTTGAATCAT GACTGA |
[0840]This encodes a protein having amino acid sequence <SEQ ID 332>:
[0000]| 1 | MSKSRRSPPL LSRPWFAFFS SMRFAVALLS LLGIASVIGT VLQQNQPQTD | |
|
| 51 | YLVKFGSFWA QIFGFLGLYD VYASAWFVVI MMFLVVSTSL CLIRNVPPFW |
|
| 101 | REMKSFREKV KEKSLAAMRH SSLLDVKIAP EVAKRYLEVQ GFQGKTINRE |
|
| 151 | DGSVLIAAKK GTMNKWGYIF AHVALIVICL GGLIDSNLLL KLGMLTGRIV |
|
| 201 | PDNQAVYAKD FKPESILGAS NLSFRGNVNI SEGQSADVVF LNADNGILVQ |
|
| 251 | DLPFEVKLKK FHIDFYNTGM PRDFASDIEV TDKATGEKLE RTIRVNHPLT |
|
| 301 | LHGITIYQAS FADGGSDLTF KAWNLGDASR EPVVLKATSI HQFPLEIGKH |
|
| 351 | KYRLEFDQFT SMNVEDMSEG AEREKSLKST LNDVRAVTQE GKKYTNIGPS |
|
| 401 | IVYRIRDAAG QAVEYKNYML PVLQEQDYFW ITGTRSGLQQ QYRWLRIPLD |
|
| 451 | KQLKADTFMA LREFLKDGEG RKRLVADATK GAPAEIREQF MLAAENTLNI |
|
| 501 | FAQKGYLGLD EFITSNIPKE QQDKMQGYFY EMLYGVMNAA LDETIRRYGL |
|
| 551 | PEWQQDEARN RFLLHSMDAY TGLTEYPAPM LLQLDGFSEV RSSGLQMTRS |
|
| 601 | PGALLVYLGS VLLVLGTVLM FYVREKRAWV LFSDGKIRFA MSSARSERDL |
|
| 651 | QKEFPKHVES LQRLGKDLNH D* |
[0841]ORF88a and ORF88-1 100.0% identity in 671 aa overlap:
[0000]
[0842]Homology with a Predicted ORF from N. gonorrhoeae
[0843]ORF88 shows 93.8% identity over a 371aa overlap with a predicted ORF (ORF88.ng) from N. gonorrhoeae:
[0000]
[0844]An ORF88ng nucleotide sequence <SEQ ID 333> was predicted to encode a protein having amino acid sequence <SEQ ID 334>:
[0000]| 1 | MVFLNADNGM LVQDLPFEVK LKKFHIDFYN TGMPRDFASD IEVTDKATGE | |
|
| 51 | KLERTIRVNH PLTLHGITIY QASFADGGSD LTFKAWNLRD ASREPVVLKA |
|
| 101 | TSIHQFPLEI GKHKYRLEFD QFTSMNVEDM SEGAEREKSL KSTLNDVRAV |
|
| 151 | TQEGKKYTNI GPSIVYRIRD AAGQAVEYKN YMLPILQDKD YFWLTGTRSG |
|
| 201 | LQQQYRWLRI PLDKQLKADT FMALREFLKD GEGRKRLVAD ATKDAPAEIR |
|
| 251 | EQFMLAAENT LNIFAQKGYL GLDEFITSNI PKGQQDKMQG YFYEMLYGVM |
|
| 301 | NAALDETIRR YGLPEWQQDE ARNRFLLHSM DAYTGLTEYP APMLLQLDGF |
|
| 351 | SEVRSSGLQM TRSPGALLVY LGSVLLVLGT VFMFYVPKKR AWVLFSNXKI |
|
| 401 | RFAMSSARSE RDLQKEFPKH VESLQRLGKD LNHD* |
[0845]Further work revealed the complete gonococcal DNA sequence <SEQ ID 335>:
[0000]| 1 | ATGAGTAAAT CCCGTATATC TCCCACACTT CTTTCCCGTC CGTGGTTCGC | |
|
| 51 | TTTTTTCAGC TCCATGCGCT TTGCGGTCGC TTTGCTCAGT CTGCTGGGTA |
|
| 101 | TTGCATCGGT TATCGGCACG GTGTTACAGC AAAACCAGCC GCAGACGGAT |
|
| 151 | TATTTGGTCA AATTCGGACC GTTTTGGACT CGGATTTTTG ATTTTTTGGG |
|
| 201 | TTTGTATGAT GTCTATGCTT CGGCATGGTT TGTCGTTATC ATGATGTTTC |
|
| 251 | TGGTGGTTTC TACCAGTTTG TGTTTAATCC GTAACGTTCC GCCGTTTTGG |
|
| 301 | CGCGAAATGA AGTCTTTCCG GGAAAAGGTT AAAGAAAAAT CTCTGGCGGC |
|
| 351 | GATGCGCCAT TCTTCGCTGT TGGATGTAAA AATTGCCCCC GAAGTTGCCA |
|
| 401 | AACGTTATCT GGAGGTGCGG GGTTTTCAGG GAAAAACCGT CAGCCGTGAG |
|
| 451 | GACGGGTCGG TTCTGATTGC CGCCAAAAAA GGCAcaatga acaaATGGGG |
|
| 501 | CTATATCTTT GCccaagtag ctTTGATTGT CATTTGCCTG GGCGGGTTGA |
|
| 551 | TAGACAGTAA CCTGCTGCTG AAGCTGGGTA TGCTGGCCGG TCGGATTGTT |
|
| 601 | CCGGACAATC AGGCGGTTTA TGCCAAGGAT TTCAAGCCCG AAAGTATTTT |
|
| 651 | GGGTGCGTCC AATCTCTCAT TTAGGGGCAA CGTCAATATT TCCGAGGGGC |
|
| 701 | AAAGTGCGGA TGTGGTTTTC CTGAATGCCG ACAACGGGAT GTTGGTTCAG |
|
| 751 | GACTTGCCTT TTGAAGTCAA ACTGAAAAAA TTCCATATCG ATTTTTACAA |
|
| 801 | TACGGGTATG CCGCGCGATT TTGCCAGCGA TATTGAAGTA ACGGACAAGG |
|
| 851 | CAACCGGTGA GAAACTCGAG CGCACCATCC GCGTGAACCA TCCTTTGACC |
|
| 901 | TTGCACGGCA TCACGATTTA TCAGGCGAGT TTTGCCGACG GCGGTTCGGA |
|
| 951 | TTTGACATTC AAGGCGTGGA ATTTGAGGGA TGCTTCGCGC GAACCTGTCG |
|
| 1001 | TGTTGAAGGC AACCTCCATA CACCAGTTTC CGTTGGAAAT CGGCAAACAC |
|
| 1051 | AAATATCGTC TTGAGTTCGA TCAGTTCACT TCTATGAATG TGGAGGACAT |
|
| 1101 | GAGCGAGGGT GCGGAACGGG AAAAAAGCCT GAAATCCACT CTGAACGATG |
|
| 1151 | TCCGCGCCGT TACTCAGGAA GGTAAAAAAT ACACCAATAT CGGCCCTTCC |
|
| 1201 | ATCGTGTACC GCATCCGTGA TGcggCAGGG CAGGCGGTCG AATATAAAAA |
|
| 1251 | CTATATGCTG CCGATTTTGC AGGACAAAGA TTATTTTTGG CTGACCGGCA |
|
| 1301 | CGCGCAGCGG CTTGCAGCAG CAATACCGCT GGCTGCGTAT CCCCTTGGAC |
|
| 1351 | AAGCAGTTGA AAGCGGACAC CTTTATGGCA TTGCGTGAGT TTTTGAAAGA |
|
| 1401 | TGGGGAAGGG CGCAAACGTC TGGTTGCCGA CGCAACCAAA GACGCACCTG |
|
| 1451 | CCGAAATCCG CGAACAATTC ATGCTGGCTG CGGAAAACAC GCTGAATATC |
|
| 1501 | TTTGCGCAAA AAGGCTATTT GGGATTGGAC GAATTTATTA CGTCCAATAT |
|
| 1551 | CCCGAAAGGG CAGCAGGATA AGATGCAGGG CTATTTCTAC GAAATGCTTT |
|
| 1601 | ACGGCGTGAT GAACGCTGCT TTGGATGAAA CCATACGCCG GTACGGCTTG |
|
| 1651 | CCCGAATGGC AGCAGGATGA AGCGCGGAAC CGTTTCCTGC TGCACAGTAT |
|
| 1701 | GGATGCCTAT ACGGGGCTGA CGGAATATCC CGCGCCTATG CTGCTCCAGC |
|
| 1751 | TTGACGGGTT TTCCGAGGTG CGTTCCTCAG GTTTGCAGAT GACCCGTTCG |
|
| 1801 | CCGGGTGCGC TTTTGGTCTA TCtcggctcg gtattgttgg TTTTGGgtac |
|
| 1851 | ggtaTttatg tTTTATGTGC GCGAAAAACG GGCGTGGgta tTGTTTTCag |
|
| 1901 | aCGGCAAAAT CCGTTTTGCT ATGtCTTcgg CCcgcagcga ACGGGATTTG |
|
| 1951 | cAGAaggaaT TTCCAAAACA CGtcgAGAGC CTGCAACggc tcggcaaggA |
|
| 2001 | CttgaaTCAT GACTga |
[0846]This corresponds to the amino acid sequence <SEQ ID 336; ORF88ng-1>:
[0000]| 1 | MSKSRISPTL LSRPWFAFFS SMRFAVALLS LLGIASVIGT VLQQNQPQTD | |
|
| 51 | YLVKFGPFWT RIFDFLGLYD VYASAWFVVI MMFLVVSTSL CLIRNVPPFW |
|
| 101 | REMKSFREKV KEKSLAAMRH SSLLDVKIAP EVAKRYLEVR GFQGKTVSRE |
|
| 151 | DGSVLIAAKK GTMNKWGYIF AQVALIVICL GGLIDSNLLL KLGMLAGRIV |
|
| 201 | PDNQAVYAKD FKPESILGAS NLSFRGNVNI SEGQSADVVF LNADNGMLVQ |
|
| 251 | DLPFEVKLKK FHIDFYNTGM PRDFASDIEV TDKATGEKLE RTIRVNHPLT |
|
| 301 | LHGITIYQAS FADGGSDLTF KAWNLRDASR EPVVLKATSI HQFPLEIGKH |
|
| 351 | KYRLEFDQFT SMNVEDMSEG AEREKSLKST LNDVRAVTQE GKKYTNIGPS |
|
| 401 | IVYRIRDAAG QAVEYKNYML PILQDKDYFW LTGTRSGLQQ QYRWLRIPLD |
|
| 451 | KQLKADTFMA LREFLKDGEG RKRLVADATK DAPAEIREQF MLAAENTLNI |
|
| 501 | FAQKGYLGLD EFITSNIPKG QQDKMQGYFY EMLYGVMNAA LDETIRRYGL |
|
| 551 | PEWQQDEARN RFLLHSMDAY TGLTEYPAPM LLQLDGFSEV RSSGLQMTRS |
|
| 601 | PGALLVYLGS VLLVLGTVFM FYVREKRAWV LFSDGKIRFA MSSARSERDL |
|
| 651 | QKEFPKHVES LQRLGKDLNH D* |
[0847]ORF88ng-1 and ORF88-1 show 97.0% identity in 671 aa overlap:
[0000]
[0848]Furthermore, ORG88ng-1 shows homology with a hypothetical protein from Aquifex aeolicus:
[0000]| gi|2984296 (AE000771) hypothetical protein [Aquifex aeolicus] | |
| Length = 537 |
| Score = 94.4 bits (231), Expect = 2e−18. |
| Identities = 91/334 (27%), Positives = 159/334 (47%), |
| Gaps = 59/334 (17%) |
|
| Query: | 16 | FAFFSSMRFAVALLSLLGIASVIG-TVLQQNQPQTDYLVKFGPFWTRIFDFLGLYDVYAS | 74 | |
| | + F +S++ A+ ++ +LGI S++G T ++QNQ YL +FG L L DV+ S | |
| Sbjct: | 80 | YDFLASLKLAIFIMLVLGILSMLGSTYIKQNQSFEWYLDQFGYDVGIWIWKLWLNDVFHS | 139 |
|
| Query: | 75 | AWFVVIMMFLVVSTSLCLIRNVPPFWREMKSFREKVKEKSLAAMRHSSLLDVKIAPEVAK | 134 |
| | ++++ ++ L V+ C I+ +P W++ S +E++ + A +H + VKI P+ K | |
| Sbjct: | 140 | WYYILFIVLLAVNLIFCSIKRLPRVWKQAFS-KERILKLDEHAEKHLKPITVKI-PDKDK | 197 |
|
| Query: | 135 | --RYLEVRGFQGKTVSREDGSVLIAAKKGTMNKWGYIFAQVALIVICLGGLIDSNLLLKL | 192 |
| | ++L +GF+ V E + + A+KG ++ G +AL+VI G LID | |
| Sbjct: | 198 | VLKFLLKKGFK-VFVEEEGNKLYVFAEKGRFSRLGVYITHIALLVIMAGALID------- | 249 |
|
| Query: | 193 | GMLAGRIVPDNQAVYAKDFKPESILGASNLSFRGNVNISEGQSADVVFLNADNGMLVQDL | 252 |
| | +I+G RG++ ++EG + DV+ + A+ L | |
| Sbjct: | 250 | ----------------------AIVGV-----RGSLIVAEGDTNDVMLVGAE--QKPYKL | 280 |
|
| Query: | 253 | PFEVKLKKFHIDFY---NTGMPRDFA-------SDIEVTDKATGEKLER--TIRVNHPLT | 300 |
| | PF V L F I Y N + + FA SDIE+ + G K+E T++VN P | |
| Sbjct: | 281 | PFAVHLIDFRIKTYAEENPNVDKRFAQAVSSYESDIEIIN---GGKVEAKGTVKVNEPFD | 337 |
|
| Query: | 301 | LHGITIYQASFA--DGGSDLTFKAWNLRDASREP | 332 |
| | ++QA++ DG S + + + A +P | |
| Sbjct: | 338 | FGRYRLFQATYGILDGTSGMGVIVVDRKKAHEDP | 371 |
[0849]Based on this analysis, including the putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 40
[0850]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 337>:
[0000]| 1 | ATGATGAGTA ATAmAATGGm ACAAAAAGGG TTTACATTGA TTGmGmTGAT | |
|
| 51 | GATAGTCGTC GCGATACTCG GCATTATCAG CGTCATTGCC ATACCTTCTT |
|
| 101 | ATCmAAGTTA TATTGAAAAA GGCTATCAGT CCCAGCTTTA TACGGAGATG |
|
| 151 | GyCGGTATCA ACAATATTTC CAAACAGTTT ATTTTGAAAA ATCCCCTGGA |
|
| 201 | CGATAATCAG ACCATCGAGA ACAAACTGGA AATATTTGTC TCAGGCTATA |
|
| 251 | AGATGAATCC GAAAATTGCC AAAAAaTATA GTGTTTCGGT AAAGTTTGTC |
|
| 301 | GATAAGGAAA AATCAAGGGC ATACAGGTTG GTCGGCGTTC CGAAGGCGGG |
|
| 351 | GACGGGTTAT ACTTTGTCGG TATGGATGAA CAGCGTGGGC GACGGATACA |
|
| 401 | AATGCCGTGA TGCCGCTTCT GCCCAAGCCC ATTTGGAGAC CTTGTCCTCA |
|
| 451 | GATGTCGGCT GTGAAGCCTT CTCTAATCGT AAAAAATAA |
[0851]This corresponds to the amino acid sequence <SEQ ID 338; ORF89>:
[0000]| 1 | MMSNXMXQKG FTLIXXMIVV AILGIISVIA IPSYXSYIEK GYQSQLYTEM | |
|
| 51 | XGINNISKQF ILKNPLDDNQ TIENKLEIFV SGYKMNPKIA KKYSVSVKFV |
|
| 101 | DKEKSRAYRL VGVPKAGTGY TLSVWMNSVG DGYKCRDAAS AQAHLETLSS |
|
| 151 | DVGCEAFSNR KK* |
[0852]Further work revealed the complete nucleotide sequence <SEQ ID 339>:
[0000]| 1 | ATGATGAGTA ATAAAATGGA ACAAAAAGGG TTTACATTGA TTGAGATGAT | |
|
| 51 | GATAGTCGTC GCGATACTCG GCATTATCAG CGTCATTGCC ATACCTTCTT |
|
| 101 | ATCAAAGTTA TATTGAAAAA GGCTATCAGT CCCAGCTTTA TACGGAGATG |
|
| 151 | GTCGGTATCA ACAATATTTC CAAACAGTTT ATTTTGAAAA ATCCCCTGGA |
|
| 201 | CGATAATCAG ACCATCGAGA ACAAACTGGA AATATTTGTC TCAGGCTATA |
|
| 251 | AGATGAATCC GAAAATTGCC AAAAAATATA GTGTTTCGGT AAAGTTTGTC |
|
| 301 | GATAAGGAAA AATCAAGGGC ATACAGGTTG GTCGGCGTTC CGAAGGCGGG |
|
| 351 | GACGGGTTAT ACTTTGTCGG TATGGATGAA CAGCGTGGGC GACGGATACA |
|
| 401 | AATGCCGTGA TGCCGCTTCT GCCCAAGCCC ATTTGGAGAC CTTGTCCTCA |
|
| 451 | GATGTCGGCT GTGAAGCCTT CTCTAATCGT AAAAAATAA |
[0853]This corresponds to the amino acid sequence <SEQ ID 340; ORF89-1>:
[0000]| 1 | MMSNKMEQKG FTLIEMMIVV AILGIISVIA IPSYQSYIEK GYQSQLYTEM | |
|
| 51 | VGINNISKQF ILKNPLDDNQ TIENKLEIFV SGYKMNPKIA KKYSVSVKFV |
|
| 101 | DKEKSRAYRL VGVPKAGTGY TLSVWMNSVG DGYKCRDAAS AQAHLETLSS |
|
| 151 | DVGCEAFSNR KK* |
[0854]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with PilE of N. gonorrhoeae (Accession Number Z69260).
[0855]ORF89 and PilE protein show 30% aa identity in 120a overlap:
[0000]| orf89 | 8 | QKGFTLIXXMIVVAILGIISVIAIPSYXSYIEKGYQSQLYTEMXGINNISKQFILKNPL- | 66 | |
| | QKGFTLI MIV+AI+GI++ +A+P+Y Y + S+ G + ++ L + + | |
| PilE | 5 | QKGFTLIELMIVIAIVGILAAVALPAYQDYTARAQVSEAILLAEGQKSAVTEYYLNHGIW | 64 |
|
| orf89 | 67 | -DDNQTIENKLEIFVSGYKMNPKIAKKYSVSVKFVDKEKSRAYRLVGVPKAGTGYTLSVW | 125 |
| | DN + +G + KI KY SV + GV K G LS+W | |
| PilE | 65 | PKDNTS---------AGVASSDKIKGKYVQSVTVAKGVVTAEMASTGVNKEIQGKKLSLW | 115 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0856]ORF89 shows 83.3% identity over a 162aa overlap with an ORF (ORF89a) from strain A of N. meningitidis.
[0000]
[0857]The complete length ORF89a nucleotide sequence <SEQ ID 341> is:
[0000]| 1 | ATGATGAGTA ATAAAATGGA ACAAAAAGGG TTTACATTGA TTGNGANGNT | |
|
| 51 | NATNGNCNTC GCGATACNCN GCNTTANCAG CGTCATTNCN ATNNNTNCNT |
|
| 101 | ATCNNAGTTA TATTGAAAAA GGCTATCAGT CCCAGCTTTA TACGGAGATG |
|
| 151 | GTCGGTATCA ACAATATTTC CAAACAGTNT ATTTTGAAAA ATCCCCTGGA |
|
| 201 | CGATAATCAG ACCATCAAGA GCAAACTGGA AATATTTGTC TCAGGCTATA |
|
| 251 | AGATGAATCC GAAAATTGCC GAAAAATATA ATGTTTCGGT GCATTTTGTC |
|
| 301 | AATGAGGAAA AACCNAGGGC ATACAGCTTG GTCGGCGTTC CAAAGACGGG |
|
| 351 | GACGGGTTAT ACTTTGTCGG TATGGATGAA CAGCGTGGGC GACGGATACA |
|
| 401 | AATGCCGTGA TGCCGCTTCT GCCCGAGCCC ATTTGGAGAC CTTGTCCTCA |
|
| 451 | GATGTCGGCT GTGAAGCCTT CTCTAATCGT AAAAAATAG |
[0858]This encodes a protein having amino acid sequence <SEQ ID 342>:
[0000]| 1 | MMSNKMEQKG FTLIXXXXXX AIXXXXSVIX XXXYXSYIEK GYQSQLYTEM | |
|
| 51 | VGINNISKQX ILKNPLDDNQ TIKSKLEIFV SGYKMNPKIA EKYNVSVHFV |
|
| 101 | NEEKPRAYSL VGVPKTGTGY TLSVWMNSVG DGYKCRDAAS ARAHLETLSS |
|
| 151 | DVGCEAFSNR KK* |
[0859]ORF89a and ORF89-1 show 83.3% identity in 162 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0860]ORF89 shows 84.6% identity over a 162aa overlap with a predicted ORF (ORF89.ng) from N. gonorrhoeae.
[0000]
[0861]The complete length ORF89ng nucleotide sequence <SEQ ID 343> is:
[0000]| 1 | aTGATGAGCA ATAAAATGGA ACAAAAAGGG TTTACATTGA TTGAGATGAT | |
|
| 51 | GATAGTTGTC ACGATACTCG GCATCATCAG CGTCATTGCC ATACCTTCTT |
|
| 101 | ATCAGAGTTA TATTGAAAAA GGCTATCAGT CCCAGCTTTA TACGGAGATG |
|
| 151 | GTCGGTATCA ACAATGTTCT CAAACAGTTT ATTTTGAAAA ATCCCCAGGA |
|
| 201 | CGATAATGAT ACCCTCAAGA GCAAACTGAA AATATTTGTC TCAGGCTATA |
|
| 251 | AGATGAATCC GAAAAttgCC AAAAAATATA GTGTTTCGGt aaggtttGTC |
|
| 301 | gatGCGGAAA AACCAAGGGC ATACAGGTTG GTCGGCGTTC CGAACGCGGG |
|
| 351 | GACGGGTTAT ACTTTGTCGG TATGGATGAA CAGCGTGGGC GACGGATACA |
|
| 401 | AATGCCGTGA TGCCACTTCT GCCCAGGCCT ATTCGGACAC CTTGTCCGCA |
|
| 451 | GATAGCGGCT GTGAAGCTTT CTCTAATCGT AAAAAATAG |
[0862]This encodes a protein having amino acid sequence <SEQ ID 344>:
[0000]| 1 | MMSNKMEQKG FTLIEMMIVV TILGIISVIA IPSYQSYIEK GYQSQLYTEM | |
|
| 51 | VGINNVLKQF ILKNPQDDND TLKSKLKIFV SGYKMNPKIA KKYSVSVRFV |
|
| 101 | DAEKPRAYRL VGVPNAGTGY TLSVWMNSVG DGYKCRDATS AQAYSDTLSA |
|
| 151 | DSGCEAFSNR KK* |
[0863]This gonococcal protein has a putative leader peptide (underlined) and N-terminal methylation site (NMePhe or type-4 pili, double-underlined). In addition, ORF89ng and ORF89-1 show 88.3% identity in 162 aa overlap:
[0000]
[0864]Based on this analysis, including the gonococcal motifs and the homology with the known PilE protein, it was predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0865]ORF89-1 (13.6 kDa) was cloned in the pGex vector and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 11A shows the results of affinity purification of the GST-fusion protein. Purified GST-fusion protein was used to immunise mice, whose sera gave a positive result in the ELISA test, confirming that ORF89-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 41
[0866]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 345>:
[0000]| 1 | ATGAAAAAAT CCTCCCTCAT CAGCGCATTG GGCATCGGTA TTTTGAGCAT | |
|
| 51 | CGGCATGGCA TTTGCCGCCC CTGCCGACGC GGTAAGCCAA ATCCGTCAAA |
|
| 101 | ACGCCACTCA AGTATTGAGC ATCTTAAAAA ACGGCGATGC CAACACCGCT |
|
| 151 | CGCCAAAAAG CCGAAGCCTA TGCGATTCCC TATTTCGATT TCCAACGTAT |
|
| 201 | GACCGCATTG GCGGTCGGCA ACCCTTGGsG CACCG.GTCC GACG.GCAAA |
|
| 251 | AACAAGCGTT GGCCn.AGAA TTTCAACCC... |
[0867]This corresponds to the amino acid sequence <SEQ ID 346; ORF91>:
[0000]| 1 | MKKSSLISAL GIGILSIGMA FAAPADAVSQ IRQNATQVLS ILKNGDANTA | |
|
| 51 | RQKAEAYAIP YFDFQRMTAL AVGNPWXTXS DXQKQALAXE FQP... |
[0868]Further work revealed the complete nucleotide sequence <SEQ ID 347>:
[0000]| 1 | ATGAAAAAAT CCTCCCTCAT CAGCGCATTG GGCATCGGTA TTTTGAGCAT | |
|
| 51 | CGGCATGGCA TTTGCCGCCC CTGCCGACGC GGTAAGCCAA ATCCGTCAAA |
|
| 101 | ACGCCACTCA AGTATTGAGC ATCTTAAAAA ACGGCGATGC CAACACCGCT |
|
| 151 | CGCCAAAAAG CCGAAGCCTA TGCGATTCCC TATTTCGATT TCCAACGTAT |
|
| 201 | GACCGCATTG GCGGTCGGCA ACCCTTGGCG CACCGCGTCC GACGCGCAAA |
|
| 251 | AACAAGCGTT GGCCAAAGAA TTTCAAACCC TGCTGATCCG CACCTATTCC |
|
| 301 | GGCACGATGC TGAAATTAAA AAACGCCAAC GTCAACGTCA AAGACAATCC |
|
| 351 | CATCGTCAAT AAAGGCGGCA AAGAAATCAT CGTCCGCGCC GAAGTCGGCG |
|
| 401 | TACCCGGGCA AAAACCCGTC AACATGGACT TCACCACCTA CCAAAGCGGC |
|
| 451 | GGTAAATACC GTACCTACAA CGTCGCCATC GAAGGCGCGA GCCTGGTTAC |
|
| 501 | CGTGTACCGC AACCAATTCG GCGAAATTAT CAAAGCGAAA GGCGTGGACG |
|
| 551 | GACTGATTGC CGAGTTGAAA GCCAAAAACG GCGGCAAATA A |
[0869]This corresponds to the amino acid sequence <SEQ ID 348; ORF91-1>:
[0000]| 1 | MKKSSLISAL GIGILSIGMA FAAPADAVSQ IRQNATQVLS ILKNGDANTA | |
|
| 51 | RQKAEAYAIP YFDFQRMTAL AVGNPWRTAS DAQKQALAKE FQTLLIRTYS |
|
| 101 | GTMLKLKNAN VNVKDNPIVN KGGKEIIVRA EVGVPGQKPV NMDFTTYQSG |
|
| 151 | GKYRTYNVAI EGASLVTVYR NQFGEIIKAK GVDGLIAELK AKNGGK* |
[0870]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0871]ORF91 shows 92.4% identity over a 92aa overlap with an ORF (ORF91a) from strain A of N. meningitidis:
[0000]
[0872]The complete length ORF91a nucleotide sequence <SEQ ID 349> is:
[0000]| 1 | ATGAAAAAAT CCTCCTTCAT CAGCGCATTG GGCATCGGTA TTTTGAGCAT | |
|
| 51 | CGGCATGGCA TTTGCCGCCC CTGCCGACGC GGTAAACCAA ATCCGTCAAA |
|
| 101 | ACGCCACTCA AGTATTGAGC ATCTTAAAAA GCGGTGATGC CAACACCGCC |
|
| 151 | CGCCAAAAAG CCGAAGCCTA TGCGATTCCC TATTTCGATT TCCAACGTAT |
|
| 201 | GACCGCATTG GCGGTCGGCA ACCCTTGGCG CACCGCGTCC GACGCGCAAA |
|
| 251 | AACAAGCGTT GGCCAAAGAA TTTCAAACCC TGCTGATCCG CACCTATTCC |
|
| 301 | GGCACGATGC TGAAATTAAA AAACGCCAAC GTCAACGTCA AAGACAATCC |
|
| 351 | CATCGTCAAT AAAGGCGGCA AAGAAATCAT CGTCCGCGCC GAAGTCGGCG |
|
| 401 | TACCCGGGCA AAAACCCGTC AACATGGACT TCACCACCTA CCAAAGCGGC |
|
| 451 | GGTAAATACC GTACCTACAA CGTCGCCATC GAAGGCGCGA GCCTGGTTAC |
|
| 501 | CGTGTACCGC AACCAATTCG GCGAAATTAT CAAAGCGAAA GGCGTGGACG |
|
| 551 | GACTGATTGC CGAGTTGAAG GCTAAAAACG GCAGCAAGTA A |
[0873]This encodes a protein having amino acid sequence <SEQ ID 350>:
[0000]| 1 | MKKSSFISAL GIGILSIGMA FAAPADAVNQ IRQNATQVLS ILKSGDANTA | |
|
| 51 | RQKAEAYAIP YFDFQRMTAL AVGNPWRTAS DAQKQALAKE FQTLLIRTYS |
|
| 101 | GTMLKLKNAN VNVKDNPIVN KGGKEIIVRA EVGVPGQKPV NMDFTTYQSG |
|
| 151 | GKYRTYNVAI EGASLVTVYR NQFGEIIKAK GVDGLIAELK AKNGSK* |
[0874]ORF91a and ORF91-1 show 98.0% identity in 196 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0875]ORF91 shows 84.8% identity over a 92aa overlap with a predicted ORF (ORF91.ng) from N. gonorrhoeae:
[0000]
[0876]The complete length ORF91ng nucleotide sequence <SEQ ID 351> is predicted to encode a protein having amino acid sequence <SEQ ID 352>:
[0000]| 1 | VKKSSFISAL GIGILSIGMA FASPADAVGQ IRQNATQVLT ILKSGDAASA | |
|
| 51 | RPKAEAYAVP YFDFQRMTAL AVGNPWRTAS DAQKQALAKE FQTLLIRTYS |
|
| 101 | GTMLKFKNAT VNVKDNPIVN KGGKEIVVRA EVGIPGQKPV NMDFTTYQSG |
|
| 151 | GKYRTYNVAI EGTSLVTVYR NQFGEIIKAK GIDGLIAELK AKNGGK* |
[0877]Further work revealed the complete nucleotide sequence <SEQ ID 353>:
[0000]| 1 | ATGAAAAAAT CCTCCTTCAT CAGCGCATTG GGCATCGGTA TTTTGAGCAT | |
|
| 51 | CGGCATGGCA TTTGCCTCCC CGGCCGACGC AGTGGGACAA ATCCGCCAAA |
|
| 101 | ACGCCACACA GGTTTTGACC ATCCTCAAAA GCGGCGACGC GGCTTCTGCA |
|
| 151 | CGCCCAAAAG CCGAAGCCTA TGCGGTTCCC TATTTCGATT TCCAACGTAT |
|
| 201 | GACCGCATTG GCGGTCGGCA ACCCTTGGCG TACCGCGTCC GACGCGCAAA |
|
| 251 | AACAAGCGTT GGCCAAAGAA TTTCAAACCC TGCTGATCCG CACCTATTCC |
|
| 301 | GGCACGATGC TGAAATTCAA AAACGCGACC GTCAACGTCA AAGACAATCC |
|
| 351 | CATCGTCAAT AAGGGCGGCA AGGAAATCGT CGTCCGTGCC GAAGTCGGCA |
|
| 401 | TCCCCGGTCA GAAGCCCGTC AATATGGACT TTACCACCTA CCAAAGCGGC |
|
| 451 | GGCAAATACC GTACCTACAA CGTCGCCATC GAAGGCACGA GCCTGGTTAC |
|
| 501 | CGTGTACCGC AACCAATTCG GCGAAATCAT CAAAGCCAAA GGCATCGACG |
|
| 551 | GGCTGATTGC CGAGTTGAAA GCCAAAAACG GCGGCAAATA A |
[0878]This corresponds to the amino acid sequence <SEQ ID 354; ORF91ng-1>:
[0000]| 1 | MKKSSFISAL GIGILSIGMA FASPADAVGQ IRQNATQVLT ILKSGDAASA | |
|
| 51 | RPKAEAYAVP YFDFQRMTAL AVGNPWRTAS DAQKQALAKE FQTLLIRTYS |
|
| 101 | GTMLKFKNAT VNVKDNPIVN KGGKEIVVRA EVGIPGQKPV NMDFTTYQSG |
|
| 151 | GKYRTYNVAI EGTSLVTVYR NQFGEIIKAK GIDGLIAELK AKNGGK* |
[0879]ORF91ng-1 and ORF91-1 show 92.3% identity in 196 aa overlap:
[0000]
[0880]In addition, ORF91ng-1 shows homology to a hypothetical E. Coli protein:
[0000]| sp|P45390|YRBC_ECOLI HYPOTHETICAL 24.0 KD PROTEIN IN MURA-RPON INTERGENIC | |
| REGION PRECURSOR (F211) >gi|606130 (U18997) ORF_f211 [Escherichia coli] |
| >gi|1789583 (AE000399) hypothetical 24.0 kD protein in murZ-rpoN intergenic |
| region [Escherichia coli]Length = 211 |
| Score = 70.6 bits (170), Expect = 6e−12 |
| Identities = 42/137 (30%), Positives = 76/137 (54%), Gaps = 6/137 (4%) |
|
| Query: | 59 | VPYFDFQRMTALAVGNPWRTASDAQKQALAKEFQTLLIRTYSGTMLKFKNATVNVKDNPI | 118 | |
| | +PY + AL +G +++A+ AQ++A F+ L + Y + + T + P |
| Sbjct: | 65 | LPYVQVKYAGALVLGQYYKSATPAQREAYFAAFREYLKQAYGQALAMYHGQTYQIA--PE | 122 |
|
| Query: | 119 | VNKGGKEIV-VRAEVGIP-GQKPVNMDFTTYQSG--GKYRTYNVAIEGTSLVTVYRNQFG | 174 |
| | G K IV +R + P G+ PV +DF ++ G ++ Y++ EG S++T +N++G |
| Sbjct: | 123 | QPLGDKTIVPIRVTIIDPNGRPPVRLDFQWRKNSQTGNWQAYDMIAEGVSMITTKQNEWG | 182 |
|
| Query: | 175 | EIIKAKGIDGLIAELKA | 191 |
| | +++ KGIDGL A+LK+ |
| Sbjct: | 183 | TLLRTKGIDGLTAQLKS | 199 |
[0881]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 42
[0882]The following DNA sequence was identified in N. meningitidis <SEQ ID 355>:
[0000]| 1 | ATGAAACACA TACTCCCCCT GATTGCCGCA TCCGCACTCT GCATTTCAAC | |
|
| 51 | CGCTTCGGCA CATCCTGCCA GCGAACCGTC CACTCAAAAC GAAACCGCTA |
|
| 101 | TGATCACGCA TACCCTCATC TCAAAATACA GTTTTGGnnn nnnnnnnnnn |
|
| 151 | nnnnnnnnnn nnGCCATAAA AAGCAAAGGG ATGGACATTT TTGCCGTCAT |
|
| 201 | CGACCATCAG GAAGCCGCAC GCCGAAACGG CTTAACGATG CAGCCGGCAA |
|
| 251 | AAGTCATCGT CTTCGGCACG CCCAAAGCCG GCACGCCGCT GATGGTCAAA |
|
| 301 | GACCCCGCCT TCGCCCTGCA ACTGCCCCTA CGCGTCCTCG TTACCGAAAC |
|
| 351 | GGACGGCAAA GTACGCGCCG CCTATACCGA TACGCGCGCC CTCATCGCCG |
|
| 401 | GCAGCCGCAT CGGTTTCGAC GAAGTGGCAA ACACTTTGGC AAACGCCGAA |
|
| 451 | AAACTGATAC AAAAAACCGT AGGCGAATAA |
[0883]This corresponds to the amino acid sequence <SEQ ID 356; ORF97>:
[0000]| 1 | MKHILPLIAA SALCISTASA HPASEPSTQN ETAMITHTLI SKYSFGXXXX | |
|
| 51 | XXXXAIKSKG MDIFAVIDHQ EAARRNGLTM QPAKVIVFGT PKAGTPLMVK |
|
| 101 | DPAFALQLPL RVLVTETDGK VRAAYTDTRA LIAGSRIGFD EVANTLANAE |
|
| 151 | KLIQKTVGE* |
[0884]Further work revealed the complete nucleotide sequence <SEQ ID 357>:
[0000]| 1 | ATGAAACACA TACTCCCCCT GATTGCCGCA TCCGCACTCT GCATTTCAAC | |
|
| 51 | CGCTTCGGCA CATCCTGCCA GCGAACCGTC CACCCAAAAC GAAACCGCTA |
|
| 101 | TGACCACGCA TACCCTCACC TCAAAATACA GTTTTGACGA AACCGTCAGC |
|
| 151 | CGCCTTGAAA CCGCCATAAA AAGCAAAGGG ATGGACATTT TTGCCGTCAT |
|
| 201 | CGACCATCAG GAAGCCGCCC GCCGAAACGG CTTAACGATG CAGCCGGCAA |
|
| 251 | AAGTCATCGT CTTCGGCACG CCCAAAGCCG GCACGCCGCT GATGGTCAAA |
|
| 301 | GACCCCGCCT TCGCCCTGCA ACTGCCCCTA CGCGTCCTCG TTACCGAAAC |
|
| 351 | GGACGGCAAA GTACGCGCCG CCTATACCGA TACGCGCGCC CTCATCGCCG |
|
| 401 | GCAGCCGCAT CGGTTTCGAC GAAGTGGCAA ACACTTTGGC AAACGCCGAA |
|
| 451 | AAACTGATAC AAAAAACCGT AGGCGAATAA |
[0885]This corresponds to the amino acid sequence <SEQ ID 358; ORF97-1>:
[0000]| 1 | MKHILPLIAA SALCISTASA HPASEPSTQN ETAMTTHTLT SKYSFDETVS | |
|
| 51 | RLETAIKSKG MDIFAVIDHQ EAARRNGLTM QPAKVIVFGT PKAGTPLMVK |
|
| 101 | DPAFALQLPL RVLVTETDGK VRAAYTDTRA LIAGSRIGFD EVANTLANAE |
|
| 151 | KLIQKTVGE* |
[0886]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0887]ORF97 shows 88.7% identity over a 159aa overlap with an ORF (ORF97a) from strain A of N. meningitidis:
[0000]
[0888]The complete length ORF97a nucleotide sequence <SEQ ID 359> is:
[0000]| 1 | ATGANACACA TACTCCCCCT GANTGNCGCA TCCGCACTCT GCATTTCAAC | |
|
| 51 | CGCTTCGGNN CATCCTGCCA GCGAACCGCA AACCCAAAAC GAAACCGCTA |
|
| 101 | TGACCACGCA TACCCTCACC TCAAAATACA GTTTTGACGA AACCGTCAGC |
|
| 151 | CGCCTTGAAA CCGCCATAAA AAGCAAAGGG ATGGACATTT TTGCCGTCAT |
|
| 201 | CGACCATCAG GAAGCCGCCC GCCGAAACGG CTTAACGATG CAGCCGGCAA |
|
| 251 | AAGTCATCGT CTTCGGCACG CCCAAAGCCG GTACGCCGCT GATGGTCAAA |
|
| 301 | GACCCCGCCT TCGCCCTGCA ACTGCCCCTG CGCGTCNTCG TTACCGAAAC |
|
| 351 | GGACGGCAAA GTACGCGCCG CCTATACCGA TACGCGCGCC CTCATCGCCG |
|
| 401 | GCAGCCGCAT CGGTTTCGAC GAAGTGGCAA ACACTTTGGC AAACGCCGAA |
|
| 451 | AAACTGATAC AAAAAACCAT AGGCGAATAA |
[0889]This encodes a protein having amino acid sequence <SEQ ID 360>:
[0000]| 1 | MXHILPLXXA SALCISTASX HPASEPQTQN ETAMTTHTLT SKYSFDETVS | |
|
| 51 | RLETAIKSKG MDIFAVIDHQ EAARRNGLTM QPAKVIVFGT PKAGTPLMVK |
|
| 101 | DPAFALQLPL RVXVTETDGK VRAAYTDTRA LIAGSRIGFD EVANTLANAE |
|
| 151 | KLIQKTIGE* |
[0890]ORF97a and ORF97-1 show 95.6% identity in 159 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0891]ORF97 shows 88.1% identity over a 159aa overlap with a predicted ORF (ORF97.ng) from N. gonorrhoeae.
[0000]
[0892]The complete length ORF97ng nucleotide sequence <SEQ ID 361> is predicted to encode a protein having amino acid sequence <SEQ ID 362>:
[0000]| 1 | MKHILPPIAA SAFCISTASA HPAGKPPTQN ETAMTTHTLT SKYSFDETVS | |
|
| 51 | RLETAIKSKG MDIFAVIDHQ EAARRNGLTM QPAKVIVFGT PKAGTPLMVK |
|
| 101 | DPAFALQLPL RVLVTETDGK VRTAYTDTRA LIVGSRISFD EVANTLANAE |
|
| 151 | KLIQKTVGE* |
[0893]Further work revealed the complete nucleotide sequence <SEQ ID 363>:
[0000]| 1 | ATGAAACACA TACTCCCcct gatcgccgca TccgcactCT GCATTTCAAC | |
|
| 51 | CGCTTCGGCA CACCCTGCCG GCAAACCGCC CACCCAAAAC GAAACCGCTA |
|
| 101 | TGACCACGCA CACCCTCACC TCGAAATACA GTTTTGACGA AACCGTCAGC |
|
| 151 | CGCCTTGAAA CCGCCATAAA AAGCAAAGGG ATGGACATTT TTGCCGTCAT |
|
| 201 | CGACCATCAG GAAGCGGCAC GCCGAAACGG CCTGACCATG CAGCCGGCAA |
|
| 251 | AAGTCATCGT CTTCGGCACG CCCAAGGCCG GTACGCCgct GATGGTCAAA |
|
| 301 | GACCCCGCCT TCGCCCTGCA ACTGCCCCTG CGCGTCCTCG TTACCGAAAC |
|
| 351 | GGACGGCAAA GTACGCACCG CCTATACCGA TACGCGCGCC CTCATCGTCG |
|
| 401 | GCAGCCGCAT CAGTTTCGAC GAAGTGGCAA ACACTTTGGC AAACGCCGAA |
|
| 451 | AAACTGATAC AAAAAACCGT AGGCGAATAA |
[0894]This corresponds to the amino acid sequence <SEQ ID 364; ORF97ng-1>:
[0000]| 1 | MKHILPLIAA SALCISTASA HPAGKPPTQN ETAMTTHTLT SKYSFDETVS | |
|
| 51 | RLETAIKSKG MDIFAVIDHQ EAARRNGLTM QPAKVIVFGT PKAGTPLMVK |
|
| 101 | DPAFALQLPL RVLVTETDGK VRTAYTDTRA LIVGSRISFD EVANTLANAE |
|
| 151 | KLIQKTVGE* |
[0895]ORF97ng-1 and ORF97-1 show 96.2% identity in 159 aa overlap:
[0000]
[0896]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0897]ORF97-1 (15.3 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIGS. 12A & 12B show, respectively, the results of affinity purification of the GST-fusion and His-fusion proteins. Purified GST-fusion protein was used to immunise mice, whose sera were used for Western Blot (FIG. 12C), ELISA (positive result), and FACS analysis (FIG. 12D). These experiments confirm that ORF97-1 is a surface-exposed protein, and that it is a useful immunogen. FIG. 12E shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF97-1.
Example 43
[0898]The following DNA, believed to be complete, sequence was identified in N. meningitidis <SEQ ID 365>:
[0000]| 1 | ATGGCTTTTA TTACGCGCTT ATTCAAAAGC AGTAAATGGC TGATTGTGCC | |
|
| 51 | GCTGATGCTC CCCGCCTTTC AGAATGTGGC GGCGGAGGGG ATAGATGTGA |
|
| 101 | GCCGTGCCGA AGCGAGGATA ACCGACGGCG GGCAGCTTTC CATCAGCAGC |
|
| 151 | CGCTTCCAAA CCGAGCTGCC CGACCAGCTC CAACAGGCGT TGCGCCGGGg |
|
| 201 | CGTGCCGCTC AACTTTACCT TAAGCTGGCA GCTTTCCGCC CCGATAATCG |
|
| 251 | CTTCTTATCG GTTTAAATTG GGGCAACTGA TTGGCGATGA CGACaATATT |
|
| 301 | GACTACAAAC TGAGTTTCCA TCCGCTGACc AaACGCTACC GCGTTACCgT |
|
| 351 | CGgCGCGTTT TCGACAGACT ACGACACCTT GGATGCGGCA TTGCGCGCGA |
|
| 401 | CCGGCGCGGT TGCCAACTGG AAAGTCCTGA ACAAAGGCGC GCTGTCCGGT |
|
| 451 | GCGGAAGCAG GGGAAACCAA GGCGGAAATC CGCCTGACGC TGTCCACTTC |
|
| 501 | AAAACTGCCC AAGCCTTTTC AAATCAATGC ATTGACTTCT CAAAACTGGC |
|
| 551 | ATTTGGATTC GGGTTGGAAA CCTCTAAACA TCATCGGGAA CAAATAA |
[0899]This corresponds to the amino acid sequence <SEQ ID 366; ORF106>:
[0000]| 1 | MAFITRLFKS SKWLIVPLML PAFQNVAAEG IDVSRAEARI TDGGQLSISS | |
|
| 51 | RFQTELPDQL QQALRRGVPL NFTLSWQLSA PIIASYRFKL GQLIGDDDNI |
|
| 101 | DYKLSFHPLT KRYRVTVGAF STDYDTLDAA LRATGAVANW KVLNKGALSG |
|
| 151 | AEAGETKAEI RLTLSTSKLP KPFQINALTS QNWHLDSGWK PLNIIGNK* |
[0900]Further work revealed the following DNA sequence <SEQ ID 367>:
[0000]| 1 | ATGGCTTTTA TTACGCGCTT ATTCAAAAGC AGTAAATGGC TGATTGTGCC | |
|
| 51 | GCTGATGCTC CCCGCCTTTC AGAATGTGGC GGCGGAGGGG ATAGATGTGA |
|
| 101 | GCCGTGCCGA AGCGAGGATA ACCGACGGCG GGCAGCTTTC CATCAGCAGC |
|
| 151 | CGCTTCCAAA CCGAGCTGCC CGACCAGCTC CAACAGGCGT TGCGCCGGGG |
|
| 201 | CGTGCCGCTC AACTTTACCT TAAGCTGGCA GCTTTCCGCC CCGATAATCG |
|
| 251 | CTTCTTATCG GTTTAAATTG GGGCAACTGA TTGGCGATGA CGACAATATT |
|
| 301 | GACTACAAAC TGAGTTTCCA TCCGCTGACC AACCGCTACC GCGTTACCGT |
|
| 351 | CGGCGCGTTT TCGACAGACT ACGACACCTT GGATGCGGCA TTGCGCGCGA |
|
| 401 | CCGGCGCGGT TGCCAACTGG AAAGTCCTGA ACAAAGGCGC GCTGTCCGGT |
|
| 451 | GCGGAAGCAG GGGAAACCAA GGCGGAAATC CGCCTGACGC TGTCCACTTC |
|
| 501 | AAAACTGCCC AAGCCTTTTC AAATCAATGC ATTGACTTCT CAAAACTGGC |
|
| 551 | ATTTGGATTC GGGTTGGAAA CCTCTAAACA TCATCGGGAA CAAATAA |
[0901]This corresponds to the amino acid sequence <SEQ ID 368; ORF106-1>:
[0000]| 1 | MAFITRLFKS SKWLIVPLML PAFQNVAAEG IDVSRAEARI TDGGQLSISS | |
|
| 51 | RFQTELPDQL QQALRRGVPL NFTLSWQLSA PIIASYRFKL GQLIGDDDNI |
|
| 101 | DYKLSFHPLT NRYRVTVGAF STDYDTLDAA LRATGAVANW KVLNKGALSG |
|
| 151 | AEAGETKAEI RLTLSTSKLP KPFQINALTS QNWHLDSGWK PLNIIGNK* |
[0902]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0903]ORF106 shows 87.4% identity over a 199aa overlap with an ORF (ORF106a) from strain A of N. meningitidis:
[0000]
[0904]Due to the K→N substitution at residue 111, the homology between ORF106a and ORF106-1 is 87.9% over the same 199 aa overlap.
[0905]The complete length ORF106a nucleotide sequence <SEQ ID 369> is:
[0000]| 1 | ATGGCTTTTA TTACGCGCTT ATTCAAAAGC ATTAAACAAT GGCTTGTGCT | |
|
| 51 | GCTGCCGATG CTTTCCGTTT TGCCGGACGC GGCGGCGGAG GGGATAGATG |
|
| 101 | TGAGCCGCGC CGAAGCGAGG ATAANCGACG GCGGGCAGCT TTCCATNAGN |
|
| 151 | AGCCGCTTCC AAACCGAGCT GCCCGACCAG CTCCAANNNG CGNNGNGCCG |
|
| 201 | GGGCGTGNCG CTCAACTNTA CCTTAAGNTG GCAGCTTTCC GCCCCGATAA |
|
| 251 | TCGCTTCTTA TCGGTTTNAA TTGGGGCAAC TGATTGGCGA TGACGACNAT |
|
| 301 | ATTGACTACA AACTGAGTTT CCATCCGCTG ACCAACCGCT ACCGCGTTAC |
|
| 351 | CGTCGGCGCG TTTTCGACAG ANTACGACAC CTTGGATGCG GCATTGCGCG |
|
| 401 | CGACCGGCGC GGTTGCCAAC TGGAAAGTCC TGAACAAAGG CGCGCTGTCC |
|
| 451 | GGTGCGGAAG CAGGGGAAAC CAAGGCGGAA ATCCGCCTGA CGCTGTCCAC |
|
| 501 | TTCAAAACTG CCCAAGCCTT TTCAAATCAA TGCATTGACT TCTCAAAACT |
|
| 551 | GGCATTTGGA TTCGGGTTGG AAACCTCTAA ACATCATCGG GAACAAATAA |
[0906]This encodes a protein having amino acid sequence <SEQ ID 370>:
[0000]| 1 | MAFITRLFKS IKQWLVLLPM LSVLPDAAAE GIDVSRAEAR IXDGGQLSXX | |
|
| 51 | SRFQTELPDQ LQXAXXRGVX LNXTLXWQLS APIIASYRFX LGQLIGDDDX |
|
| 101 | IDYKLSFHPL TNRYRVTVGA FSTXYDTLDA ALRATGAVAN WKVLNKGALS |
|
| 151 | GAEAGETKAE IRLTLSTSKL PKPFQINALT SQNWHLDSGW KPLNIIGNK* |
|
Homology with a Predicted ORF from
N. gonorrhoeae [0907]ORF106 shows 90.5% identity over a 199aa overlap with a predicted ORF (ORF106.ng) from N. gonorrhoeae:
[0000]
[0908]Due to the K→N substitution at residue 111, the homology between ORF106ng and ORF106-1 is 91.0% over the same 199 aa overlap.
[0909]The complete length ORF106ng nucleotide sequence <SEQ ID 371> is:
[0000]| 1 | ATGGCTTTTA TTACGCGCTT ATTCAAAAGC ATTAAACAAT GGCTTGTGCT | |
|
| 51 | GTTGCCGATA CTCTCCGTTT TGCCGGACGC GGCGGCGGAG GGCATTGCCG |
|
| 101 | CGACCCGCGC CGAAGCGAGG ATAACCGACG GCGGGCGGCT TTCCATCAGC |
|
| 151 | AGCCGCTTCC AAACCGAGCT GCCCGACCAG CTCCAACAGG CGTTGCGCCG |
|
| 201 | GGGCGTACCG CTCAACTTTA CCTTAAGCTG GCAGCTTTCC GCCCCGACAA |
|
| 251 | TCGCTTCTTA TCGGTTTAAA TTGGGGCAAC TGATTGGCGA TGACGACAAT |
|
| 301 | ATTGACTACA AACTAAGTTT CCATCCGCTG ACCAACCGCT ACCGCGTTAC |
|
| 351 | CGTCGGCGCA TTTTCCACCG ATTACGACAC TTTGGATGCG GCATTGCGCG |
|
| 401 | CGACCGGCGC GGTTGCCAAC TGGAAAGTCC TGAACAAAGG CGCGTTGTCC |
|
| 451 | GGTGCGGAAG CAGGGGAAAC CAAGGCGGAA ATCCGCCTGA CGCTGTCCAC |
|
| 501 | TTCAAAACTG CCCAAGCCTT TCCAAATCAA CGCATTGACT TCTCAAAACT |
|
| 551 | GGCATTTGGA TTCGGGTTGG AAACCTCTAA ACATCATCGG GAACAAATAA |
[0910]This encodes a protein having amino acid sequence <SEQ ID 372>:
[0000]| 1 | MAFITRLFKS IKQWLVLLPI LSVLPDAAAE GIAATRAEAR ITDGGRLSIS | |
|
| 51 | SRFQTELPDQ LQQALRRGVP LNFTLSWQLS APTIASYRFK LGQLIGDDDN |
|
| 101 | IDYKLSFHPL TNRYRVTVGA FSTDYDTLDA ALRATGAVAN WKVLNKGALS |
|
| 151 | GAEAGETKAE IRLTLSTSKL PKPFQINALT SQNWHLDSGW KPLNIIGNK* |
[0911]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[0912]ORF106-1 (18 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 13A shows the results of affinity purification of the His-fusion protein, and FIG. 13B shows the results of expression of the GST-fusion in E. coli. Purified His-fusion protein was used to immunise mice, whose sera were used for FACS analysis (FIG. 13C) These experiments confirm that ORF106-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 44
[0913]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 373>:
[0000]| 1 | ATGGACACAA AAGAAATCCT CGG.TACGCG GcAGGcTCGA TCGGCAGCGC | |
|
| 51 | GGTTTTAGCC GTCATCATCc TGCCGCTGCT GTCGTGGTAT TTCCCCGCCG |
|
| 101 | ACGACATCGG GCGCATCGTG CTGATGCAGA CGGCGGCGGG GCTgACGGTG |
|
| 151 | TCGGTGTTGT GCCTCGGGCT GGATCAGGCA TACGTCCGCG AATACTATGC |
|
| 201 | CACCGCCGAC AAAGACAcCT TGTTCAAAAC CCTGTTCCTG CCGCCGCTGC |
|
| 251 | TGTCTGCCGC CGCGATAGCC GCCCTGCTGC TTTCCCGCCC GTCCCTGCCG |
|
| 301 | TCTGAAATCC TGTTTTCACT CGACGATGCC gCCGCCGGCa TCGGGCTGGT |
|
| 351 | GCTGTTTGAA CtGAGCTTCC TGCCCATCCG cTTTCTCTTA CTGGTTTTGC |
|
| 401 | GTATGGAAGG ACGCGCCcTT GCCTTTTCGT CCGCGCAACT CGTGCcCAAG |
|
| 451 | CTCGCCATCC TGCTGCTG.T GCCGCTGACG GTCGGGCTGC TGCACTTTCC |
|
| 501 | AGCGAACACC GCCGTCCTGA CCGCCGTTTA CGCGCTGGCA AACCTTGCCG |
|
| 551 | CCGCCGCCTT TTTGCTGTTT CAAAACCGAT GCCGTCTGAA GGCCGTCCGG |
|
| 601 | CACGCACCGT TTTCGCCCGC CGTCCTGCAC CGGGGG.TGC GCTACGGCAT |
|
| 651 | ACCGATCGCA CTGAGCAGCA TCGCCTATTG GGGGCTGGCA TCCGCCGACC |
|
| 701 | GTTTGTTCCT GAAAAAATAT GCCGGCCTGG AACAGCTCGG CGTTTATTCG |
|
| 751 | ATGGGTATTT CGTTCGGCGG GGCGGCATTA TTGTTCCAAA GCATCTTTTC |
|
| 801 | AACGGTCTGG ACACCGTATA TTTTCCGCGC AATCGAAGAA AACGCCCCGC |
|
| 851 | CCGCTCGCCT CTCGGCAACG GCAGAATCCG CCGCCGCCCT GCTTGCCTCC |
|
| 901 | GCCCTCTGC. TGACCGGCAT TTTCTCGCCC CTTGCCTCCC TCCTGCTGCC |
|
| 951 | GGAAAACTAC GCCGCCGTCC GGTTTATCGT CGTATCGTGT ATG.TGCCGC |
|
| 1001 | CGCTGTTTTG CACGCTGGCG GAAATCAGCG GCATCGGTTT GAACGTCGTT |
|
| 1051 | CGCAAAACGC GCCCGATCGC GCTCGCCACC TTGGGCGCGC TGGCGGCAAA |
|
| 1101 | CCTGCTGCTG CTGGGGCTTG ACCGTGCCGT ACCGGCGAGG CCGCC.GGCG |
|
| 1151 | CGGCGGTTGC CTGTGCCGCC TCATTCTGGC TGTTTTTTGC CTTCAAGACC |
|
| 1201 | GAAAGCTCyT GCCGCCTGTG GCAGCCGCTC AAACGCCTGC CGCTTTATCT |
|
| 1251 | GCACACATTG TTCTGCCTGA CCTCCTCGGC GGCCTACACC TGCTTCGGCA |
|
| 1301 | CGCCGGCAAA CTATCCCCTG TTTGCCGGCG TATGGGCGGC ATATCTGGCA |
|
| 1351 | GGCTGCATCC TGCGCCACCG GAAAGATTTG CACAAACTGT TTCATTATTT |
|
| 1401 | GAAAAAACAA GGTTTCCCAT TATGA |
[0914]This corresponds to the amino acid sequence <SEQ ID 374; ORF10>:
[0000]| 1 | MDTKEILXYA AGSIGSAVLA VIILPLLSWY FPADDIGRIV LMQTAAGLTV | |
|
| 51 | SVLCLGLDQA YVREYYATAD KDTLFKTLFL PPLLSAAAIA ALLLSRPSLP |
|
| 101 | SEILFSLDDA AAGIGLVLFE LSFLPIRFLL LVLRMEGRAL AFSSAQLVPK |
|
| 151 | LAILLLXPLT VGLLHFPANT AVLTAVYALA NLAAAAFLLF QNRCRLKAVR |
|
| 201 | HAPFSPAVLH RGXRYGIPIA LSSIAYWGLA SADRLFLKKY AGLEQLGVYS |
|
| 251 | MGISFGGAAL LFQSIFSTVW TPYIFRAIEE NAPPARLSAT AESAAALLAS |
|
| 301 | ALCXTGIFSP LASLLLPENY AAVRFIVVSC MXPPLFCTLA EISGIGLNVV |
|
| 351 | RKTRPIALAT LGALAANLLL LGLDRAVPAR PXGAAVACAA SFWLFFAFKT |
|
| 401 | ESSCRLWQPL KRLPLYLHTL FCLTSSAAYT CFGTPANYPL FAGVWAAYLA |
|
| 451 | GCILRHRKDL HKLFHYLKKQ GFPL* |
[0915]Further sequence analysis revealed the complete DNA sequence <SEQ ID 375> to be:
[0000]| 1 | ATGGACACAA AAGAAATCCT CGGCTACGCG GCAGGCTCGA TCGGCAGCGC | |
|
| 51 | GGTTTTAGCC GTCATCATCC TGCCGCTGCT GTCGTGGTAT TTCCCCGCCG |
|
| 101 | ACGACATCGG GCGCATCGTG CTGATGCAGA CGGCGGCGGG GCTGACGGTG |
|
| 151 | TCGGTGTTGT GCCTCGGGCT GGATCAGGCA TACGTCCGCG AATACTATGC |
|
| 201 | CACCGCCGAC AAAGACACCT TGTTCAAAAC CCTGTTCCTG CCGCCGCTGC |
|
| 251 | TGTCTGCCGC CGCGATAGCC GCCCTGCTGC TTTCCCGCCC GTCCCTGCCG |
|
| 301 | TCTGAAATCC TGTTTTCACT CGACGATGCC GCCGCCGGCA TCGGGCTGGT |
|
| 351 | GCTGTTTGAA CTGAGCTTCC TGCCCATCCG CTTTCTCTTA CTGGTTTTGC |
|
| 401 | GTATGGAAGG ACGCGCCCTT GCCTTTTCGT CCGCGCAACT CGTGCCCAAG |
|
| 451 | CTCGCCATCC TGCTGCTGCT GCCGCTGACG GTCGGGCTGC TGCACTTTCC |
|
| 501 | AGCGAACACC GCCGTCCTGA CCGCCGTTTA CGCGCTGGCA AACCTTGCCG |
|
| 551 | CCGCCGCCTT TTTGCTGTTT CAAAACCGAT GCCGTCTGAA GGCCGTCCGG |
|
| 601 | CACGCACCGT TTTCGCCCGC CGTCCTGCAC CGGGGGCTGC GCTACGGCAT |
|
| 651 | ACCGATCGCA CTGAGCAGCA TCGCCTATTG GGGGCTGGCA TCCGCCGACC |
|
| 701 | GTTTGTTCCT GAAAAAATAT GCCGGCCTGG AACAGCTCGG CGTTTATTCG |
|
| 751 | ATGGGTATTT CGTTCGGCGG GGCGGCATTA TTGTTCCAAA GCATCTTTTC |
|
| 801 | AACGGTCTGG ACACCGTATA TTTTCCGCGC AATCGAAGAA AACGCCCCGC |
|
| 851 | CCGCCCGCCT CTCGGCAACG GCAGAATCCG CCGCCGCCCT GCTTGCCTCC |
|
| 901 | GCCCTCTGCC TGACCGGCAT TTTCTCGCCC CTTGCCTCCC TCCTGCTGCC |
|
| 951 | GGAAAACTAC GCCGCCGTCC GGTTTATCGT CGTATCGTGT ATGCTGCCGC |
|
| 1001 | CGCTGTTTTG CACGCTGGCG GAAATCAGCG GCATCGGTTT GAACGTCGTC |
|
| 1051 | CGCAAAACGC GCCCGATCGC GCTCGCCACC TTGGGCGCGC TGGCGGCAAA |
|
| 1101 | CCTGCTGCTG CTGGGGCTTG CCGTGCCGTC CGGCGGCGCG CGCGGCGCGG |
|
| 1151 | CGGTTGCCTG TGCCGCCTCA TTCTGGCTGT TTTTTGCCTT CAAGACCGAA |
|
| 1201 | AGCTCCTGCC GCCTGTGGCA GCCGCTCAAA CGCCTGCCGC TTTATCTGCA |
|
| 1251 | CACATTGTTC TGCCTGACCT CCTCGGCGGC CTACACCTGC TTCGGCACGC |
|
| 1301 | CGGCAAACTA TCCCCTGTTT GCCGGCGTAT GGGCGGCATA TCTGGCAGGC |
|
| 1351 | TGCATCCTGC GCCACCGGAA AGATTTGCAC AAACTGTTTC ATTATTTGAA |
|
| 1401 | AAAACAAGGT TTCCCATTAT GA |
[0916]This corresponds to the amino acid sequence <SEQ ID 376; ORF10-1>:
[0000]| 1 | MDTKEILGYA AGSIGSAVLA VIILPLLSWY FPADDIGRIV LMQTAAGLTV | |
|
| 51 | SVLCLGLDQA YVREYYATAD KDTLFKTLFL PPLLSAAAIA ALLLSRPSLP |
|
| 101 | SEILFSLDDA AAGIGLVLFE LSFLPIRFLL LVLRMEGRAL AFSSAQLVPK |
|
| 151 | LAILLLLPLT VGLLHFPANT AVLTAVYALA NLAAAAFLLF QNRCRLKAVR |
|
| 201 | HAPFSPAVLH RGLRYGIPIA LSSIAYWGLA SADRLFLKKY AGLEQLGVYS |
|
| 251 | MGISFGGAAL LFQSIFSTVW TPYIFRAIEE NAPPARLSAT AESAAALLAS |
|
| 301 | ALCLTGIFSP LASLLLPENY AAVRFIVVSC MLPPLFCTLA EISGIGLNVV |
|
| 351 | RKTRPIALAT LGALAANLLL LGLAVPSGGA RGAAVACAAS FWLFFAFKTE |
|
| 401 | SSCRLWQPLK RLPLYLHTLF CLTSSAAYTC FGTPANYPLF AGVWAAYLAG |
|
| 451 | CILRHRKDLH KLFHYLKKQG FPL* |
[0917]Computer analysis of this amino acid sequence gave the following results:
Prediction
[0918]ORF10-1 is predicted to be the precursor of an integral membrane protein, since it comprises several (12-13) potential transmembrane segments, and a probable cleavable signal peptide
[0000]Homology with EpsM from Streptococcus thermophilus (Accession Number U40830).
[0919]ORF10 shows homology with the epsM gene of S. thermophilus, which encodes a protein of a size similar to ORF10 and is involved in expolysaccharide synthesis. Other homologies are with prokaryotic membrane proteins:
[0000]| Identities = (25%) | |
| Query: | 213 | LRYGIPLALSSLAYWGLASADRLFLKKYAGLEQLGVYSMGISFGGAALLLQSIFSTVW | 270 | |
| | L Y +PL SS+ +W L ++ R F+ + G G+ ++ + +IF+ W |
| Sbjct: | 210 | LYYALPLIPSSILWWLLNASSRYFVLFFLGAGANGLLAVATKIPSIISIFNTIFTQAW | 267 |
|
| Identities = 15/57 (26%), Positives = 31/57 (54%) | |
|
| Query: | 7 | LGYAAGSIGSAVLAVIILPLLSWYFPADDIGRIVLMQTAAGLTVSVLCLGLDQAYVR | 63 | |
| | L + G++GS +L +++PL ++ + G L QT A L + ++ + + A +R |
| Sbjct: | 12 | LVFTIGNLGSKLLVFLLVPLYTYAMTPQEYGMADLYQTTANLLLPLITMNVFDATLR | 68 |
|
| Identities = 16/96 (16%), Positives = 36/96 (37%) | |
|
| Query: | 307 | IFSPLASLLLPENYAAVRFTVVSCMLPPLFYTLTEISGIGLNVVRKTRPIXXXXXXXXXX | 366 | |
| | + P+ ++ +YA+ V ML LF + ++ G ++T+ + |
| Sbjct: | 305 | VLKPIVEKVVSSDYASSWQYVPFFMLSMLFSSFSDFFGTNYIAAKQTKGVFMTSIYGTIV | 364 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0920]ORF10 shows 95.4% identity over a 475aa overlap with an ORF (ORF10a) from strain A of N. meningitidis:
[0000]
[0921]The complete length ORFLOa nucleotide sequence <SEQ ID 377> is:
[0000]| 1 | ATGGACACAA AAGAAATCCT CGGCTACGCG GCAGGCTCGA TCGGCAGCGC | |
|
| 51 | GGTTTTAGCC GTCATCATCC TGCCGCTGCT GTCGTGGTAT TTCCCTGCCG |
|
| 101 | ACGACATCGG ACGCATCGTG CTGATGCAGA CGGCGGCGGG GCTGACGGTG |
|
| 151 | TCGGTGTTGT GCCTCGGGCT GGATCAGGCA TACGTCCGCG AATACTATGC |
|
| 201 | CGCCGCCGAC AAAGACACTT TGTTCAAAAC CCTGTTCCTG CCGCCGCTGC |
|
| 251 | TGTCTGCCGC CGCGATAGCC GCCCTGCTGC TTTCCCGCCC ATCCCTGCCG |
|
| 301 | TCTGAAATCC TGTTTTCGCT CGACGATGCC GCCGCCGGCA TCGGGCTGGT |
|
| 351 | GCTGTTTGAA CTGAGCTTCC TGCCCATCCG CTTTCTCTTA CTGGTTTTGC |
|
| 401 | GTATGGAAGG ACGCGCCCTT GCCTTTTCGT CCGCGCAACT CGTGTCCAAG |
|
| 451 | CTCGCCATCC TGCTGCTGCT GCCGCTGACG GTCGGGCTGC TGCACTTTCC |
|
| 501 | GGCGAACACC GCCGTCCTGA CCGCCGTTTA CGCGCTGGCA AACCTTGCCG |
|
| 551 | CCGCCGCCTT TTTGCTGTTT CAAAACCGAT GCCGTCTGAA GGCCGTCCGG |
|
| 601 | CGCGCACCGT TTTCATCCGC CGTCCTGCAT CGCGGCCTGC GCTACGGCAT |
|
| 651 | ACCGATCGCA CTAAGCAGCA TCGCCTATTG GGGGCTGGCA TCCGCCGACC |
|
| 701 | GTTTGTTCCT GAAAAAATAT GCCGGCCTAG AACAGCTCGG CGTTTATTCG |
|
| 751 | ATGGGTATTT CGTTCGGCGG AGCGGCATTA TTGTTCCAAA GCATCTTTTC |
|
| 801 | AACGGTCTGG ACACCGTATA TTTTCCGCGC AATCGAAGCA AACGCCCCGC |
|
| 851 | CCGCCCGCCT CTCGGCAACG GCAGAATCCG CCGCCGCCCT GCTTGCCTCC |
|
| 901 | GCCCTCTGCC TGACCGGCAT TTTCTCGCCC CTCGCCTCCC TCCTGCTGCC |
|
| 951 | GGAAAACTAC GCCGCCGTCC GGTTTATCGT CGTATCGTGT ATGCTGCCTC |
|
| 1001 | CGCTGTTTTG CACGCTGGTA GAAATCAGCG GCATCGGTTT GAACGTCGTC |
|
| 1051 | CGAAAAACAC GCCCGATCGC GCTCGCCACC TTGGGCGCGC TGGCGGCAAA |
|
| 1101 | CCTGCTGCTG CTGGGGCTTG CCGTACCGTC CGGCGGCGCG CGCGGCGCGG |
|
| 1151 | CGGTTGCCTG TGCCGCCTCA TTTTGGCTGT TTTTTGTTTT CAAGACCGAA |
|
| 1201 | AGCTCCTGCC GCCTGTGGCA GCCGCTCAAA CGCCTGCCGC TTTATATGCA |
|
| 1251 | CACATTGTTC TGCCTGGCCT CCTCGGCGGC CTACACCTGC TTCGGCACTC |
|
| 1301 | CGGCAAACTA CCCCCTGTTT GCCGGCGTAT GGGCGGTATA TCTGGCAGGC |
|
| 1351 | TGCATCCTGC GCCACCGGAA AGATTTGCAC AAACTGTTTC ATTATTTGAA |
|
| 1401 | AAAACAAGGT TTCCCATTAT GA |
[0922]This encodes a protein having amino acid sequence <SEQ ID 378>:
[0000]| 1 | MDTKEILGYA AGSIGSAVLA VIILPLLSWY FPADDIGRIV LMQTAAGLTV | |
|
| 51 | SVLCLGLDQA YVREYYAAAD KDTLFKTLFL PPLLSAAAIA ALLLSRPSLP |
|
| 101 | SEILFSLDDA AAGIGLVLFE LSFLPIRFLL LVLRMEGRAL AFSSAQLVSK |
|
| 151 | LAILLLLPLT VGLLHFPANT AVLTAVYALA NLAAAAFLLF QNRCRLKAVR |
|
| 201 | RAPFSSAVLH RGLRYGIPIA LSSIAYWGLA SADRLFLKKY AGLEQLGVYS |
|
| 251 | MGISFGGAAL LFQSIFSTVW TPYIFRAIEA NAPPARLSAT AESAAALLAS |
|
| 301 | ALCLTGIFSP LASLLLPENY AAVRFIVVSC MLPPLFCTLV EISGIGLNVV |
|
| 351 | RKTRPIALAT LGALAANLLL LGLAVPSGGA RGAAVACAAS FWLFFVFKTE |
|
| 401 | SSCRLWQPLK RLPLYMHTLF CLASSAAYTC FGTPANYPLF AGVWAVYLAG |
|
| 451 | CILRHRKDLH KLFHYLKKQG FPL* |
[0923]ORF10a and ORF10-1 show 95.4% identity in 475 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0924]ORF10 shows 94.1% identity over a 475aa overlap with a predicted ORF (ORF10.ng) from N. gonorrhoeae:
[0000]
[0925]The complete length ORF10ng nucleotide sequence <SEQ ID 379> is:
[0000]| 1 | ATGGACACAA AAGAAATCCT CGGCTACGCG GCAGGCTCGA TCGGCAGCGC | |
|
| 51 | GGTTTTAGCC GTCATCATCC TGCCGCTGCT GTCGTGGTAT TTCcccgCCG |
|
| 101 | ACGACATCGG GCGCATCGTG CTGATGCAGA CGGCGGCGGG ACTGACGGTG |
|
| 151 | TCGGTATTGT GCCTCGGGCT GGATCAGGCA TACGTCCGCG AATACTATGC |
|
| 201 | CGCCGCCGAC AAAGACACTT TGTTCAAAAC CCTGTTCCTG CCGCCGCTGC |
|
| 251 | TGTTTTCCGC CGCGATAGCC GCCCTGCTGC TTTCCCGCCC GTCCCTGCCG |
|
| 301 | TCTGAAATCC TGTTTTCGCT CGACGATGCC GCCGCCGGCA TCGGGCTGGT |
|
| 351 | GCTGTTTGAA CTGAGCTTCC TGCCCATCCG CTTTCTCTTA CTGGTTTTGC |
|
| 401 | GTATGGAAGG GCGCGCCCTT GCCTTTTCGT CCGCGCAACT CGTGCCCAAA |
|
| 451 | CTCGCCATTC TGCTGCTGTT GCCGCTGACG GTCGGGCTGC TGCACTTTCC |
|
| 501 | GGCGAACACC TCCGTCCTGA CCGCCGTTTA CGCGCTGGCA AACCTTGCCG |
|
| 551 | CCGCCGCCTT TTTGCTGTTT CAAAACCGAT GCCGTCTGAA GGCCGTCCGG |
|
| 601 | CGCGCGCCGT TTTCGCCCGC CGTCCTGCAC CGGGGGCTGC GCTACGGCAT |
|
| 651 | ACCGCTCGCA CTGAGCAGCC TTGCCTATTG GGGGCTGGCA TCCGCCGACC |
|
| 701 | GTTTGTTCCT GAAAAAATAT GCGGGCCTGG AACAGCTCGG CGTTTATTCG |
|
| 751 | ATGGGTATTT CGTTCGGCGG GGCGGCATTA TTGCTCCAAA GCATCTTTTC |
|
| 801 | AACGGTCTGG ACACCGTATA TTTTCCGTGC AATCGAAGAA AACGCCACGC |
|
| 851 | CCGCCCGCCT CTCGGCAACG GCAGAATCCG CCGCCGCCCT GCTTGCCTCC |
|
| 901 | GCCCTCTGCC TGACCGGAAT TTTCTCGCCC CTCGCCTCCC TCCTGCTGCC |
|
| 951 | GGAAAACTAC GCCGCCGTCC GGTTTACCGT CGTATCGTGT ATGCTGccgc |
|
| 1001 | cgctGTTTTA CACGCTGACC GAAATCAGCG GCATCGGTTT GAACGTCGTC |
|
| 1051 | CGCAAAACGC GTCCGATCGC GCTTGCCACC TTGGGCGCGC TGGCGGCAAA |
|
| 1101 | CCTGCTGCTG CTGGGGCTTG CCGTACCGTC CGGCGGCACG CGCGGCGCGG |
|
| 1151 | CGGTTGCCTG TGCCGCCTCA TTCTGGTTGT TTTTTGTTTT CAAGACAGAA |
|
| 1201 | AGCTCCTGCC GCCTGTGGCA GCCGCTCAAA CGCCTGCCGC TTTATATGCA |
|
| 1251 | CACATTGTTC TGCCTgGCCT CCTCGGCGGC CTACACCTGC TTCGGCACAC |
|
| 1301 | CGGCAAACTA CCCcctgttt gccggcgtAT GGGCGGCATA TCTGGCAGGC |
|
| 1351 | TGCATCCTGC GCCACCGGAA AAATTTGCAC AAACTGTTTC ATTATTTGAA |
|
| 1401 | AAAACAAGGT TTCCCATTAT GA |
[0926]This encodes a protein having amino acid sequence <SEQ ID 380>:
[0000]| 1 | MDTKEILGYA AGSIGSAVLA VIILPLLSWY FPADDIGRIV LMQTAAGLTV | |
|
| 51 | SVLCLGLDQA YVREYYAAAD KDTLFKTLFL PPLLFSAAIA ALLLSRPSLP |
|
| 101 | SEILFSLDDA AAGIGLVLFE LSFLPIRFLL LVLRMEGRAL AFSSAQLVPK |
|
| 151 | LAILLLLPLT VGLLHFPANT SVLTAVYALA NLAAAAFLLF QNRCRLKAVR |
|
| 201 | RAPFSPAVLH RGLRYGIPLA LSSLAYWGLA SADRLFLKKY AGLEQLGVYS |
|
| 251 | MGISFGGAAL LLQSIFSTVW TPYIFRAIEE NATPARLSAT AESAAALLAS |
|
| 301 | ALCLTGIFSP LASLLLPENY AAVRFTVVSC MLPPLFYTLT EISGIGLNVV |
|
| 351 | RKTRPIALAT LGALAANLLL LGLAVPSGGT RGAAVACAAS FWLFFVFKTE |
|
| 401 | SSCRLWQPLK RLPLYMHTLF CLASSAAYTC FGTPANYPLF AGVWAAYLAG |
|
| 451 | CILRHRKNLH KLFHYLKKQG FPL* |
[0927]ORF10ng and ORF10-1 show 96.4% identity in 473 aa overlap:
[0000]
[0928]Based on this analysis, including the presence of a putative leader peptide and several transmembrane segments and the presence of a leucine-zipper motif (4 Leu residues spaced by 6 aa, shown in bold), it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 45
[0929]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 381>:
[0000]| 1 | ..ATCCTGAAAC CGCATAACCA GCTTAAGGAA GACATCCAAC CTGATCCGGC | |
|
| 51 | CGATCAAAAC GCCTTGTCCG AACCGGATGC TGCGACAGAG GCAGAGCAGT |
|
| 101 | CGGATGCGGA AAATGCTGCC GACAAGCAGC CCGTTGCCGA TAAAGCCGAC |
|
| 151 | GAGGTTGAAG AAAAGGCGGG CGAGCCGGAA CGGGAAGAGC CGGACGGACA |
|
| 201 | GGCAGTGCGT AAGAAAGCGC TGACGGAAGA GCGTGAACAA ACCGTCAGGG |
|
| 251 | AAAAAGCGCA GAAGAAAGAT GCCGAAACGG TTAAAATACA AGCGGTAAAA |
|
| 301 | CCGTCTAAAG AAACAGAGAA AAAAGCTTCA AAAGAAGAGA AAAAGGCGGC |
|
| 351 | GAAGGAAAAA GTTGCACCCA AACCAACCCC GGAACAAATC CTCAACAGCG |
|
| 401 | GCAgCATCGA AAAmGCGCGC AgTGCCGCCG CCAAAGAAGT GCAGAAAATG |
|
| 451 | AA.AACGTCC GACAAGGCGG AAGC.AACGC ATTATCTGCA AATGGGCGCG |
|
| 501 | TATGCCGACC GTCAGAGCGC GGAAGGGCAG CGTGCCAAAC TGGCAATCTT |
|
| 551 | GGGCATATCT TCCAAGGTGG TCGGTTATCA GGCGGGACAT AAAACGCTTT |
|
| 601 | ACCGGGTGCA AAGCGGCAAT ATGTCTGCCG ATGCGGTGA |
[0930]This corresponds to the amino acid sequence <SEQ ID 382; ORF65>:
[0000]| 1 | ..ILKPHNQLKE DIQPDPADQN ALSEPDAATE AEQSDAENAA DKQPVADKAD | |
|
| 51 | EVEEKAGEPE REEPDGQAVR KKALTEEREQ TVREKAQKKD AETVKIQAVK |
|
| 101 | PSKETEKKAS KEEKKAAKEK VAPKPTPEQI LNSGSIEXAR SAAAKEVQKM |
|
| 151 | XNVRQGGSXR IICKWARMPT VRARKGSVPN WQSWAYLPRW SVIRRDIKRF |
|
| 201 | TGCKAAICLP MR* |
[0931]Further work revealed the complete nucleotide sequence <SEQ ID 383>:
[0000]| 1 | ATGTTTATGA ACAAATTTTC CCAATCCGGA AAAGGTCTGT CCGGTTTTTT | |
|
| 51 | CTTCGGTTTG ATACTGGCGA CGGTCATTAT TGCCGGTATT TTGTTTTATC |
|
| 101 | TGAACCAGAG CGGTCAAAAT GCGTTCAAAA TCCCGGCTTC GTCGAAGCAG |
|
| 151 | CCTGCAGAAA CGGAAATCCT GAAACCGAAA AACCAGCCTA AGGAAGACAT |
|
| 201 | CCAACCTGAA CCGGCCGATC AAAACGCCTT GTCCGAACCG GATGCTGCGA |
|
| 251 | CAGAGGCAGA GCAGTCGGAT GCGGAAAAAG CTGCCGACAA GCAGCCCGTT |
|
| 301 | GCCGATAAAG CCGACGAGGT TGAAGAAAAG GCGGGCGAGC CGGAACGGGA |
|
| 351 | AGAGCCGGAC GGACAGGCAG TGCGTAAGAA AGCGCTGACG GAAGAGCGTG |
|
| 401 | AACAAACCGT CAGGGAAAAA GCGCAGAAGA AAGATGCCGA AACGGTTAAA |
|
| 451 | AAACAAGCGG TAAAACCGTC TAAAGAAACA GAGAAAAAAG CTTCAAAAGA |
|
| 501 | AGAGAAAAAG GCGGCGAAGG AAAAAGTTGC ACCCAAACCA ACCCCGGAAC |
|
| 551 | AAATCCTCAA CAGCGGCAGC ATCGAAAAAG CGCGCAGTGC CGCCGCCAAA |
|
| 601 | GAAGTGCAGA AAATGAAAAC GTCCGACAAG GCGGAAGCAA CGCATTATCT |
|
| 651 | GCAAATGGGC GCGTATGCCG ACCGTCAGAG CGCGGAAGGG CAGCGTGCCA |
|
| 701 | AACTGGCAAT CTTGGGCATA TCTTCCAAGG TGGTCGGTTA TCAGGCGGGA |
|
| 751 | CATAAAACGC TTTACCGGGT GCAAAGCGGC AATATGTCTG CCGATGCGGT |
|
| 801 | GAAAAAAATG CAGGACGAGT TGAAAAAACA TGAAGTCGCC AGCCTGATCC |
|
| 851 | GTTCTATCGA AAGCAAATAA |
[0932]This corresponds to the amino acid sequence <SEQ ID 384; ORF65-1>:
[0000]| 1 | MFMNKFSQSG KGLSGFFFGL ILATVIIAGI LFYLNQSGQN AFKIPASSKQ | |
|
| 51 | PAETEILKPK NQPKEDIQPE PADQNALSEP DAATEAEQSD AEKAADKQPV |
|
| 101 | ADKADEVEEK AGEPEREEPD GQAVRKKALT EEREQTVREK AQKKDAETVK |
|
| 151 | KQAVKPSKET EKKASKEEKK AAKEKVAPKP TPEQILNSGS IEKARSAAAK |
|
| 201 | EVQKMKTSDK AEATHYLQMG AYADRQSAEG QRAKLAILGI SSKVVGYQAG |
|
| 251 | HKTLYRVQSG NMSADAVKKM QDELKKHEVA SLIRSIESK* |
[0933]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0934]ORF65 shows 92.0% identity over a 150aa overlap with an ORF (ORF65a) from strain A of N. meningitidis.
[0000]
[0935]The complete length ORF65a nucleotide sequence <SEQ ID 385> is:
[0000]| 1 | ATGTTTATGA ACAAATTTTC CCAATCCGGA AAAGGTCTGT CCGGTTTTTT | |
|
| 51 | CTTCGGTTTG ATACTGGCGA CGGTCATTAT TGCCGGTATT TTGTTTTATC |
|
| 101 | TGAACCAGAG CGGTCAAAAT GCGTTCAAAA TCCCGGTTCC GTCGAAGCAG |
|
| 151 | CCTGCAGAAA CGGAAATCCT GAAACCGAAA AACCAGCCTA AGGAAGACAT |
|
| 201 | CCAACCTGAA CCGGCCGATC AAAACGCCTT GTCCGAACCG GATGCTGCGA |
|
| 251 | AAGAGGCAGA GCAGTCGGAT GCGGAAAAAG CTGCCGACAA GCAGCCCGTT |
|
| 301 | GCCGACAAAG CCGACGAGGT TGAGGAAAAG GCGGACGAGC CGGAGCGGGA |
|
| 351 | AAAGTCGGAC GGACAGGCAG TGCGCAAGAA AGCACTGACG GAAGAGCGTG |
|
| 401 | AACAAACCGT CGGGGAAAAA GCGCAGAAGA AAGATGCCGA AACGGTTAAA |
|
| 451 | AAACAAGCGG TAAAACCATC TAAAGAAACA GAGAAAAAAG CTTCAAAAGA |
|
| 501 | AGAGAAAAAG GCGGAGAAGG AAAAAGTTGC ACCCAAACCG ACCCCGGAAC |
|
| 551 | AAATCCTCAA CAGCGGCAGC ATCGAAAAAG CGCGCAGTGC CGCTGCCAAA |
|
| 601 | GAAGTGCAGA AAATGAAAAC GCCCGACAAG GCGGAAGCAA CGCATTATCT |
|
| 651 | GCAAATGGGC GCGTATGCCG ACCGCCGGAG CGCGGAAGGG CAGCGTGCCA |
|
| 701 | AACTGGCAAT CTTGGGCATA TCTTCCAAGG TGGTCGGTTA TCAGGCGGGA |
|
| 751 | CATAAAACGC TTTACCGGGT GCAAAGCGGC AATATGTCTG CCGATGCGGT |
|
| 801 | GAAAAAAATG CAGGACGAGT TGAAAAAACA TGAAGTCGCC AGCCTGATCC |
|
| 851 | GTTCTATCGA AAGCAAATAA |
[0936]This encodes a protein having amino acid sequence <SEQ ID 386>:
[0000]| 1 | MFMNKFSQSG KGLSGFFFGL ILATVIIAGI LFYLNQSGQN AFKIPVPSKQ | |
|
| 51 | PAETEILKPK NQPKEDIQPE PADQNALSEP DAAKEAEQSD AEKAADKQPV |
|
| 101 | ADKADEVEEK ADEPEREKSD GQAVRKKALT EEREQTVGEK AQKKDAETVK |
|
| 151 | KQAVKPSKET EKKASKEEKK AEKEKVAPKP TPEQILNSGS IEKARSAAAK |
|
| 201 | EVQKMKTPDK AEATHYLQMG AYADRRSAEG QRAKLAILGI SSKVVGYQAG |
|
| 251 | HKTLYRVQSG NMSADAVKKM QDELKKHEVA SLIRSIESK* |
[0937]ORF65a and ORF65-1 show 96.5% identity in 289 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0938]ORF65 shows 89.6% identity over a 212aa overlap with a predicted ORF (ORF65.ng) from N. gonorrhoeae:
[0000]
[0939]An ORF65ng nucleotide sequence <SEQ ID 387> was predicted to encode a protein having amino acid sequence <SEQ ID 388>:
[0000]| 1 | MFMNKFSQSG KGLSGFFFGL ILATVIIAGI LLYLNQGGQN AFKIPAPSKQ | |
|
| 51 | PAETEILKLK NQPKEDIQPE PADQNALSEP DVAKEAEQSD AEKAADKQPV |
|
| 101 | ADKADEVEEK AGEPEREEPD GQAVRKKALT EEREQTVREK AQKKDAETVK |
|
| 151 | KKAVKPSKET EKKASKEEKK AAKEKVAPKP TPEQILNSRS IEKARSAAAK |
|
| 201 | EVQKMKNFGQ GGSQRIICKW ARMPNPGARK GSVPNWQSWA YLPKWSAIRR |
|
| 251 | DIKRFTACKA AICPPMR* |
[0940]After further analysis, the complete gonococcal DNA sequence <SEQ ID 389> was found to be:
[0000]| 1 | ATGTTTATGA ACAAATTTTC CCAATCCGGA AAAGGTCTGT CCGGTTTCTT | |
|
| 51 | CTTCGGTTTG ATACTGGCAA CGGTCATTAT TGCCGGTATT TTGCTTTATC |
|
| 101 | TGAACCAGGG CGGTCAAAAT GCGTTCAAAA TCCCGGCTCC GTCGAAGCAG |
|
| 151 | CCTGCAGAAA CGGAAATCCT GAAACTGAAA AACCAGCCTA AGGAAGACAT |
|
| 201 | CCAACCTGAA CCGGCCGATC AAAACGCCTT GTCCGAACCG GATGTTGCGA |
|
| 251 | AAGAGGCAGA GCAGTCGGAT GCGGAAAAAG CTGCCGACAA GCAGCCCGTT |
|
| 301 | GCCGACAAag ccgacgAGGT TGAAGAAAag GcGGgcgAgc cggaACGGga |
|
| 351 | aGAGCCGGAC ggACAGGCAG TGCGCAAGAA AGCACTGAcg gAAGAgcGTG |
|
| 401 | AACAAACcgt cagggAAAAA GCGCagaaga AAGATGCCGA AACGgTTAAA |
|
| 451 | AAacaaGCgg tAaaaccgtc tAAAGAAACa gagaaaaaag cTtcaaaaga |
|
| 501 | agagaaaaag gcggcgaaag aaaAAGttgc acccaaaccg accccggaaC |
|
| 551 | aaatcctcaa cagccgCagc atcgaaaaag cgcgtagtgc cgctgccaaa |
|
| 601 | gaAgtgcaGA AAatgaaaaa ctTtgggcaa ggcgGaagcc aacgcattaT |
|
| 651 | CTGcaaatgg gcgcgtatgc cgaccgtccg gagcgcggaA gggcagcgtg |
|
| 701 | ccaaACtggc aAtcttgGgc atatctTccg aagtggtcgG CTATCAGGCG |
|
| 751 | GGACATAAAA CGCTTTACCG CGTGCAAagc GGCAatatgt ccgccgatgc |
|
| 801 | gGTGAAAAAA ATGCAGGACG AGTTGAAAAA GCATGGGGtt gcCAGCCTGA |
|
| 851 | TCCGTGcgAT TGAAGGCAAA TAA |
[0941]This encodes the following amino acid sequence <SEQ ID 390>:
[0000]| 1 | MFMNKFSQSG KGLSGFFFGL ILATVIIAGI LLYLNQGGQN AFKIPAPSKQ | |
|
| 51 | PAETEILKLK NQPKEDIQPE PADQNALSEP DVAKEAEQSD AEKAADKQPV |
|
| 101 | ADKADEVEEK AGEPEREEPD GQAVRKKALT EEREQTVREK AQKKDAETVK |
|
| 151 | KQAVKPSKET EKKASKEEKK AAKEKVAPKP TPEQILNSRS IEKARSAAAK |
|
| 201 | EVQKMKNFGQ GGSQRIICKW ARMPTVRSAE GQRAKLAILG ISSEVVGYQA |
|
| 251 | GHKTLYRVQS GNMSADAVKK MQDELKKHGV ASLIRAIEGK * |
[0942]ORF65ng-1 and ORF65-1 show 89.0% identity in 290 aa overlap:
[0000]
[0943]On this basis, including the presence of a putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 46
[0944]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 391>:
[0000]| 1 | ATGAACCACG ACATCACTTT CCTCACCCTG TTCCTACTCG GTkTCTTCGG | |
|
| 51 | CGGAAcGCAC TGCATCGGTA TGTGCGGCGG ATTAAGCAGC GcGTTTGs.s |
|
| 101 | TCCAACTCCC CCCGCATATC AACCGCTTTT GGCTGATCCT GCTGCTTAAC |
|
| 151 | ACAGGACGGG TAAGCAGCTA TACGGCAAtC GGCCTGATAC TCGGATTAAT |
|
| 201 | CGGACAGGTC GGCGTTTCAC TCGAcCAaAC CCGCGTCCTG CAGAATATTT |
|
| 251 | TATACACGGC CGCCAACCTC CTGCTGCTCT TTTTAGGCTT ATACTTGAGC |
|
| 301 | GGTATTTCTT CCTTGGCGGC AAAAATCGAG AAaATCGGCA AACCGATATG |
|
| 351 | GCGGAACCTG AACCCGATAC TCAACCGGCT GTTACCCATA AAATCCATAC |
|
| 401 | CCGCCTGCCT tGCGgTCGGA ATATTATGGG GCTGGCTGCC GTGCGGACTG |
|
| 451 | GTTTACAGCG CGTCGCTTTA CGCGCTGGGA AgCGGTAGTG CGGCAACGGG |
|
| 501 | CGGGTTATAT ATGCTTGCCT TTGCACTGGG TACGCTGCCC AATCTTtTAG |
|
| 551 | CAATCGGCAT TTTtTCCCTG CAACTGAAwA AAATCATGCA AAACCGATAT |
|
| 601 | ATCCGCCTGT GTACGGGATT ATCCGTATCA TTATGGGCAT TATGGAAACT |
|
| 651 | TGCCGTCCTG TGGCTGTAA |
[0945]This corresponds to the amino acid sequence <SEQ ID 392; ORF103>:
[0000]| 1 | MNHDITFLTL FLLGXFGGTH CIGMCGGLSS AFXXQLPPHI NRFWLILLLN | |
|
| 51 | TGRVSSYTAI GLILGLIGQV GVSLDQTRVL QNILYTAANL LLLFLGLYLS |
|
| 101 | GISSLAAKIE KIGKPIWRNL NPILNRLLPI KSIPACLAVG ILWGWLPCGL |
|
| 151 | VYSASLYALG SGSAATGGLY MLAFALGTLP NLLAIGIFSL QLXKIMQNRY |
|
| 201 | IRLCTGLSVS LWALWKLAVL WL* |
[0946]Further work elaborated the DNA sequence <SEQ ID 393> as:
[0000]| 1 | ATGAACCACG ACATCACTTT CCTCACCCTG TTCCTACTCG GTTTCTTCGG | |
|
| 51 | CGGAACGCAC TGCATCGGTA TGTGCGGCGG ATTAAGCAGC GCGTTTGCGC |
|
| 101 | TCCAACTCCC CCCGCATATC AACCGCTTTT GGCTGATCCT GCTGCTTAAC |
|
| 151 | ACAGGACGGG TAAGCAGCTA TACGGCAATC GGCCTGATAC TCGGATTAAT |
|
| 201 | CGGACAGGTC GGCGTTTCAC TCGACCAAAC CCGCGTCCTG CAGAATATTT |
|
| 251 | TATACACGGC CGCCAACCTC CTGCTGCTCT TTTTAGGCTT ATACTTGAGC |
|
| 301 | GGTATTTCTT CCTTGGCGGC AAAAATCGAG AAAATCGGCA AACCGATATG |
|
| 351 | GCGGAACCTG AACCCGATAC TCAACCGGCT GTTACCCATA AAATCCATAC |
|
| 401 | CCGCCTGCCT TGCGGTCGGA ATATTATGGG GCTGGCTGCC GTGCGGACTG |
|
| 451 | GTTTACAGCG CGTCGCTTTA CGCGCTGGGA AGCGGTAGTG CGGCAACGGG |
|
| 501 | CGGGTTATAT ATGCTTGCCT TTGCACTGGG TACGCTGCCC AATCTTTTAG |
|
| 551 | CAATCGGCAT TTTTTCCCTG CAACTGAAAA AAATCATGCA AAACCGATAT |
|
| 601 | ATCCGCCTGT GTACGGGATT ATCCGTATCA TTATGGGCAT TATGGAAACT |
|
| 651 | TGCCGTCCTG TGGCTGTAA |
[0947]This corresponds to the amino acid sequence <SEQ ID 394; ORF103-1>:
[0000]| 1 | MNHDITFLTL FLLGFFGGTH CIGMCGGLSS AFALQLPPHI NRFWLILLLN | |
|
| 51 | TGRVSSYTAI GLILGLIGQV GVSLDQTRVL QNILYTAANL LLLFLGLYLS |
|
| 101 | GISSLAAKIE KIGKPIWRNL NPILNRLLPI KSIPACLAVG ILWGWLPCGL |
|
| 151 | VYSASLYALG SGSAATGGLY MLAFALGTLP NLLAIGIFSL QLKKIMQNRY |
|
| 201 | IRLCTGLSVS LWALWKLAVL WL* |
[0948]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0949]ORF103 shows 93.8% identity over a 222aa overlap with an ORF (ORF103a) from strain A of N. meningitidis:
[0000]
[0950]The complete length ORF103a nucleotide sequence <SEQ ID 395> is:
[0000]| 1 | ATGAACCANG ACATCACTTT CCTCACCCTG TTCCTACTCG GTTTCTTCGG | |
|
| 51 | CGGAACGCAC TGCATCGGTA TGTGCGGCGG ATTAAGCAGC GCGTTTGCGC |
|
| 101 | TCCAACTCCC CCCGCATATC AACCGCTTNT GGCTGATCCT GCTGCTTAAC |
|
| 151 | ACAGGACGGG TAAGCAGCTA TACGGCAATC GGCCTGATAC TCGGATTAAT |
|
| 201 | CGGACAGGTC GGCGTTTCAC TCGACCAAAC CCGCGTCNTG CAGAATATTT |
|
| 251 | TATACACGGC CGCCAACCTC CTGCTGCTCT TTTTAGGCTT ATACTTGAGC |
|
| 301 | GGTATTTCTT CCTTGGCGGC AAAAATCGAG AAAATCGGCA AACCGATATG |
|
| 351 | GCGGAACCTG AACCCGATAC TCAACCGGCT GTTACCCATA AAATCCATAC |
|
| 401 | CCGCCTGCCT TGCGGTCGGA ATATTATGGG GCTGGCTGCC GTGCGGACTA |
|
| 451 | GTTTACAGCG CGTCGCTTTA CGCGCTGGGA AGCGGTAGTG CGGCAACGGG |
|
| 501 | CGGGTTATAT ATGCTTGCCT TTGCACTGGG TACGCTGCCC AATCTTTNGG |
|
| 551 | CAATCGGCAT TTTTTCCCTG CAACTGNAAA AAATCATGCA AAACCGATAT |
|
| 601 | ATCCGCCTGT GTACGGGATT ATCCGTATCA TTATGGGCAT TATGGAAACT |
|
| 651 | TGCCGTCCTG TGGCTGTAA |
[0951]This encodes a protein having amino acid sequence <SEQ ID 396>:
[0000]| 1 | MNXDITFLTL FLLGFFGGTH CIGMCGGLSS AFALQLPPHI NRXWLILLLN | |
|
| 51 | TGRVSSYTAI GLILGLIGQV GVSLDQTRVX QNILYTAANL LLLFLGLYLS |
|
| 101 | GISSLAAKIE KIGKPIWRNL NPILNRLLPI KSIPACLAVG ILWGWLPCGL |
|
| 151 | VYSASLYALG SGSAATGGLY MLAFALGTLP NLXAIGIFSL QLXKIMQNRY |
|
| 201 | IRLCTGLSVS LWALWKLAVL WL* |
[0952]ORF103a and ORF103-1 show 97.7% identity in 222 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0953]ORF103 shows 95.5% identity over a 222aa overlap with a predicted ORF (ORF103.ng) from N. gonorrhoeae:
[0000]
[0954]The complete length ORF103ng nucleotide sequence <SEQ ID 397> is:
[0000]| 1 | ATGAACCACG ACATCACTTT CCTCACCCTG TTCCTGCTCG GTTTCTTCGG | |
|
| 51 | CGGAACTCAC TGCATCGGTA TGTGCGGCGG ATTAAGCAGC GCGTTTGCGC |
|
| 101 | TCCAACTCCC CCCGCATATC AACCGCTTTT GGCTGATTCT GCTGCTTAAC |
|
| 151 | ACAGGACGGA TAAGCAGCTA TACGGCAATC GGCCTGATGC TCGGATTAAT |
|
| 201 | CGGACAACTC GGCATTTCAC TCGACCAAAc ccgcgTCCTG CAAAATATTT |
|
| 251 | tatacacagc ctccaaCCTC CTGCTGCTCT TTTTAGGCTT ATACTTGAGC |
|
| 301 | GGTATTTCTT CCTTGGCGGC AAAAATCGAG AAAATCGGCA AACCGATATG |
|
| 351 | GCGCAACCTG AACCCGATAC TCAACCGGCT GCTGCCCATA AAATCCATAC |
|
| 401 | CCGCCTGCCT TGCTGTCGGA ATATTATGGG GCTGGCTGCC GTGCGGACTG |
|
| 451 | GTTTACAGCG CATCACTTTA CGCGCTGGGA AGCGGTAGTG CGACAACCGG |
|
| 501 | CGGACTGTAT ATGCTTGCCT TTGCACTGGG TACGCTGCCC AATCTTTTGG |
|
| 551 | CAATCGGCAT TTTTTCCCTG CAACTGAAAA AAATCATGCA AAACCGATAT |
|
| 601 | ATCCGCCTGT GTACAGGATT ATCCGTATCA TTATGGGCAT TATGGAAGCT |
|
| 651 | TGCCGTCCTG TGGCTGTAA |
[0955]This encodes a protein having amino acid sequence <SEQ ID 398>:
[0000]| 1 | MNHDITFLTL FLLGFFGGTH CIGMCGGLSS AFALQLPPHI NRFWLILLLN | |
|
| 51 | TGRISSYTAI GLMLGLIGQL GISLDQTRVL QNILYTASNL LLLFLGLYLS |
|
| 101 | GISSLAAKIE KIGKPIWRNL NPILNRLLPI KSIPACLAVG ILWGWLPCGL |
|
| 151 | VYSASLYALG SGSATTGGLY MLAFALGTLP NLLAIGIFSL QLKKIMQNRY |
|
| 201 | IRLCTGLSVS LWALWKLAVL WL* |
[0956]In addition, ORF103ng and ORF103-1 show 97.3% identity in 222 aa overlap:
[0000]
[0957]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 47
[0958]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 399>:
[0000]| 1 | ATGGAAAACC AAAGGCCGCT CCTAGGCTTT CGCTTGGCAC TTTTGGCGGC | |
|
| 51 | GATGACGTGG GGAACGCTGC CGAT.TCCGT GCGGCAGGTA TTGAAGTTTG |
|
| 101 | TCGATGCGCC GACGCTGGTG TGGGTGCGTT TTACCGTGGC GGCGGCGGTA |
|
| 151 | TTGTTTGTTT TGCTGGCACT GGGCGGGCGG CTGCcGAAGC GGCGaGGATT |
|
| 201 | TTTCTTGGTG CTCATTCAGG CTGCTGCTGC TCGGCGTGGC GGGCATTTCG |
|
| 251 | GCAAACTTTG TGCTGATTGC CCAAGGGCTG CATTATATTT CGCCGACCAC |
|
| 301 | GACGCAGGTT TTGTGGCAGA TTTCGCCGTT TACGATGATT GTwGTCGGTG |
|
| 351 | TGTTGGTGTT TAAAGACCGG ATGACTGCCG CTCAGAAAAT CGGCTTGGTT |
|
| 401 | TTGCTGCTTG CCGGTTTGCT TATGTATTTT AACGATAAAT TCGGCGAGTT |
|
| 451 | GTCGGGTTTG GGCGCGTATG C.AAGGGCGT GTTGCTGTGT GCGGCAGGCA |
|
| 501 | GTATGGCATG GGTGTGTAAT GCCGTGGCGC AAAAGCTGCT GTCGGCGCAA |
|
| 551 | TTCGGGCCGC AACAGATTCT GCTGTTGATT TATGCGGCAA GTGCCGCCGT |
|
| 601 | GTTCCTGCCG TTTGCCGAAC CGGCACACAT CGGAAGTATG GACGGTACGT |
|
| 651 | TGGCGTGGGT ATGTATTGCG TATTGCTGCT TGAATACGTT AATCGGTTAC |
|
| 701 | GGCTCGTTCG GCGAGGCGTT GAAACATTGG GAGGCTTCCA AAGTCAGCGC |
|
| 751 | GGTAACAACC TTGCTCCCCG TGTTTACCGT AATAAATACT TTGCTCGGGC |
|
| 801 | ATTATGTGAT GCCTGAAACT TTTGCCGCGC CGGA.. |
[0959]This corresponds to the amino acid sequence <SEQ ID 400; ORF104>:
[0000]| 1 | MENQRPLLGF RLALLAAMTW GTLPXSVRQV LKFVDAPTLV WVRFTVAAAV | |
|
| 51 | LFVLLALGGR LPKRRDFSWC SFRLLLLGVA GISANFVLIA QGLHYISPTT |
|
| 101 | TQVLWQISPF TMIVVGVLVF KDRMTAAQKI GLVLLLAGLL MYFNDKFGEL |
|
| 151 | SGLGAYXKGV LLCAAGSMAW VCNAVAQKLL SAQFGPQQIL LLIYAASAAV |
|
| 201 | FLPFAEPAHI GSMDGTLAWV CIAYCCLNTL IGYGSFGEAL KHWEASKVSA |
|
| 251 | VTTLLPVFTV INTLLGHYVM PETFAAP... |
[0960]Further work revealed further partial DNA sequence <SEQ ID 401>:
[0000]| 1 | ATGGAAAACC AAAGGCCGCT CCTAGGCTTC GCGTTGGCAC TTTTGGCGGC | |
|
| 51 | GATGACGTGG GGAACGCTGC CGATTGCCGT GCGGCAGGTA TTGAAGTTTG |
|
| 101 | TCGATGCGCC GACGCTGGTG TGGGTGCGTT TTACCGTGGC GGCGGCGGTA |
|
| 151 | TTGTTTGTTT TGCTGGCACT GGGCGGGCGG CTGCCGAAGC GGCGGGATTT |
|
| 201 | TTCTTGGTGC TCATTCAGGC TGCTGCTGCT CGGCGTGGCG GGCATTTCGG |
|
| 251 | CAAACTTTGT GCTGATTGCC CAAGGGCTGC ATTATATTTC GCCGACCACG |
|
| 301 | ACGCAGGTTT TGTGGCAGAT TTCGCCGTTT ACGATGATTG TTGTCGGTGT |
|
| 351 | GTTGGTGTTT AAAGACCGGA TGACTGCCGC TCAGAAAATC GGCTTGGTTT |
|
| 401 | TGCTGCTTGC CGGTTTGCTT ATGTTTTTTA ACGATAAATT CGGCGAGTTG |
|
| 451 | TCGGGTTTGG GCGCGTATGC GAAGGGCGTG TTGCTGTGTG CGGCAGGCAG |
|
| 501 | TATGGCATGG GTGTGTTATG CCGTGGCGCA AAAGCTGCTG TCGGCGCAAT |
|
| 551 | TCGGGCCGCA ACAGATTCTG CTGTTGATTT ATGCGGCAAG TGCCGCCGTG |
|
| 601 | TTCCTGCCGT TTGCCGAACC GGCACACATC GGAAGTTTGG ACGGTACGTT |
|
| 651 | GGCGTGGGTT TGTTTTGCGT ATTGCTGCTT GAATACGTTA ATCGGTTACG |
|
| 701 | GCTCGTTCGG CGAGGCGTTG AAACATTGGG AGGCTTCCAA AGTCAGCGCG |
|
| 751 | GTAACAACCT TGCTCCCCGT GTTTACCGTA ATAwTwwCTT TGCTCGGGCA |
|
| 801 | TTATGTGATG CCTGAAACTT TTGCCGCGCC GGA... |
[0961]This corresponds to the amino acid sequence <SEQ ID 402; ORF104-1>:
[0000]| 1 | MENQRPLLG ALALLAAMTW GTLPIAVRQV LKFVDAPTLV WVRFTVAAAV | |
|
| 51 | LFVLLALGGR LPKRRDFSWC SFRLLLLGVA GISANFVLIA QGLHYISPTT |
|
| 101 | TQVLWQISPF TMIVVGVLVF KDRMTAAQKI GLVLLLAGLL MFFNDKFGEL |
|
| 151 | SGLGAYAKGV LLCAAGSMAW VCYAVAQKLL SAQFGPQQIL LLIYAASAAV |
|
| 201 | FLPFAEPAHI GSLDGTLAWV CFAYCCLNTL IGYGSFGEAL KHWEASKVSA |
|
| 251 | VTTLLPVFTV IXXLLGHYVM PETFAAP... |
[0962]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical HI0878 Protein of H. influenzae (Accession Number U32769)
[0963]ORF104 and HI0878 show 40% aa identity in 277aa overlap:
[0000]| orf104 | 4 | QRPLLGFRLALLAAMTWGTLPXSVRQVLKFVDAPTLVWXXXXXXXXXXXXXXXXXXXXP- | 62 | |
| | Q+PLLGF AL+ AM WG+LP +++QVL ++A T+VW P |
| HI0878 | 3 | QQPLLGFTFALITAMAWGSLPIALKQVLSVMNAQTIVWYRFIIAAVSLLALLAYKKQLPE | 62 |
|
| orf104 | 63 | --KRRDFSWCSFALLLLGVAGISANFVLIAQGLHYISPTTTQVLWQISPFTMIVVGVLVF | 120 |
| | K R ++W ++L+GV G+++NF+L + L+YI P+ Q+ +S F M++ GVL+F |
| HI0878 | 63 | LMKVRQYAW----IMLIGVIGLTSNFLLFSSSLNYIEPSVAQIFIHLSSFGMLICGVLIF | 118 |
|
| orf104 | 121 | KDRMTAAOKIXXXXXXXXXXMYFNDKFGELSGLGAYXKGVLLCAAGSMAWVCNAVAQKLL | 180 |
| | K+++ QKI ++FND+F +GL Y GV+L G++ WV +AQKL+ |
| HI0878 | 119 | KEKLGLHQKIGLFLLLIGLGLFFNDRFDAFAGLNQYSTGVILGVGGALIWVAYGMAQKLM | 178 |
|
| orf104 | 181 | SAQFGPQQILLLIYAASAAVFLPFAEPAHIGSMDGTLAWVCIAYCCLNTLIGYGSFGEAL | 240 |
| | +F QQILL++Y A F+P A+ + + + LA +C YCCLNTLIGYGS+ EAL |
| HI0878 | 179 | LRKFNSQQILLMMYLGCAIAFMPMADFSQVQELT-PLALICFIYCCLNTLIGYGSYAEAL | 237 |
|
| orf104 | 241 | KHWEASKVSAVTTLLPVFTVINTLLGHYVMPETFAAP | 277 |
| | W+ SKVS V TL+P+FT++ + + HY P FAAP |
| HI0878 | 238 | NRWDVSKVSVVITLVPLFTILFSHIAHYFSPADFAAP | 274 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[0964]ORF104 shows 95.3% identity over a 277aa overlap with an ORF (ORF104a) from strain A of N. meningitidis:
[0000]
[0965]The complete length ORF104a nucleotide sequence <SEQ ID 403> is:
[0000]| 1 | ATGGAAAACC AAAGGCCGCT CCTAGGCTTC GCGTTGGCAC TTTTGGCGGC | |
|
| 51 | GATGACGTGG GGAACGCTGC CGATTGCCGT GCGGCAGGTA TTGAAGTTTG |
|
| 101 | TCGATGCGCC GACGCTGGTG TGGGTGCGTT TTACCGTGGC GGCGGCGGTA |
|
| 151 | TTGTTTGTTT TGCTGGCATT GGGCGGGCGG CTGCCGAAGT GGCGGGATTT |
|
| 201 | TTCTTGGTGC TCATTCAGGC TGCTGCTGCT CGGCGTGGCG GGCATTTCGG |
|
| 251 | CAAACTTTGT GCTGATTGCC CAAGGGCTGC ATTATATTTC GCCGACCACG |
|
| 301 | ACGCAGGTTT TGTGGCAGAT TTCGCCGTTT ACGATGATTG TTGTCGGTGT |
|
| 351 | GTTGGTGTTT AAAGACCGGA TGACTGCCGC TCAGAAAATC GGCTTGGTTT |
|
| 401 | TGCTGCTTGC CGGTTTGCTT ATGTTTTTTA ACGATAAATT CGGCGAGTTG |
|
| 451 | TCGGGTTTGG GCGCGTATGC GAAGGGCGTG TTGCTGTGTG CGGCAGGCAG |
|
| 501 | TATGGCATGG GTGTGTTATG CCGTGGCGCA AAAGCTGCTG TCGGCGCAAT |
|
| 551 | TCGGGCCGCA ACAGATTCTG CTGTTGATTT ATGCGGCAAG TGCCGCCGTG |
|
| 601 | TTCCTGCCGT TTGCCGAACT GGCACACATC GGAAGTTTGG ACGGTACGTT |
|
| 651 | GGCGTGGGTT TGTTTTGCGT ATTGCTGCTT GAATACGTTA ATCGGTTACG |
|
| 701 | GCTCGTTCGG CGAGGCGTTG AAACATTGGG AGGCTTCCAA AGTCAGCGCG |
|
| 751 | GTAACAACCT TGCTCCCCGT GTTTACCGTA ATATTTTCTT TGCTCGGGCA |
|
| 801 | TTATGTGATG CCTGATACTT TTGCCGCGCC GGATATGAAC GGTTTGGGTT |
|
| 851 | ATGCCGGCGC ACTGGTCGTG GTCGGGGGTG CGGTTACGGC GGCGGTGGGG |
|
| 901 | GACAGGCTGT TCAAACGCCG CTAG |
[0966]This encodes a protein having amino acid sequence <SEQ ID 404>:
[0000]| 1 | MENQRPLLGF ALALLAAMTW GTLPIAVRQV LKFVDAPTLV WVRFTVAAAV | |
|
| 51 | LFVLLALGGR LPKWRDFSWC SFRLLLLGVA GISANFVLIA QGLHYISPTT |
|
| 101 | TQVLWQISPF TMIVVGVLVF KDRMTAAQKI GLVLLLAGLL MFFNDKFGEL |
|
| 151 | SGLGAYAKGV LLCAAGSMAW VCYAVAQKLL SAQFGPQQIL LLIYAASAAV |
|
| 201 | FLPFAELAHI GSLDGTLAWV CFAYCCLNTL IGYGSFGEAL KHWEASKVSA |
|
| 251 | VTTLLPVFTV IFSLLGHYVM PDTFAAPDMN GLGYAGALVV VGGAVTAAVG |
|
| 301 | DRLFKRR* |
[0967]ORF104a and ORF104-1 show 98.2% identity in 277 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0968]ORF104 shows 93.9% identity over a 277aa overlap with a predicted ORF (ORF104.ng) from N. gonorrhoeae:
[0000]
[0969]The complete length ORF104ng nucleotide sequence <SEQ ID 405> is predicted to encode a protein having amino acid sequence <SEQ ID 406>:
[0000]| 1 | MENQRPLLGF ALALLAAMTW GTLPIAVRQV LKFVDAPTLV WVRFTVAAAV | |
|
| 51 | LFVLLALGGR LPKRRDFSWH SFRLLLLGVT GISANFVLIA QGLHYISPTT |
|
| 101 | TQVLWQISPF TMIVVGVLVF KDRMTAAQKI GLVLLLVGLL MFFNDKFGEL |
|
| 151 | SGLGAYAKGV LLCAAGSMAW VCYAVAQKLL SAQFGPQQIL LLIYAASAAV |
|
| 201 | FLLXAEPAHI GSLDGTLAWV CFVYCCLNTL IGYGSFGEAL KHWEASKVSA |
|
| 251 | VTTLLPVFTV IFSLLGHYVM PDTFAAPDMN GLGYVGALVV VGGAVTAAVG |
|
| 301 | DRPFKRR* |
[0970]Further work revealed the complete gonococcal nucleotide sequence <SEQ ID 407>:
[0000]| 1 | ATGGAAAACC AAAGGCCGCT CCTAGGCTTC GCGTTGGCAC TTTTGGCGGC | |
|
| 51 | GATGACGTGG GGGACGCTGC CGATTGCCGT GCGGCAGGTA TTGAAGTTTG |
|
| 101 | TCGATGCGCC GACGCTGGTG TGGGTGCGTT TTACCGTGGC GGCGGCGGTA |
|
| 151 | TTGTTTGTTT TGCTGGCATT GGGCGGGCGG CTGCCGAAGC GGCGGGATTT |
|
| 201 | TTCTTGGCAT TCATTCAGGC TGCTGCTGCT CGGCGTGACG GGCATTTCGG |
|
| 251 | CAAACTTTGT GCTGATTGCC CAAGGGCTGC ATTATATTTC GCCGACCACG |
|
| 301 | ACGCAGGTTT TGTGGCAGAT TTCGCCGTTT ACGATGATTG TTGTCGGCGT |
|
| 351 | GTTGGTGTTT AAAGACCGGA tgaCTGCCGC GCAGAAAATC GGTTTGGTTT |
|
| 401 | TGCTGCttgT CGGTttgCTT ATGTTTTtta ACGACAAATT CGGCGAGTTG |
|
| 451 | TCGGGTTTGG GCGCGTATGC GAAGGGCGTG TTGCTGTGTG CGGCAGGCAG |
|
| 501 | TATGGCCTGG GTGTGTTATG CCGTGGCGCA AAAGCTGCTG TCGGCGCAAT |
|
| 551 | TCGGGCCGCA ACAGATTCTG CTGTTGATTT ATGCGGcaag tgccgccGTG |
|
| 601 | TTCCtgccgT TTGccgaaCC GGCACACATC GGAAGTTTgg aCGGTACGtt |
|
| 651 | GGCGTGGGTT TGTTTTGTGT ATTGCTGCTT GAATACGTTA ATCGGTTACG |
|
| 701 | GCTCGTTCGG CGAGGCGTTG AAACATTGGG AGGCTTCCAA AGTCAGCGCG |
|
| 751 | GTAACAACCT TGCTCCCCGT GTTTACCGTA ATATTTTCTT TGCTCGGGCA |
|
| 801 | TTATGTGATG CCTGATACTT TTGCCGCGCC GGATATGAAC GGTTTGGGTT |
|
| 851 | ATGTCGGCGC ACTGGTCGTG GTCGGGGGTG CGGTTACGGC GGCGGTGGGG |
|
| 901 | GACAGGCCGT TCAAACGCCG CTAG |
[0971]This corresponds to the amino acid sequence <SEQ ID 408; ORF104ng-1>:
[0000]| 1 | MENQRPLLGF ALALLAAMTW GTLPIAVRQV LKFVDAPTLV WVRFTVAAAV | |
|
| 51 | LFVLLALGGR LPKRRDFSWH SFRLLLLGVT GISANFVLIA QGLHYISPTT |
|
| 101 | TQVLWQISPF TMIVVGVLVF KDRMTAAQKI GLVLLLVGLL MFFNDKFGEL |
|
| 151 | SGLGAYAKGV LLCAAGSMAW VCYAVAQKLL SAQFGPQQIL LLIYAASAAV |
|
| 201 | FLPFAEPAHI GSLDGTLAWV CFVYCCLNTL IGYGSFGEAL KHWEASKVSA |
|
| 251 | VTTLLPVFTV IFSLLGHYVM PDTFAAPDMN GLGYVGALVV VGGAVTAAVG |
|
| 301 | DRPFKRR* |
[0972]ORF104ng-1 and ORF104-1 show 97.5% identity in 277 aa overlap:
[0000]
[0973]In addition, ORF104ng-1 shows significant homology with a hypothetical H. influenzae protein:
[0000]| gi|1573895 (U32769) hypothetical [Haemophilus influenzae] Length = 306 | |
| Score = 237 bits (598), Expect = 8e−62 |
| Identities = 114/280 (40%), Positives = 168/280 (59%), Gaps = 8/280 (2%) |
|
| Query: | 30 | QRPXXXXXXXXXXXMTWGTLPIAVRQVLKFVDAPTLVWXXXXXXXXXXXXXXXXXXXXP- | 88 | |
| | Q+P M WG+LPIA++QVL ++A T+VW P |
| Sbjct: | 3 | QQPLLGFTFALITAMAWGSLPIALKQVLSVMNAQTIVWYRFIIAAVSLLALLAYKKQLPE | 62 |
|
| Query: | 89 | --KRRDFSWHSFRLLLLGVTGISANFVLIAQGLHYISPTTTQVLWQISPFTMIVVGVLVF | 146 |
| | K R ++W ++L+GV G+++NF+L + L+YI P+ Q+ +S F M++ GVL+F |
| Sbjct: | 63 | LMKVRQYAW----IMLIGVIGLTSNFLLFSSSLNYIEPSVAQIFIHLSSFGMLICGVLIF | 118 |
|
| Query: | 147 | KDRMTAAQKIXXXXXXXXXXMFFNDKFGELSGLGAYAKGVLLCAAGSMAWVCYAVAQKLL | 206 |
| | K+++ QKI +FFND+F +GL Y+GV+L G++ WV Y +AQKL+ |
| Sbjct: | 119 | KEKLGLHQKIGLFLLLIGLGLFFNDRFDAFAGLNQYSTGVILGVGGALIWVAYGMAQKLM | 178 |
|
| Query: | 207 | SAQFGPQQILLLIYAASAAVFLPFAEPAHIGSLDGTLAWVCFVYCCLNTLIGYGSFGEAL | 266 |
| | +F QQILL++Y A F+P A+ + + L LA +CF+YCCLNTLIGYGS+ EAL |
| Sbjct: | 179 | LRKFNSQQILLMMYLGCAIAFMPMADFSQVQELT-PLALICFIYCCLNTLIGYGSYAEAL | 237 |
|
| Query: | 267 | KHWEASKVSAVTTLLPVFTVIFSLLGHYVMPDTFAAPDMN | 306 |
| | W+ SKVS V TL+P+FT++FS + HY P FAAP++N |
| Sbjct: | 238 | NRWDVSKVSVVITLVPLFTILFSHIANYFSPADFAAPELN | 277 |
[0974]Based on this analysis, including the presence of a putative leader sequence and several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 48
[0975]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 409>:
[0000]| 1 | ATGGTAGCTC GTCGGGCTCA TAACCCGAAG GTCGTAGGTT CGAATCCTGT | |
|
| 51 | .CCCGCAACC TAATTTCAAA CCCCTCGGTT CAATGCCGAG GG.GTTTTGT |
|
| 101 | T.TTGCCTGT TTCCTGTTTC CTGTTTCCTG CCGCCTCCGT TTTTTGCCGG |
|
| 151 | ATTTTCCTTC CGGCCGCAAT ATCGGAACGG CAGACCGCCG TCTGTTTGCG |
|
| 201 | GTTGCAAATT CAGGCAGTTT GGCTACAATC TTCCGCATTG TCTTCAAGAA |
|
| 251 | AGCCAACCAT GCCGACCGTC CGTTTTACCG AATCCGTCAG CAAACAAGAC |
|
| 301 | CTTGATGCTC TGTTCGAGTG GGCAAAAGCA AGTTACGGTG CAGAAAGTTG |
|
| 351 | CTGGAAAACG CTGTATCTGA ACGGTCysCC TTTGGGCAAC CTGTCGCCGG |
|
| 401 | AATGGGTGGA ACGCGTsmmA AAAGACTGGG AGGCAGGCTG CyCGGAGTCT |
|
| 451 | TCAGACGGCA TTTTTCTGAA TgCGGACGGc TGgCctGATA TGGgCGGAcg |
|
| 501 | cTTACAGCAC CTCGCCCTCG GTTGGCACTG TGCGGGGCTG TTGGACGgsT |
|
| 551 | GGCGCAACGA GTGTTTCGAC CTGACCGACG GCGGCGGCAA CCCCTTGTTC |
|
| 601 | ACGCTCGaAc GCGCCGyTTT mCGTCCTkTC GGACTGCTCA GCCGCGCCGT |
|
| 651 | CCATCTCAAC GGTCTGACCG AATCGGACGG CCGATGGCAT TTCTGGATAG |
|
| 701 | GCAGGCGCAG TCCGCACAAA GCAGTCGATC CCAACAAACT CGACAATACT |
|
| 751 | rCCGCCGGCG GTGTTTCCGG CGGCGAAATG CCGTCTGAAG CCGTGTGTCG |
|
| 801 | CGAAAGCAGC GAAGAAGCCG GTTTGGATAA AACGCTGcTT CCGCTCATCC |
|
| 851 | GCCCGGTATC GCAGCTGCAC AGCCTGCGCT CCGTCAGCCG GGGTGTACAC |
|
| 901 | AATGAAATCC TGTATGTATT CGATGCCGTC CTGCCG... |
[0976]This corresponds to the amino acid sequence <SEQ ID 410; ORF105>:
[0000]| 1 | MVARRAHNPK VVGSNPXPAT XFQTPRFNAE XVLXLPVSCF LFPAASVFCR | |
|
| 51 | IFLPAAISER QTAVCLRLQI QAVWLQSSAL SSRKPTMPTV RFTESVSKQD |
|
| 101 | LDALFEWAKA SYGAESCWKT LYLNGXPLGN LSPEWVERVX KDWEAGCXES |
|
| 151 | SDGIFLNADG WPDMGGRLQH LALGWHCAGL LDGWRNECFD LTDGGGNPLF |
|
| 201 | TLERAXXRPX GLLSRAVHLN GLTESDGRWH FWIGRRSPHK AVDPNKLDNT |
|
| 251 | XAGGVSGGEM PSEAVCRESS EEAGLDKTLL PLIRPVSQLH SLRSVSRGVH |
|
| 301 | NEILYVFDAV LP... |
[0977]Further work revealed the complete nucleotide sequence <SEQ ID 411>:
[0000]| 1 | ATGCCGACCG TCCGTTTTAC CGAATCCGTC AGCAAACAAG ACCTTGATGC | |
|
| 51 | TCTGTTCGAG TGGGCAAAAG CAAGTTACGG TGCAGAAAGT TGCTGGAAAA |
|
| 101 | CGCTGTATCT GAACGGTCTG CCTTTGGGCA ACCTGTCGCC GGAATGGGTG |
|
| 151 | GAACGCGTCA AAAAAGACTG GGAGGCAGGC TGCTCGGAGT CTTCAGACGG |
|
| 201 | CATTTTTCTG AATGCGGACG GCTGGCCTGA TATGGGCGGA CGCTTACAGC |
|
| 251 | ACCTCGCCCT CGGTTGGCAC TGTGCGGGGC TGTTGGACGG CTGGCGCAAC |
|
| 301 | GAGTGTTTCG ACCTGACCGA CGGCGGCGGC AACCCCTTGT TCACGCTCGA |
|
| 351 | ACGCGCCGCT TTCCGTCCTT TCGGACTGCT CAGCCGCGCC GTCCATCTCA |
|
| 101 | ACGGTCTGAC CGAATCGGAC GGCCGATGGC ATTTCTGGAT AGGCAGGCGC |
|
| 451 | AGTCCGCACA AAGCAGTCGA TCCCAACAAA CTCGACAATA CTGCCGCCGG |
|
| 501 | CGGTGTTTCC GGCGGCGAAA TGCCGTCTGA AGCCGTGTGT CGCGAAAGCA |
|
| 551 | GCGAAGAAGC CGGTTTGGAT AAAACGCTGC TTCCGCTCAT CCGCCCGGTA |
|
| 601 | TCGCAGCTGC ACAGCCTGCG CTCCGTCAGC CGGGGTGTAC ACAATGAAAT |
|
| 651 | CCTGTATGTA TTCGATGCCG TCCTGCCCGA AACCTTCCTG CCTGAAAATC |
|
| 701 | AGGATGGCGA AGTGGCGGGT TTTGAGAAAA TGGACATCGG CGGTCTGTTG |
|
| 751 | GATGCCATGT TGTCGGGAAA CATGATGCAC GACGCGCAAC TGGTTACGCT |
|
| 801 | GGACGCGTTT TGCCGTTACG GTCTGATTGA TGCCGCCCAT CCGCTGTCCG |
|
| 851 | AGTGGCTGGA CGGCATACGT TTATAG |
[0978]This corresponds to the amino acid sequence <SEQ ID 412; ORF105-1>:
[0000]| 1 | MPTVRFTESV SKQDLDALFE WAKASYGAES CWKTLYLNGL PLGNLSPEWV | |
|
| 51 | ERVKKDWEAG CSESSDGIFL NADGWPDMGG RLQHLALGWH CAGLLDGWRN |
|
| 101 | ECFDLTDGGG NPLFTLERAA FRPFGLLSRA VHLNGLTESD GRWHFWIGRR |
|
| 151 | SPHKAVDPNK LDNTAAGGVS GGEMPSEAVC RESSEEAGLD KTLLPLIRPV |
|
| 201 | SQLHSLRSVS RGVHNEILYV FDAVLPETFL PENQDGEVAG FEKMDIGGLL |
|
| 251 | DAMLSGNMMH DAQLVTLDAF CRYGLIDAAH PLSEWLDGIR L* |
[0979]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0980]ORF105 shows 89.4% identity over a 226aa overlap with an ORF (ORF105a) from strain A of N. meningitidis.
[0000]
[0981]The complete length ORF105a nucleotide sequence <SEQ ID 413> is:
[0000]| 1 | ATGCCGACCG TCCGTTTTAC CGAATCCGTC AGCAAACACG ACCTTGATGC | |
|
| 51 | CCTATTCGAG TGGGCAAAGG CAAGTTACGG TGCGGAAAGT TGCTGGAAAA |
|
| 101 | CGCTGTATCT GAACGGTCTG CCTTTGGGCA ATCTGTCGCC GGAATGGGCG |
|
| 151 | GAGCGCGTCA AAAAAGACTG GGAGGCAGGC TGCTCGGAGT CTTCAGACGG |
|
| 201 | CATTTTCCTG AATGCGGACG GCTGGCCAGA TATGGGCAGA CGCTTGCAGC |
|
| 251 | ACCTCGCCCG AATATGGAAA GAAGCGGGAC TGCTTCACGG CTGGCGCGAC |
|
| 301 | GAGTGTTTCG ACCTGACCGA CGGCGGCAGC AATCCCTTGT TCGCGCTCGA |
|
| 351 | ACGCGCCGCT TTCCGTCCGT TCGGACTGCT CAGCCGCGCC GTCCATCTCA |
|
| 401 | ACGGTTTGGT CGAATCGGAC GGCCGATGGC ATTTCTGGAT AGGCAGGCGC |
|
| 451 | AGTCCGCACA AAGCAGTCGA TCCCGACAAA CTCGACAATA CTGCCGCCGG |
|
| 501 | CGGTGTTTCC AGCGGTGAAT TGCCGTCTGA AACCGTGTGT CGCGAAAGCA |
|
| 551 | GCGAAGAAGC CGGTTTGGAT AAAACGCTGC TTCCGCTCAT CCGCCCGGTA |
|
| 601 | TCGCAGCTGC ACAGCCTGCG CCCCGTCAGC CGGGGTGTGC ACAATGAAAT |
|
| 651 | CCTGTATGTA TTCGATGCCG TCCTGCCCGA AACCTTCCTG CCTGAAAATC |
|
| 701 | AGGATGGCGA AGTGGCGGGT TTTGAGAAAA TGGACATCGG CGGTCTGTTG |
|
| 751 | GCTGCCATGT TGTCGGGAAA CATGATGCAC GACGCGCAAC TGGTTACGCT |
|
| 801 | GGACGCGTTT TGCCGTTACG GTCTGATTGA TGCCGCCCAT CCGCTGTCCG |
|
| 851 | AGTGGCTGGA CGGCATACGT TTATAG |
[0982]This encodes a protein having amino acid sequence <SEQ ID 414>:
[0000]| 1 | MPTVRFTESV SKHDLDALFE WAKASYGAES CWKTLYLNGL PLGNLSPEWA | |
|
| 51 | ERVKKDWEAG CSESSDGIFL NADGWPDMGR RLQHLARIWK EAGLLHGWRD |
|
| 101 | ECFDLTDGGS NPLFALERAA FRPFGLLSRA VHLNGLVESD GRWHFWIGRR |
|
| 151 | SPHKAVDPDK LDNTAAGGVS SGELPSETVC RESSEEAGLD KTLLPLIRPV |
|
| 201 | SQLHSLRPVS RGVHNEILYV FDAVLPETFL PENQDGEVAG FEKMDIGGLL |
|
| 251 | AAMLSGNMMH DAQLVTLDAF CRYGLIDAAH PLSEWLDGIR L* |
[0983]ORF105a and ORF105-1 show 93.8% identity in 291 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[0984]ORF105 shows 87.5% identity over a 312aa overlap with a predicted ORF (ORF105.ng) from N. gonorrhoeae:
[0000]
[0985]A complete length ORF105ng nucleotide sequence <SEQ ID 415> was predicted to encode a protein having amino acid sequence <SEQ ID 416>:
[0000]| 1 | MVARRAHNPK VVGSNPAPAT KYQTPRFNAE GVLFFLFPAA SVFCRIFLPA | |
|
| 51 | AISERQAAVC LRLQIQAVWL QSSALCSRKP AMPTVRFTES VSKQDLDALF |
|
| 101 | ERAKASYGAE SCWKTLYLNR LPLGNLSPEW AERIKKDWEA GCSESSNGIF |
|
| 151 | LNADGWPDMG GRLQHLARTW NKAGLLHGWR NECFDLTDGG GNPLFTLERA |
|
| 201 | AFRPFGLLIR AVHLNGLVES NGRWHFWIGR RSPHKAVDPG KLDNIAGGGV |
|
| 251 | SGGEMPSEAV CRESSEEAGL DKTLFPLIRP VSRLHSLRPV SRGVHNEILY |
|
| 301 | VFDAVLPETF LPENQDGEVA GFEKMDIGGL LDAMLSKNMM HDAQLVTLDA |
|
| 351 | FYRYGLIDAA HPLSEWLDGI RL* |
[0986]Further work revealed the complete nucleotide sequence <SEQ ID 417>:
[0000]| 1 | ATGCCGACCG TCCGTTTTAC CGAATCCGTC AGCAAACAAG ACCTTGATGC | |
|
| 51 | CCTGTTCGAG CGGGCAAAAG CAAGTTACGG TGCCGAAAGT TGCTGGAAAA |
|
| 101 | CGCTGTATCT GAACCGTCTT CCTTTGGGCA ATCTGTCGCC GGAATGGGCT |
|
| 151 | GAGCGCATCA AAAAAGACTG GGAGGCAGGC TGCTCCGAGT CTTCAGACGG |
|
| 201 | CATTTTTCTG AATGCGGACG GCTGGCCGGA TATGGGCGGA CGCTTGCAGC |
|
| 251 | ACCTCGCCCG CACATGGAAC AAGGCGGGGC TGCTTCACGG ATGGCGCAAC |
|
| 301 | GAGTGTTTCG ACCTGACCGA CGGCGGCGGC AACCCCTTGT TCACGCTCGA |
|
| 351 | ACGCGCCGCT TTCCGTCCGT TCGGACTACT CAGCCGCGCC GTCCATCTCA |
|
| 401 | ACGGTTTGGT CGAATCGAAC GGCAGATGGC ATTTTTGGAT AGGCAGGCGC |
|
| 451 | AGTCCGCACA AAGCAGTCGa tcCCGGCAAG CTCGACAATA TTGCCGGCGG |
|
| 501 | CGGTGTTTCC GGCGGCGAAA TGCCGTCTGA AGCCGTGTGC CGCGAAAGCA |
|
| 551 | GCGAAGAAGC CGGTTTGGAT AAAACGCTGT TTCCGCTCAT CCGCCCAGTA |
|
| 601 | TCGCGGCTGC ACAGCCTTCG CCCCGTCAGC CGAGGTGTGC ACAATGAAAT |
|
| 651 | CCTGTATGTG TTCGATGCCG TCCTGCCCGA AACCTTCCTG CCTGAAAATC |
|
| 701 | AGGATGGCGA GGTAGCGGGT TTTGAAAAGA TGGACATTGG CGGCCTATTG |
|
| 751 | GATGCCATGT TGTCGAAAAA CATGATGCAC GACGCGCAAC TGGTTACGCT |
|
| 801 | GGACGCGTTT TACCGTTACG GTCTGATTGA TGCCGCCCAT CCGCTGTCCG |
|
| 851 | AGTGGCTGGA CGGCATACGT TTATAG |
[0987]This corresponds to the amino acid sequence <SEQ ID 418; ORF105ng-1>:
[0000]| 1 | MPTVRFTESV SKQDLDALFE RAKASYGAES CWKTLYLNRL PLGNLSPEWA | |
|
| 51 | ERIKKDWEAG CSESSDGIFL NADGWPDMGG RLQHLARTWN KAGLLHGWRN |
|
| 101 | ECFDLTDGGG NPLFTLERAA FRPFGLLSRA VHLNGLVESN GRWHFWIGRR |
|
| 151 | SPHKAVDPGK LDNIAGGGVS GGEMPSEAVC RESSEEAGLD KTLFPLIRPV |
|
| 201 | SRLHSLRPVS RGVHNEILYV FDAVLPETFL PENQDGEVAG FEKMDIGGLL |
|
| 251 | DAMLSKNMMH DAQLVTLDAF YRYGLIDAAH PLSEWLDGIR L* |
[0988]ORG105ng-1 and ORF105-1 show 93.5% identity in 291 aa overlap:
[0000]
[0989]Furthermore, ORF105ng-1 shows homology with a yeast enzyme:
[0000]| sp|P41888|TNR3_SCHPO THIAMIN PYROPHOSPHOKINASE (TPK) (THIAMIN KINASE) | |
| >gi|1076928|pir||S52350 thiamin pyrophosphokinase (EC 2.7.6.2) - fission |
| yeast (Schizosaccharomyces pombe) >gi|666111 (X84417) thiamin |
| pyrophosphokinase [Schizosaccharomyces pombe] >gi|2330852|gnl|PID|e334056 |
| (Z98533) thiamin pyrophosphokinase [Schizosaccharomyces pombe] |
| Length = 569 Score = 105 bits (259), Expect = 4e−22 |
| Identities = 64/192 (33%), Positives = 94/192 (48%), Gaps = 3/192 (1%) |
|
| Query: | 268 | NKAGLLHGWRNECFDLTDGGGNPLFTLERAAFRPFGLLSRAVHLNGLVESNGRW--HFWI | 441 | |
| | N G+ WRNE + + P+ +ER F FG LS VH + + W+ |
| Sbjct: | 96 | NTFGIADQWRNELYTVYGKSKKPVLAVERGGFWLFGFLSTGVHCTMYIPATKEHPLRIWV | 155 |
|
| Query: | 442 | GRRSPHKAVDPGKLDNIAGGGVSGGEMPSEAVCRESSEEAGLDKTLFPLIRPVSRLHSLR | 621 |
| | RRSP K P LDN GG++ G+ + +E SEEA LD + LI P + ++ |
| Sbjct: | 156 | PRRSPTKQTWPNYLDNSVAGGIAHGDSVIGTMIKEFSEEANLDVSSMNLI-PCGTVSYIK | 214 |
|
| Query: | 622 | PVSRG-VHNEILYVFDAVLPETFLPENQDGEVAGFEKMDIGGLLDAMLSKNMMHDAQLVT | 798 |
| | R + E+ YVFD + + +P DGEVAGF + + +L + K+ + LV |
| Sbjct: | 215 | MEKRHWIQPELQYVFDLPVDDLVIPRINDGEVAGFSLLPLNQVLHELELKSFKPNCALVL | 274 |
|
| Query: | 799 | LDAFYRYGLIDAAHP | 843 |
| | LD R+G+I HP |
| Sbjct: | 275 | LDFLIRHGIITPQHP | 289 |
[0990]Based on this analysis, including the presence of a putative transmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 49
[0991]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 419>:
[0000]| 1 | ATGAATAGAC CCAAGCAACC CTTCTTCCGT CCCGAAGTCG CCGTTGCCCG | |
|
| 51 | CCAAACCAGC CTGACGGGTA AAGTGATTCT GACACGACCG TTGTCATTTT |
|
| 101 | CCCTATGGAC GACATTTGCA TCGATATCTG CGTTATTGAT TATCCTGTTT |
|
| 151 | TTGATATTTG GTAACTATAC GCGAAAGACA ACAGTGGAGG GACAAATTTT |
|
| 201 | ACCTGCATCG GGCGTAATCA GGGTGTATGC ACCGgATACG rGkACAATTA |
|
| 251 | CAGCGAAATT CGTGGAAGAT GGmsAAAAGG TTAAGGCTGG CGACAAGCTA |
|
| 301 | TTTGCGCTTT CGACCTCACG TTTCGGCGCA GGAGGTAGCG TGCAGCAGCA |
|
| 351 | GTTGAAAACG GAGGCAGTTT TGAAGAAAAC GTTGGCAGAA CAGGAACTGG |
|
| 401 | GTCGTCTGAA GCTGATACAC GGGAATGAAA CGCGCAgCcT TAAAGCAACT |
|
| 451 | GTCGAACGTT TGGAAAACCA GGAACTCCAT ATTTCGCAAC AGATAGACGG |
|
| 501 | TCAGAAAAGG CGCATTAGAC TTGCGGAAGA AATGTTGCAG AAATATCGTT |
|
| 551 | TCCTATCCGC .CAATGA |
[0992]This corresponds to the amino acid sequence <SEQ ID 420; ORF107>:
[0000]| 1 | MNRPKQPFFR PEVAVARQTS LTGKVILTRP LSFSLWTTFA SISALLIILF | |
|
| 51 | LIFGNYTRKT TVEGQILPAS GVIRVYAPDT XTITAKFVED GXKVKAGDKL |
|
| 101 | FALSTSRFGA GGSVQQQLKT EAVLKKTLAE QELGRLKLIH GNETRSLKAT |
|
| 151 | VERLENQELH ISQQIDGQKR RIRLAEEMLQ KYRFLSXQ* |
[0993]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[0994]ORF107 shows 97.8% identity over a 186aa overlap with an ORF (ORF107a) from strain A of N. meningitidis:
[0000]
[0995]The complete length ORF107a nucleotide sequence <SEQ ID 421> is:
[0000]| 1 | ATGAATAGAC CCAAGCAACC NTTCTTCCGT CCCGAAGTCG CCGTTGCCCG | |
|
| 51 | CCAAACCAGC CTGACGGGTA AAGTGATTCT GACACGACCG TTGTCATTTT |
|
| 101 | CCCTATGGAC GACATTTGCA TCGATATCTG CGTTATTGAT TATCCTGTTT |
|
| 151 | TTGATATTTG GTAACTATAC GCGAAAGACA ACAGTGGAGG GACAAATTTT |
|
| 201 | ACCTGCATCG GGCGTAATCA GGGTGTATGC ACCGGATACG GGGACAATTA |
|
| 251 | CNGCGAAATT CNTGGAAGAT GGAGAAAAGG TTAAGGCTGG CGACAAGCTA |
|
| 301 | TTTGCGCTTT CGACCTCACG TTTCGGCGCA GGAGATAGCG TGCAGCAGCA |
|
| 351 | GTTGAAAACG GAGGCAGTTT TGAAGAAAAC GTTGGCAGAA CAGGAACTGG |
|
| 401 | GTCGTCTGAA GCTGATACAC GGGAATGAAA CGCGCAGCCT TAAAGCAACT |
|
| 451 | GTCGAACGTT TGGAAAACCA GGAACTCCAT ATTTCGCAAC AGATAGACGG |
|
| 501 | TCAGAAAAGG CGCATTAGAC TTGCGGAAGA AATGTTGCAG AAATATCGTT |
|
| 551 | TCCTATCCGC CAATGATGCA GTGCCAAAAC AAGAAATGAT GAATGTCAAG |
|
| 601 | GCAGAGCTTT TAGAGCAGAA AGCCAAACTT GATGCCTACC GCCGAGAAGA |
|
| 651 | AGTCGGGCTG CTTCAGGAAA TCCGCACGCA GAATCTGACA TTGGNNAGCC |
|
| 701 | TCCCCCAAGC GGCATGA |
[0996]This encodes a protein having amino acid sequence <SEQ ID 422>:
[0000]| 1 | MNRPKQPFFR PEVAVARQTS LTGKVILTRP LSFSLWTTFA SISALLIILF | |
|
| 51 | LIFGNYTRKT TVEGQILPAS GVIRVYAPDT GTITAKFXED GEKVKAGDKL |
|
| 101 | FALSTSRFGA GDSVQQQLKT EAVLKKTLAE QELGRLKLIH GNETRSLKAT |
|
| 151 | VERLENQELH ISQQIDGQKR RIRLAEEMLQ KYRFLSANDA VPKQEMMNVK |
|
| 201 | AELLEQKAKL DAYRREEVGL LQEIRTQNLT LXSLPQAA* |
Homology with a Predicted ORF from
N. gonorrhoeae [0997]ORF107 shows 95.7% identity over a 188aa overlap with a predicted ORF (ORF107.ng) from N. gonorrhoeae:
[0000]
[0998]The complete length ORF107ng nucleotide sequence <SEQ ID 423> is predicted to encode a protein having amino acid sequence <SEQ ID 424>:
[0000]| 1 | MNRPKQPFFR PEVAIARQTS LTGKVILTRP LSFSLWTTFA SISALLIILF | |
|
| 51 | LIFGNYTRKT TMEGQILPAS GVIRVYAPDT GTITAKFVED GEKVKAGDKL |
|
| 101 | FALSTSRFGA GGSVQQQLKT EAVLKKTLAE QELGRLKLIH ENETRSLKAT |
|
| 151 | VERLENQKLH ISQQIDGQKR RIRLAEEMLR KYRFLSAQ* |
[0999]Based on the presence of a putative ransmembrane domain in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 50
[1000]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 425>:
[0000]| 1 | ATGCTGAATA CTTTTTTTGC CGTATTGGGC GGCTGCCTGC TGCT.TTGCC | |
|
| 51 | GTGCGGCAAA TCCGTAAATA CGGCGGTACA GCCGCAAAAC GCGGTACAAA |
|
| 101 | GCGCGCCGAA ACCGGTTTTC AAAGTCATAT ATATCGACAA TACGGCGATT |
|
| 151 | GCCGGTTTGG ATTTGGGACA AAGCAGCGAA GGCAAAACCA ACGACGGCAA |
|
| 201 | AAAACAAATC AGTTATCCGA TTAAAGGCTT GCCGGAACAA AATGTTATCC |
|
| 251 | GACTGATCGG CAAGCATCCC GGCGACTTGG AAGCCGTCAG CGGCAAATGT |
|
| 301 | ATGGAAACCG ATGATAAGGA CAGTCCGGCA GGTTGGGCAG AAAACGGCGT |
|
| 351 | GTGCCATACC TTGTTTGCCA AACTGGTGGG CAATATCGCC GAAGACGGCG |
|
| 401 | GCAAACTGAC GGATTACCTA GTTTCGCATG CCGCCCTGCA ACCCTATCAG |
|
| 451 | GCAGGCAAAA GCGGCTATGC CGCCGTGCAG AACGGACGCT ATGTGCTGGA |
|
| 501 | AATCGACAGC GAAGGGGCGT TTTATTTCCG CCGCCGCCAT TATTGA |
[1001]This corresponds to the amino acid sequence <SEQ ID 426; ORF108>:
[0000]| 1 | MLNTFFAVLG GCLLXLPCGK SVNTAVQPQN AVQSAPKPVF KVIYIDNTAI | |
|
| 51 | AGLDLGQSSE GKTNDGKKQI SYPIKGLPEQ NVIRLIGKHP GDLEAVSGKC |
|
| 101 | METDDKDSPA GWAENGVCHT LFAKLVGNIA EDGGKLTDYL VSHAALQPYQ |
|
| 151 | AGKSGYAAVQ NGRYVLEIDS EGAFYFRRRH Y* |
[1002]Further work revealed the following DNA sequence <SEQ ID 427>:
[0000]| 1 | ATGCTGAAAA CATCTTTTGC CGTATTGGGC GGCTGCCTGC TGCTTGCCGC | |
|
| 51 | CTGCGGCAAA TCCGAAAATA CGGCGGAACA GCCGCAAAAC GCGGTACAAA |
|
| 101 | GCGCGCCGAA ACCGGTTTTC AAAGTCAAAT ATATCGACAA TACGGCGATT |
|
| 151 | GCCGGTTTGG ATTTGGGACA AAGCAGCGAA GGCAAAACCA ACGACGGCAA |
|
| 201 | AAAACAAATC AGTTATCCGA TTAAAGGCTT GCCGGAACAA AATGTTATCC |
|
| 251 | GACTGATCGG CAAGCATCCC GGCGACTTGG AAGCCGTCAG CGGCAAATGT |
|
| 301 | ATGGAAACCG ATGATAAGGA CAGTCCGGCA GGTTGGGCAG AAAACGGCGT |
|
| 351 | GTGCCATACC TTGTTTGCCA AACTGGTGGG CAATATCGCC GAAGACGGCG |
|
| 401 | GCAAACTGAC GGATTACCTA GTTTCGCATG CCGCCCTGCA ACCCTATCAG |
|
| 451 | GCAGGCAAAA GCGGCTATGC CGCCGTGCAG AACGGACGCT ATGTGCTGGA |
|
| 501 | AATCGACAGC GAAGGGGCGT TTTATTTCCG CCGCCGCCAT TATTGA |
[1003]This corresponds to the amino acid sequence <SEQ ID 428; ORF108-1>:
[0000]| 1 | MLKTSFAVLG GCLLLAACGK SENTAEQPQN AVQSAPKPVF KVKYIDNTAI | |
|
| 51 | AGLDLGQSSE GKTNDGKKQI SYPIKGLPEQ NVIRLIGKHP GDLEAVSGKC |
|
| 101 | METDDKDSPA GWAENGVCHT LFAKLVGNIA EDGGKLTDYL VSHAALQPYQ |
|
| 151 | AGKSGYAAVQ NGRYVLEIDS EGAFYFRRRH Y* |
[1004]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1005]ORF108 shows 88.4% identity over a 18 laa overlap with a predicted ORF (ORF108.ng) from N. gonorrhoeae:
[0000]
[1006]ORF108-1 shows 92.3% identity with ORF108ng over the same 181 aa overlap:
[0000]
[1007]The complete length ORF108ng nucleotide sequence <SEQ ID 429> is:
[0000]| 1 | ATGCTGAAAa tacctTTTGC CGTGTtgggc ggCtgcctGC TGCTTGCCGC | |
|
| 51 | CTGCGGCAAA TCCGAAAATa cggcggaACA GCCGCAAAAT gcggCACAAA |
|
| 101 | GCGCGCCGAA ACCGGTTTTC AAAGTCAAAT ACATCGACAA TACGGCGATT |
|
| 151 | GCCGGTTTGG CTTTGGGACA AAGTAGCGAA GGCAAAACCA acgacgGCAA |
|
| 201 | AAAACAAATC AGTTATccgA TTAAAGGCTT GCCGGAACAA Aacgccgtcc |
|
| 251 | gGCTGACCGG AAAGCATCCC AACGACTTGG AagccgtcgT CGGCAAATGT |
|
| 301 | ATGGAAACCG ACGGAAAGGA CGCGCCTTCG GGCTGGGCGG AAAACGGCGT |
|
| 351 | GTGCCATACC TTGTTTGCCA AACTGGTGGG CAATATCGCC GAAGACGGCG |
|
| 401 | GCAAACTGAC TGATTACCTG ATTTCGCATT CCGCCCTGCA ACCCTATCAG |
|
| 451 | GCAGGCAAAA GCGGCTATGC CGCCGTGCAG AACGGACGCT ATGTGCTGGA |
|
| 501 | AATCGACAGC GagggGGCGT TTTATttccg ccgccgccat tattgA |
[1008]This encodes a protein having amino acid sequence <SEQ ID 430>:
[0000]| 1 | MLKIPFAVLG GCLLLAACGK SENTAEQPQN AAQSAPKPVF KVKYIDNTAI | |
|
| 51 | AGLALGQSSE GKTNDGKKQI SYPIKGLPEQ NAVRLTGKHP NDLEAVVGKC |
|
| 101 | METDGKDAPS GWAENGVCHT LFAKLVGNIA EDGGKLTDYL ISHSALQPYQ |
|
| 151 | AGKSGYAAVQ NGRYVLEIDS EGAFYFRRRH Y* |
[1009]Based on this analysis, including the presence of a predicted prokaryotic membrane lipoprotein lipid attachment site (underlined) and a putative ATP/GTP-binding site motif A (P-loop, double-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 51
[1010]The following DNA sequence was identified in N. meningitidis <SEQ ID 431>:
[0000]| 1 | ATGGAAGATT TATATATAAT ACTCGCTTTG GGTTTGGTTG CGATGATTGC | |
|
| 51 | CGgATTTATC GATgcgatTg cGggCGGGGG TGGTTTGATT ACGCTGCCCG |
|
| 101 | CACTCTTGTT GGCAGGTATT CCTCCCGTGT CGGCAATTGC CACCAACAAG |
|
| 151 | CTGCAAgCAG CCGCTGCTAC GTTTTCAGCT ACGGTTTCTT TTGCACGCAA |
|
| 201 | AGGTTTGATT GATTGGAAGA AAGGTCTCCC GATTGCCGCA GCATCGTTTG |
|
| 251 | TAGGCGGCGT GGcCGGTGCA TTATCGGTCA GCTTGGTTTC CAAAGATATT |
|
| 301 | CTgCTgGCGG TCGTGCCGGT TTTGTTGATA TTTGTCGCAC TGTATTTTGT |
|
| 351 | GTTTTCGCCC AAGCTCGACG GCAGTAAGGA AGGCAAAGCC AGAATGTCTT |
|
| 401 | TTTTTCTGTT cGGGCTGACG GTCGC.ACCG CTTTTGGGTT TTTACGACGG |
|
| 451 | TGTGTTCGGA CCGGGTGTCG GCTCGTTTTT TCTGATTGCC TTTATTGTTT |
|
| 501 | TGCTCGGCTG CAAgCTGTTG AACGCGATGT CTTACACCAA ATTGGCGAAC |
|
| 551 | GTTGCCTGCA ATCTTGGTTC GCTATCGGTA TTCCTGCTGC ACGGTTCGAT |
|
| 601 | TATTTTCCCG ATTGCGGCAA CGaTGGCGGT CGGTGCGTTT GTCGGtGCGA |
|
| 651 | ATTTAgGTGC GAGATTTGCC GTaCgctTCG GTTCGAAGCT GATTAA |
[1011]This corresponds to the amino acid sequence <SEQ ID 432; ORF109>:
[0000]| 1 | MEDLYIILAL GLVAMIAGFI DAIAGGGGLI TLPALLLAGI PPVSAIATNK | |
|
| 51 | LQAAAATFSA TVSFARKGLI DWKKGLPIAA ASFVGGVAGA LSVSLVSKDI |
|
| 101 | LLAVVPVLLI FVALYFVFSP KLDGSKEGKA RMSFFLFGLT VXTAFGFLRR |
|
| 151 | CVRTGCRLVF SDCLYCFARL QAVERDVLHQ IGERCLQSWF AIGIPAARFD |
|
| 201 | YFPDCGNDGG RCVCRCEFRC EICRTLRFEA D* |
[1012]Further work revealed the following DNA sequence <SEQ ID 433>:
[0000]| 1 | ATGGAAGATT TATATATAAT ACTCGCTTTG GGTTTGGTTG CGATGATTGC | |
|
| 51 | CGGATTTATC GATGCGATTG CGGGCGGGGG TGGTTTGATT ACGCTGCCCG |
|
| 101 | CACTCTTGTT GGCAGGTATT CCTCCCGTGT CGGCAATTGC CACCAACAAG |
|
| 151 | CTGCAAGCAG CCGCTGCTAC GTTTTCAGCT ACGGTTTCTT TTGCACGCAA |
|
| 201 | AGGTTTGATT GATTGGAAGA AAGGTCTCCC GATTGCCGCA GCATCGTTTG |
|
| 251 | TAGGCGGCGT GGCCGGTGCA TTATCGGTCA GCTTGGTTTC CAAAGATATT |
|
| 301 | CTGCTGGCGG TCGTGCCGGT TTTGTTGATA TTTGTCGCAC TGTATTTTGT |
|
| 351 | GTTTTCGCCC AAGCTCGACG GCAGTAAGGA AGGCAAAGCC AGAATGTCTT |
|
| 401 | TTTTTCTGTT CGGGCTGACG GTCGCACCGC TTTTGGGTTT TTACGACGGT |
|
| 451 | GTGTTCGGAC CGGGTGTCGG CTCGTTTTTT CTGATTGCCT TTATTGTTTT |
|
| 501 | GCTCGGCTGC AAGCTGTTGA ACGCGATGTC TTACACCAAA TTGGCGAACG |
|
| 551 | TTGCCTGCAA TCTTGGTTCG CTATCGGTAT TCCTGCTGCA CGGTTCGATT |
|
| 601 | ATTTTCCCGA TTGCGGCAAC GATGGCGGTC GGTGCGTTTG TCGGTGCGAA |
|
| 651 | TTTAGGTGCG AGATTTGCCG TCCGCTTCGG TTCGAAGCTG ATTAAGCCGC |
|
| 701 | TGCTGATTGT CATCAGCATT TCGATGGCTG TGAAATTGTT GATAGACGAG |
|
| 751 | AGAAATCCGC TGTATCAGAT GATTGTTTCG ATGTTTTAA |
[1013]This corresponds to the amino acid sequence <SEQ ID 434; ORF109-1>:
[0000]| 1 | MEDLYIILAL GLVAMIAGFI DAIAGGGGLI TLPALLLAGI PPVSAIATNK | |
|
| 51 | LQAAAATFSA TVSFARKGLI DWKKGLPIAA ASFVGGVAGA LSVSLVSKDI |
|
| 101 | LLAVVPVLLI FVALYFVFSP KLDGSKEGKA RMSFFLFGLT VAPLLGFYDG |
|
| 151 | VFGPGVGSFF LIAFIVLLGC KLLNAMSYTK LANVACNLGS LSVFLLHGSI |
|
| 201 | IFPIAATMAV GAFVGANLGA RFAVRFGSKL IKPLLIVISI SMAVKLLIDE |
|
| 251 | RNPLYQMIVS MF* |
[1014]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1015]ORF109 shows 95.9% identity over a 147aa overlap with an ORF (ORF109a) from strain A of N. meningitidis.
[0000]
[1016]The complete length ORF109a nucleotide sequence <SEQ ID 435> is:
[0000]| 1 | ATGGAAGATT TATACATAAT ACTCGCTTTG GGTTTGGTTG CGATGATTGC | |
|
| 51 | CGGATTTATC GATGCGATTG CGGGTGGGGG TGGTTTGATT ACGCTGCCTG |
|
| 101 | CACTCTTGTT GGCAGGTATT CCTCCCGTGT CGGCAATTGC CACCAACAAG |
|
| 151 | CTGCAAGCAG CCGCTGCTAC GTTTTCGGCT ACGGTTTCTT TTGCACGCAA |
|
| 201 | AGGTTTGATT GATTGGAAGA AAGGTCTCCC GATTGCGGCA GCATCGTTTG |
|
| 251 | CAGGCGGCGT GGTCGGTGCA TTATCGGTCA GCTTGGTTTC CAAAGATATT |
|
| 301 | CTGCTGGCGG TCGTGCCGGT TTTGTTGATA TTTGTCGCGC TGTATTTTGT |
|
| 351 | GTTTTCGCCC AAGCTCGACG GCAGTAAGGA AGGCAAAGCC AGAATGTCTT |
|
| 401 | TTTTTCTGTT CGGTCTGACG GTTGCACCAC TTTTGGGTTT TTACGACGGT |
|
| 451 | GTGTTCGGAC CGGGTGTCGG CTCGTTTTTT CTGATTGCCT TTATTGTTTT |
|
| 501 | GCTCGGCTGC AAGCTGTTGA ACGCGATGTC TTACACCAAA TTGGCGAACG |
|
| 551 | TTGCCTGCAA TCTTGGTTCG CTATCGGTAT TCCTGCTGCA CGGTTCGATT |
|
| 601 | ATTTTCCCGA TTGCGGCAAC GATGGCGGTC GGTGCGTTTG TCGGTGCGAA |
|
| 651 | TTTAGGTGCG AGATTTGCCG TCCGCTTCGG TTCGAAGCTG ATTAAGCCGC |
|
| 701 | TGCTGATTGT CATCAGCATT TCGATGGCTG TGAAATTGTT GATAGACGAG |
|
| 751 | AGAAATCCGC TGTATCAGAT GATTGTTTCG ATGTTTTAA |
[1017]This encodes a protein having amino acid sequence <SEQ ID 436>:
[0000]| 1 | MEDLYIILAL GLVAMIAGFI DAIAGGGGLI TLPALLLAGI PPVSAIATNK | |
|
| 51 | LQAAAATFSA TVSFARKGLI DWKKGLPIAA ASFAGGVVGA LSVSLVSKDI |
|
| 101 | LLAVVPVLLI FVALYFVFSP KLDGSKEGKA RMSFFLFGLT VAPLLGFYDG |
|
| 151 | VFGPGVGSFF LIAFIVLLGC KLLNAMSYTK LANVACNLGS LSVFLLHGSI |
|
| 201 | IFPIAATMAV GAFVGANLGA RFAVRFGSKL IKPLLIVISI SMAVKLLIDE |
|
| 251 | RNPLYQMIVS MF* |
[1018]ORF109a and ORF109-1 show 99.2% identity in 262 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1019]ORF109 shows 98.3% identity over a 231aa overlap with a predicted ORF (ORF109.ng) from N. gonorrhoeae:
[0000]
[1020]An ORF109ng nucleotide sequence <SEQ ID 437> was predicted to encode a protein having amino acid sequence <SEQ ID 438>:
[0000]| 1 | MEDLYIILAL GLVAMIAGFI DAIAGGGGLI TLPALLLAGI PPVSAIATNK | |
|
| 51 | LQAAAATFSA TVSFARKGLI DWKKGLPIAA ASFAGGVVGA LSVSLVSKDI |
|
| 101 | LLAVVPVLLI FVALYFVFSP KLDGSKEGKA RMSFFLFGLT VATAFGFLRR |
|
| 151 | CVRTGCRLVF SDCLYCFARL QAVERDVLHQ IGERCLQSWF AIGIPAARFD |
|
| 201 | YFPDCGNDGG RCVCRCEFRC EICRPLRFEA D* |
[1021]Further work revealed the following gonococcal DNA sequence <SEQ ID 439>:
[0000]| 1 | ATGGAAGATT TATACATAAT ACTCGCTTTG GGTTTGGTTG CGATGATCGC | |
|
| 51 | CGGATTTATC GATGCGATTG CGGGCGGGGG TGGTTTGATT ACGCTGCCTG |
|
| 101 | CACTCTTGTT GGCAGGTATT CCTCCCGTGT CGGCAATTGC CACCAACAAG |
|
| 151 | CTGCAAGCAG CCGCTGCTAC GTTTTCGGCT ACGGTTTCTT TTGCACGCAA |
|
| 201 | AGGTTTGATT GATTGGAAGA AAGGTCTCCC GATTGCCGCA GCATCGTTTG |
|
| 251 | CAGGCGGCGT GGTCGGTGCA TTATCGGTCA GCTTGGTTTC CAAAGATATT |
|
| 301 | TTGCTGGCGG TCGTGCCGGT TTTGTTGATA TTTGTCGCGC TGTATTTTGT |
|
| 351 | GTTTTCGCCC AAGCTCGACG GCAGTAAGGA AGGCAAAGCC AGAATGTCTT |
|
| 401 | TTTTTCTATT CGGGCTGACG GTTGCACCGC TTTTGGGTTT TTACGACGGT |
|
| 451 | GTGTTCGGAC CGGGTGTCGG CTCGTTTTTT CTGATTGCCT TTATTGTTTT |
|
| 501 | GCTCGGCTGC AAGCTGTTGA ACGCGATGTC TTACACCAAA TTGGCGAACG |
|
| 551 | TTGCTTGCAA TCTTGGTTCG CTATCGGTAT TCCTGCTGCA CGGTTCGATT |
|
| 601 | ATTTTCCCGA TTGTGGCAAC GATGGCGGTC GGTGCGTTTG TCGGTGCGAA |
|
| 651 | TTTAGGTGCG AGATTTGCCG TCCGCTTCGG TTCGAAGCTG ATTAAGCCGC |
|
| 701 | TGCTGATTGT CATCAGCATT TCGATGGCTG TGAAATTGTT GATAGACGAG |
|
| 751 | AGAAATCCGC TGTATCAGAT GATTGTTTCG ATGTTTTAA |
[1022]This corresponds to the amino acid sequence <SEQ ID 440; ORF109ng-1>:
[0000]| 1 | MEDLYIILAL GLVAMIAGFI DAIAGGGGLI TLPALLLAGI PPVSAIATNK | |
|
| 51 | LQAAAATFSA TVSFARKGLI DWKKGLPIAA ASFAGGVVGA LSVSLVSKDI |
|
| 101 | LLAVVPVLLI FVALYFVFSP KLDGSKEGKA RMSFFLFGLT VAPLLGFYDG |
|
| 151 | VFGPGVGSFF LIAFIVLLGC KLLNAMSYTK LANVACNLGS LSVFLLHGSI |
|
| 201 | IFPIVATMAV GAFVGANLGA RFAVRFGSKL IKPLLIVISI SMAVKLLIDE |
|
| 251 | RNPLYQMIVS MF* |
[1023]ORF109ng-1 and ORF109-1 show 98.9% identity in 262 aa overlap:
[0000]
[1024]In addition, ORF109ng-1 shows homology to a hypothetical Pseudomonas protein:
[0000]| sp|P29942|YCB9_PSEDE HYPOTHETICAL 27.4 KD PROTEIN IN COBO 3′REGION (ORF9) | |
| >gi|94984|pir||I38164 hypothetical protein 9 - Pseudomonas sp >gi|551929 |
| (M62866) ORF9 [Pseudomonas denitrificans] Length = 261 |
| Score = 175 bits (439), Expect = 3e−43 |
| Identities = 83/214 (38%), Positives = 131/214 (60%), Gaps = 1/214 (0%) |
|
| Query: | 41 | PPVSAIATNKLQXXXXXXXXXXXXXRKGLIDWKKGLPIXXXXXXXXXXXXXXXXXXXKDI | 100 | |
| | PP+ + TNKLQ R+G ++ K+ LP+ D+ |
| Sbjct: | 43 | PPLQTLGTNKLQGLFGSGSATLSYARRGHVNLKEQLPMALMSAAGAVLGALLATIVPGDV | 102 |
|
| Query: | 101 | LLAVVPVLLIFVALYFVFSPKLDGSKEGKARMSFFLFGLTVAPLLGFYDGVFGPGVGSFF | 160 |
| | L A++P LLI +ALYF P + G + +R++ F+F LT+ PL+GFYDGVFGPG GSFF |
| Sbjct: | 103 | LKAILPFLLIAIALYFGLKPNM-GDVDQHSRVTPFVFTLTLVPLIGFYDGVFGPGTGSFF | 161 |
|
| Query: | 161 | LIAFIVLLGCKLLNAMSYTKLANVACNLGSLSVFLLHGSIIFPIVATMAVGAFVGANLGA | 220 |
| | ++ F+ L G +L A ++TK N N+G+ VFL G++++ + M +G F+GA +G+ |
| Sbjct: | 162 | MLGFVTLAGFGVLKATAHTKFLNFGSNVGAFGVFLFFGAVLWKVGLLMGLGQFLGAQVGS | 221 |
|
| Query: | 221 | RFAVRFGSKLIKPLLIVISISMAVKLLIDERNPL | 254 |
| | R+A+ G+K+IKPLL+++SI++A++LL D +PL |
| Sbjct: | 222 | RYAMAKGAKIIKPLLVIVSIALAIRLLADPTHPL | 255 |
[1025]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 52
[1026]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 441>:
[0000]| 1 | ..CTGCTAGGGT ATTGCATCGG TTATCGGTAC GGCTGTTGCA GCAAAACCAG | |
|
| 51 | CCGCAGACGG ATTATTTGGT CAAATTCGGA TCGTTTTGGG CGAG.ATTTT |
|
| 101 | TGGTTTTCTG GGACTGTATG ACGTCTATGC TTCGGCATGG TTTGTCGTTA |
|
| 151 | TCATGATGTT TTTGGTGGTT TCTACCAGTT TGTGCCTGAT TCGCAATGTG |
|
| 201 | CCGCCGTTCT GGCGCGAAAT GAAGTCTTTT CGGGAAAAGG TTAAAGAAAA |
|
| 251 | ATCTCTGGCG GCGATGCGCC ATTCTTCGCT GTTGGATGTA AAAATTGCGC |
|
| 301 | CCGAGGTTGC CAAACGTTAT CTGGAAGTAC AAGGTTTTCA GGGGAAAACC |
|
| 351 | ATTAACCGTG AAGACGGGTC GGTTCTGATT GCCGCCAAAA AAGGCACAAT |
|
| 401 | GAACAAATGG GGCTATATCT TTGCCCATGT TGCTTTGATT GTCATTTGCC |
|
| 451 | TGGGCGGGTT GATAGACAGT AACCTGCTGT TGAAACTGGG TATGCTGACC |
|
| 501 | GGTCGGATTG TTCCGGACAA TCAGGCGGTT TATGCCAAGG ATTTC.AAGC |
|
| 551 | CCGAAAGTAT .TTTGGGTGC gTCCAATCTC TCATTTAGGG GCAACGTCAA |
|
| 601 | TATTTCCG.A GGGGCAGAgT GCGGATGTGG TTTTCCTGA |
[1027]This corresponds to the amino acid sequence <SEQ ID 442; ORF110>:
[0000]| 1 | ..LLGIASVIGT LLQQNQPQTD YLVKFGSFWA XIFGFLGLYD VYASAWFVVI | |
|
| 51 | MMFLVVSTSL CLIRNVPPFW REMKSFREKV KEKSLAAMRH SSLLDVKIAP |
|
| 101 | EVAKRYLEVQ GFQGKTINRE DGSVLIAAKK GTMNKWGYIF AHVALIVICL |
|
| 151 | GGLIDSNLLL KLGMLTGRIF RTIRRFMPRI XKPESXFGCV QSLI*GQRQY |
|
| 201 | FXRGRVRMWF S* |
[1028]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with ORF88a from N. meningitidis (Strain A)
[1029]ORF110 shows 91.5% identity over a 188aa overlap with ORF88a from strain A of N. meningitidis:
[0000]
[1030]However, ORF88 and ORF110 do not align, because they represent two different fragments of the same protein.
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1031]ORF110 shows 88.6% identity over a 21 laa overlap with a predicted ORF (ORF110.ng) from N. gonorrhoeae:
[0000]
[1032]The complete length ORF110ng nucleotide sequence <SEQ ID 443> is predicted to encode a protein having amino acid sequence <SEQ ID 444>:
[0000]| 1 | MSKSRISPTL LSRPWFAFFS SMRFAVALLS LLGIASVIGT VLQQNQPQTD | |
|
| 51 | YLVKFGPFWT RIFDFLGLYD VYASAWFVVI MMFLVVSTSL CLIRNVPPFW |
|
| 101 | REMKSFREKV KEKSLAAMRH SSLLDVKIAP EVAKRYLEVR GFQGKTVSRE |
|
| 151 | DGSVLIAAKK GTMNKWGYIX AHVALIVICL GRLINXNLLL KLGMLAGSIF |
|
| 201 | RNNRRVMPRI SKPESIWGGV QSLIKGQRQY FQRGKVRMWF S* |
[1033]Based on the putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 53
[1034]The following DNA sequence was identified in N. meningitidis <SEQ ID 445>:
[0000]| 1 | ATGCCGTCTG AAACACGCCT GCCGAACTTT ATCCGCGTCT TGATATTTGC | |
|
| 51 | CCTGGGTTTC ATCTTCCTGA ACGCCTGTTC GGAACAAACC GCGCAAACCG |
|
| 101 | TTACCCTGCA AGGCGAAACG ATGGGCACGA CCTATACCGT CAAATACCTT |
|
| 151 | TCAAATAATC GGGACAAACT CCCCTCACCT GCCGAAATAC AAAAACGCAT |
|
| 201 | CGATGACGCG CTTAAAGAAG TCAACCGGCA GATGTCCACC TATCAGCCCG |
|
| 251 | ACTCCGAAAT CAGCCGGTTC AACCAACACA CAGCCGGCAA GCCCCTCCGC |
|
| 301 | ATTTCAAGCG ACTTCGCACA CGTTACTGCC GAAGCCGTCC GCCTGAACCG |
|
| 351 | CCTGACACAC GGCGCGCTGG ACGTAACCGT CGGCCCCTTG GTCAACCTTT |
|
| 401 | GGGGATTCGG CCCCGACAAA TCCGTTACCC GTGAACCGTC GCCGGAACAA |
|
| 451 | ATCAAACAGG CGGCATCTTA TACGGGCATA GACAAAATCA TTTTGAAACA |
|
| 501 | AGGCAAAGAT TACGCTTCCT TGAGCAAAAC CCACCCCAAG GCCTATTTGG |
|
| 551 | ATTTATCTTC GATTGCCAAA GGCTTCGGCG TTGATAAAGT TGCGGGCGAA |
|
| 601 | CTGGAAAAAT ACGGCATTCA AAATTATCTG GTCGAAATCG GCGGCGAGTT |
|
| 651 | GCACGGCAAA GGCAAAAACG CGCGCGGCGA ACCGTGGCGC ATCGGTATCG |
|
| 701 | AGCAGCCCAA TATCGTCCAA GGCGGCAATA CGCAGATTAT CGTCCCGCTG |
|
| 751 | AACAACCGTT CGCTTGCCAC TTCCGGCGAT TACCGTATTT TCCACGTCGA |
|
| 801 | TAAAAACGGC AAACGCCTCT CCCATATCAT CAACCCGAAC AACAAACGAC |
|
| 851 | CCATCAGCCA CAACCTCGCC TCCATCAGCG TGGTCGCAGA CAGTGCGATG |
|
| 901 | ACGGCGGACG GCTTGTCCAC AGGATTATTC GTATTGGGCG AAACCGAAGC |
|
| 951 | CTTAAAGCTG GCAGAGCGCG AAAAACTCGC TGTTTTCCTG ATTGTCAGGG |
|
| 1001 | ATAAAGGCGG CTACCGCACC GCCATGTCTT CCGAATTTGA AAAACTGCTC |
|
| 1051 | CGCTAA |
[1035]This corresponds to the amino acid sequence <SEQ ID 446; ORF111>:
[0000]| 1 | MPSETRLPNF IRVLIFALGF IFLNACSEQT AQTVTLQGET MGTTYTVKYL | |
|
| 51 | SNNRDKLPSP AEIQKRIDDA LKEVNRQMST YQPDSEISRF NQHTAGKPLR |
|
| 101 | ISSDFAHVTA EAVRLNRLTH GALDVTVGPL VNLWGFGPDK SVTREPSPEQ |
|
| 151 | IKQAASYTGI DKIILKQGKD YASLSKTHPK AYLDLSSIAK GFGVDKVAGE |
|
| 201 | LEKYGIQNYL VEIGGELHGK GKNARGEPWR IGIEQPNIVQ GGNTQIIVPL |
|
| 251 | NNRSLATSGD YRIFHVDKNG KRLSHIINPN NKRPISHNLA SISVVADSAM |
|
| 301 | TADGLSTGLF VLGETEALKL AEREKLAVFL IVRDKGGYRT AMSSEFEKLL |
|
| 351 | R* |
[1036]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1037]ORF111 shows 96.9% identity over a 351 aa overlap with an ORF (ORF111a) from strain A of N. meningitidis:
[0000]
[1038]The complete length ORF111a nucleotide sequence <SEQ ID 447> is:
[0000]| 1 | ATGCCGTCTG AAACACGCCT GCCGAACTTT ATCCGCACCT TGATATTTGC | |
|
| 51 | CCTGAGTTTT ATCTTCCTGA ACGCCTGTTC GGAACAAACC GCGCAAACCG |
|
| 101 | TTACCCTGCA AGGTGAAACG ATGGGCACGA CCTATACCGT CAAATACCTT |
|
| 151 | TCAAATAATC GGGACNAACT CCCNTCACCT GCCGAAATAC AAAANCGCAT |
|
| 201 | CGATGACGCG CTTAAAGAAG TCAACCGGCA GATGTCCACC TATCAGCCCG |
|
| 251 | ACTCCGAAAT CAGCCGGTTC AACCAACACA CAGCCGGCAA GCCCCTCCGC |
|
| 301 | ATTTCAAGCG ACTTCGCACA CGTTACTGCC GAAGCCGTCC ACCTGAACCG |
|
| 351 | CCTGACACAC GGCGCGCTGG ACGTAACCGT CGGCCCCTTG GTCAACCTTT |
|
| 401 | GGGGATTCGG CCCCGACAAA TCCGTTACCC GTGAACCGTC GCCGGAACAA |
|
| 451 | ATCAAACAAG CAGCATCTTA TACGGGCATA GACAAAATCA TTTTGAAACA |
|
| 501 | AGGCAAAGAT TACGCTTCCT TGAGCAAAAC CCACCCCAAG GCCTATTTGG |
|
| 551 | ATTTATCTTC GATTGCCAAA GGCTTCGGCG TTGATNANGT TGCGGGCGAA |
|
| 601 | CTGGAAAAAT ACGGCATTCA AAATTATCTG GTCGAAATCG GCGGNGAGTT |
|
| 651 | GCACGGCAAA GNCAAAAACG CGCGCGGCGA ACCTTGGCGC ATCGGCATCG |
|
| 701 | AACAGCCCAA CATCGTCCAA GGCGGCAATA CGCAGATTAT CGTCCCGCTG |
|
| 751 | AACAACCGTT CGNTTGCCAC TTCCGGCGAT TACCGTATTT TCCACGTCGA |
|
| 801 | TAAAAGCGGC AAACGCCTCT CCCATATCAT TAATCCGAAC AACAAACGAC |
|
| 851 | CCATCAGCCA CAACCTCGCC TCCATCAGCG TGNTCGCAGA CAGTGCGATG |
|
| 901 | ACGGCGGACG GCTTNTCCAC AGGATTATTC GTATTGGGCG AAACCGAAGC |
|
| 951 | CTTAAAGCTG GCAGAGCGCG AAAAACTCGC TGTTTTCCTG ATTGTCAGGG |
|
| 1001 | ATAAAGGCGG CTACCGCACC GCCATGTCTT CCGAATTTGA AAAACTGCTC |
|
| 1051 | CGCTAA |
[1039]This encodes a protein having amino acid sequence <SEQ ID 448>:
[0000]| 1 | MPSETRLPNF IRTLIFALSF IFLNACSEQT AQTVTLQGET MGTTYTVKYL | |
|
| 51 | SNNRDXLPSP AEIQXRIDDA LKEVNRQMST YQPDSEISRF NQHTAGKPLR |
|
| 101 | ISSDFAHVTA EAVHLNRLTH GALDVTVGPL VNLWGFGPDK SVTREPSPEQ |
|
| 151 | IKQAASYTGI DKIILKQGKD YASLSKTHPK AYLDLSSIAK GFGVDXVAGE |
|
| 201 | LEKYGIQNYL VEIGGELHGK XKNARGEPWR IGIEQPNIVQ GGNTQIIVPL |
|
| 251 | NNRSXATSGD YRIFHVDKSG KRLSHIINPN NKRPISHNLA SISVXADSAM |
|
| 301 | TADGXSTGLF VLGETEALKL AEREKLAVFL IVRDKGGYRT AMSSEFEKLL |
|
| 351 | R* |
Homology with a Predicted ORF from
N. gonorrhoeae [1040]ORF111 shows 96.6% identity over a 351aa overlap with a predicted ORF (ORF111.ng) from N. gonorrhoeae.
[0000]
[1041]The complete length ORF111ng nucleotide sequence <SEQ ID 449> is:
[0000]| 1 | ATGCCGTCTG AAACACGCCT GCCGAACCTT ATCCGCGCCT TGATATTTGC | |
|
| 51 | CCTGGGTTTC ATCTTCCTGA ACGCCTGTTC GGaacaaacC GCGCAaaccg |
|
| 101 | TTACCCTGCA AGGCGAAAcg aTGGGTACGA CCTATACCGT CAAATACCTT |
|
| 151 | TCAAATAATC GGGACAAACT CCCCTCCCCT GCCAAAATAC AAAAGCGCAT |
|
| 201 | TGATGATGCG CTTAAAGAAG TCAACCGGCA GATGTCCACC TACCAGACCG |
|
| 251 | ATTCCGAAAT CAGCCGGTTC AACCAACACA CAGCCGGCAA GCCCCTCCGC |
|
| 301 | ATTTCAAGCG ATTTCGCACA CGTTACCGCC GAAGCCGTCC GCCTGAACCG |
|
| 351 | CCTGACTCAC GGCGCACTGG ACGTAACCGT CGGCCCTTTG GTCAACCTTT |
|
| 401 | GGGGGTTCGG CCCCGACAAA TCCGTTACCC GTGAACCGTC GCCGGAACAA |
|
| 451 | ATCAAACAGG CGGCATCTTA TACGGGCATA GACAAAATCA TTTTGCAACA |
|
| 501 | AGGCAAAGAT TACGCTTCCT TGAGCAAAAC CCACCCCAAA GCCTATTTGG |
|
| 551 | ATTTATCTTC GATTGCCAAA GGCTTCGGCG TTGATAAAGT TGCGGGCGAA |
|
| 601 | CTGGAAAAAT ACGGCATTCA AAATTATCTG GTCGAAAtcg gcggcGAGTT |
|
| 651 | GCACGGCAAA GGCAAAAATG CGCACGGCGA ACCGTGGCGC ATCGGTATAG |
|
| 701 | AGCAACCCAA TATCATCCAA GgcgGCAata CGCAGATTAt cgtcccgctg |
|
| 751 | aaCaaccgtt cgctTGCCAC TTCCGGCGAT TAccgtaTTT tccacgtcgA |
|
| 801 | TAAAAAcggc aaacgccttt cccacaTCAT CAATCCCaAC aacAAACgac |
|
| 851 | ccATCAGcca caacctcgcc tccatcagcg tggtctcAGA CAGTGCAATG |
|
| 901 | ACGGCGGACG GTTtatCCAC AGGATTATTT GTTTTAGGCG AAACCGAAGC |
|
| 951 | CTTAAGGCTG GCAGAACAAG AAAAACTCGC TGTTTTCCTA ATTGTCCGGG |
|
| 1001 | ATAAGGACGG CTACCGCACC GCCATGTCTT CCGAATTTGC CAAGCTGCTC |
|
| 1051 | CGCTAA |
[1042]This encodes a protein having amino acid sequence <SEQ ID 450>:
[0000]| 1 | MPSETRLPNL IRALIFALGF IFLNACSEQT AQTVTLQGET MGTTYTVKYL | |
|
| 51 | SNNRDKLPSP AKIQKRIDDA LKEVNRQMST YQTDSEISRF NQHTAGKPLR |
|
| 101 | ISSDFAHVTA EAVRLNRLTH GALDVTVGPL VNLWGFGPDK SVTREPSPEQ |
|
| 151 | IKQAASYTGI DKIILQQGKD YASLSKTHPK AYLDLSSIAK GFGVDKVAGE |
|
| 201 | LEKYGIQNYL VEIGGELHGK GKNAHGEPWR IGIEQPNIIQ GGNTQIIVPL |
|
| 251 | NNRSLATSGD YRIFHVDKNG KRLSHIINPN NKRPISHNLA SISVVSDSAM |
|
| 301 | TADGLSTGLF VLGETEALRL AEQEKLAVFL IVRDKDGYRT AMSSEFAKLL |
|
| 351 | R* |
[1043]This protein shows homology with a hypothetical lipoprotein precursor from H. influenzae:
[0000]| sp|P44550|YOJL_HAEIN HYPOTHETICAL LIPOPROTEIN | |
| HI0172 PRECURSOR >gi|1074292|pir|4 |
| hypothetical protein HI0172 - Haemophilus influenzae (strain Rd KW20) |
| >gi|1573128 (U32702) hypothetical [Haemophilus influenzae] |
| Length = 346 |
| Score = 353 bits (896), Expect = 9e−97 |
| Identities = 181/344 (52%), Positives = 247/344 (71%), |
| Gaps = 4/344 (1%) |
|
| Query: | 7 | LPNLIRALIFALGFIFLNACSEQTAQTVTLQGETMGTTYTVKYLSNNRDKLPSPAKIQKR | 66 | |
| | + LI +I + L AC ++T + ++L G+TMGTTY VKYL + S K + |
| Sbjct: | 1 | MKKLISGIIAVAMALSLAACQKET-KVISLSGKTMGTTYHVKYLDDGSITATSE-KTHEE | 58 |
|
| Query: | 67 | IDDALKEVNRQMSTYQTDSEISRFNQHT-AGKPLRISSDFAHVTAEAVRLNRLTHGALDV | 125 |
| | I+ LK+VN +MSTY+ DSE+SRFNQ+T P+ IS+DFA V AEA+RLN++T GALDV |
| Sbjct: | 59 | IEAILKDVNAKMSTYKKDSELSRFNQNTQVNTPIEISADFAKVLAEAIRLNKVTEGALDV | 118 |
|
| Query: | 126 | TVGPLVNLWGFGPDKSVTREPSPEQIKQAASYTGIDKIILQQGKDYASLSKTHPKAYLDL | 185 |
| | TVGP+VNLWGFGP+K ++P+PEQ+ + ++ GIDKI L K+ A+LSK P+ Y+DL |
| Sbjct: | 119 | TVGPVVNLWGFGPEKRPEKQPTPEQLAERQAWVGIDKITLDTNKEKATLSKALPQVYVDL | 178 |
|
| Query: | 186 | SSIAKGFGVDKVAGELEKYGIQNYLVEIGGELHGKGKNAHGEPWRIGIEQPNIIQGGNTQ | 245 |
| | SSIAKGFGVD+VA +LE+ QNY+VEIGGE+ KGKN G+PW+I IE+P + |
| Sbjct: | 179 | SSIAKGFGVDQVAEKLEQLNAQNYMVEIGGEIRAKGKNIEGKPWQIAIEKPTTTGERAVE | 238 |
|
| Query: | 246 | IIVPLNNRSLATSGDYRIFHVDKNGKRLSHIINPNNKRPISHNLASISVVSDSAMTADGL | 305 |
| | ++ LNN +A+SGDYRI+ ++NGKR +H I+P PI H+LASI+V++ ++MTADGL |
| Sbjct: | 239 | AVIGLNNMGMASSGDYRIY-FEENGKRFAHEIDPKTGYPIQHHLASITVLAPTSMTADGL | 297 |
|
| Query: | 306 | STGLFVLGETEALRLAEQEKLAVFLIVRDKDGYRTAMSSEFAKL | 349 |
| | STGLFVLGE +AL +AE+ LAV+LI+R +G+ T SS F KL |
| Sbjct: | 298 | STGLFVLGEDKALEVAEKNNLAVYLIIRTDNGFVTKSSSAFKKL | 341 |
[1044]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 54
[1045]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 451>:
[0000]| 1 | ..CCGTGCCGCC GACAGGGCGA CGACGTGTAT GCGGCGCACG CGTCCCGTCA | |
|
| 51 | AAAATTGTGG CTGCGCTTCA TCGGCGGCCG GTCGCATCAA AATATACGGG |
|
| 101 | GCGGCGCGGC TGCGGACGGG TGGCGCAAAG GCGTGCAAAT CGGCGGCGAG |
|
| 151 | GTGTTTGTAC GGCAAAATGA AGGCAGCCkA yTGGCAATCG GCGTGATGGG |
|
| 201 | CGGCAGGGCC GGCCAGCACG CwTCAGTCAA CGGCAAAGGC GGTGCGGCAG |
|
| 251 | gCAGTGATTT GTATGGTTAT GgCGGGGgTG TTTATGCTgC GTGGCATCAG |
|
| 301 | TTGCGCGATA AACAAACGGG TgCGTATTTG GACGGCTGGT TGCAATACCA |
|
| 351 | ACGTTTCAAA CACCGCATCA ATGATGAAAA CCGTGCGGAA CgCTACAAAA |
|
| 401 | CCAAAGGTTG GACGGCTTCT GTCGAAGGCG GCTACAACGC GCTTGTGGCG |
|
| 451 | GAAGGCATTG TCGGAAAAGG CAATAATGTG CGGTTTTACC TACAACCGCA |
|
| 501 | GgCGCAGTTT ACCTACTTGG GCGTAAACGG CGGCTTTACC GACAGCGAGG |
|
| 551 | GGACGGCGGT CGGACTGCTC GGCAGCGGTC AGTGGCAAAG CCGCGCCGGC |
|
| 601 | AtTCGGGCAA AAACCCGTTT TGCTTTGCGT AACGGTGTCA ATCTTCAGCC |
|
| 651 | TTTTGCCGCT TTTAATGTtt TGCACAGGTC AAAATCTTTC GGCGTGGAAA |
|
| 701 | TGGACGGCGA AAAACAGACG CTGGCAGGCA GGACGGCACT CGAAGGGCGG |
|
| 751 | TTCGGTATTG AAGCCGGTTG GAAAGGCCAT ATGTCCGCA.. |
[1046]This corresponds to the amino acid sequence <SEQ ID 452; ORF35>:
[0000]| 1 | ..PCRRQGDDVY AAHASRQKLW LRFIGGRSHQ NIRGGAAADG WRKGVQIGGE | |
|
| 51 | VFVRQNEGSX LAIGVMGGRA GQHASVNGKG GAAGSDLYGY GGGVYAAWHQ |
|
| 101 | LRDKQTGAYL DGWLQYQRFK HRINDENRAE RYKTKGWTAS VEGGYNALVA |
|
| 151 | EGIVGKGNNV RFYLQPQAQF TYLGVNGGFT DSEGTAVGLL GSGQWQSRAG |
|
| 201 | IRAKTRFALR NGVNLQPFAA FNVLHRSKSF GVEMDGEKQT LAGRTALEGR |
|
| 251 | FGIEAGWKGH MSA.. |
[1047]Computer analysis of this amino acid sequence gave the following results:
[1048]Homology with Putative Secreted VirG-Homologue of N. meningitidis (Accession Number A32247)
[1049]ORF and virg-h protein show 51% aa identity in 261aa overlap:
[0000]| Orf35 | 5 | QGDDVYAAHASRQKLWLRFIGGRSHQNIRGGAA-ADGWRKGVQIGGEVFVRQNEGSXLAI | 63 | |
| | + D++ R+ LWLR I G S+Q ++G A +G+RKGVQ+GGEVF QNE + L+I |
| virg-h | 396 | KNSDIFDRTLPRKGLWLRVIDGHSNQWVQGKTAPVEGYRKGVQLGGEVFTWQNESNQLSI | 455 |
|
| Orf35 | 64 | GVMGGRAGQHASVNGKG--GAAGSDLYGYGGGVYAAWHQLRDKQTGAYLDGWLQYQRFKH | 121 |
| | G+MGG+A Q ++ + ++ G+G GVYA WHQL+DKQTGAY D W+QYQRF+H |
| virg-h | 456 | GLMGGQAEQRSTFHNPDTDNLTTGNVKGFGAGVYATWHQLQDKQTGAYADSWMQYQRFRH | 515 |
|
| Orf35 | 122 | RINDENRAERYKTKGWTASVEGGYNALVAEGIVGKGNNVRFYLQPQAQFTYLGVNGGFTD | 181 |
| | RIN E+ ER+ +KG TAS+E GYNAL+AE KGN++R YLQPQAQ TYLGVNG F+D |
| virg-h | 516 | RINTEDGTERFTSKGITASIEAGYNALLAEHFTKKGNSLRVYLQPQAQLTYLGVNGKFSD | 575 |
|
| Orf35 | 182 | SEGTAVGLLGSGQWQSRAGIRAKTRFALRNGVNLQPFAAFNVLHRSKSFGVEMDGEKQTL | 241 |
| | SE V LLGS Q Q+R G++AK +F+L + ++PFAA N L+ +K FGVEMDGE++ + |
| virg-h | 576 | SENAHVNLLGSRQLQTRVGVQAKAQFSLYKNIAIEPFAAVNALYHNKPFGVEMDGERRVI | 635 |
|
| Orf35 | 242 | AGRTALEGRFGIEAGWKGHMS | 262 |
| | +TA+E + G+ K H++ |
| virg-h | 636 | NNKTAIESQLGVAVKIKSHLT | 656 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1050]ORF35 shows 96.9% identity over a 259aa overlap with an ORF (ORF35a) from strain A of N. meningitidis.
[0000] | 10 20 30 | |
| orf35.pep | PCRRQGDDVYAAHASRQKLWLRFIGGRSHQNIRG |
| :||||||| |||||||||||||||||||| |
| orf35a | QRLAIPEAEAVLYAQQAYAANTLFGLRAADRGDDVYAADPSRQKLWLRFIGGRSHQNIRG |
| 310 320 330 340 350 360 |
|
| 40 50 60 70 80 90 |
| orf35.pep | GAAADGWRKGVQIGGEVFVRQNEGSXLAIGVMGGRAGQHASVNGKGGAAGSDLYGYGGGV |
| |||||| |||||||||||||||||| ||||||||||||||||||||||||| |:|||||| |
| orf35a | GAAADGRRKGVQIGGEVFVRQNEGSRLAIGVMGGRAGQHASVNGKGGAAGSYLHGYGGGV |
| 370 380 390 400 410 420 |
|
| 100 110 120 130 140 150 |
| orf35.pep | YAAWHQLRDKQTGAYLDGWLQYQRFKHRINDENRAERYKTKGWTASVEGGYNALVAEGIV |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||:| |
| orf35a | YAAWHQLRDKQTGAYLDGWLQYQRFKHRINDENRAERYKTKGWTASVEGGYNALVAEGVV |
| 430 440 450 460 470 480 |
|
| 160 170 180 190 200 210 |
| orf35.pep | GKGNNVRFYLQPQAQFTYLGVNGGFTDSEGTAVGLLGSGQWQSRAGIRAKTRFALRNGVN |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |
| orf35a | GKGNNVRFYLQPQAQFTYLGVNGGFTDSEGTAVGLLGSGQWQSRAGIRAKTRFALRNGVN |
| 490 500 510 520 530 540 |
|
| 220 230 240 250 260 |
| orf35.pep | LQPFAAFNVLHRSKSFGVEMDGEKQTLAGRTALEGRFGIEAGWKGHMSA |
| ||||||||||||||||||||||||||||||||||||||||||||||||| |
| orf35a | LQPFAAFNVLHRSKSFGVEMDGEKQTLAGRTALEGRFGIEAGWKGHMSARIGYGKRTDGD |
| 550 560 570 580 590 600 |
|
| orf35a | KEAALSLKWLFX |
| 610 620 |
[1051]The complete length ORF35a nucleotide sequence <SEQ ID 453> is:
[0000]| 1 | ATGTTCAGAG CTCAGCTTGG TTCAAATACT CGTTCTACCA AAATCGGCGA | |
|
| 51 | CGATGCCGAT TTTTCATTTT CAGACAAGCC GAAACCCGGC ACTTCCCATT |
|
| 101 | ATTTTTCCAG CGGTAAAACC GATCAAAATT CATCCGAATA TGGGTATGAC |
|
| 151 | GAAATCAATA TCCAAGGTAA AAACTACAAT AGCGGCATAC TCGCCGTCGA |
|
| 201 | TAATATGCCC GTTGTTAAGA AATATATTAC AGATACTTAC GGGGATAATT |
|
| 251 | TAAAGGATGC GGTTAAGAAG CAATTACAGG ATTTATACAA AACAAGACCC |
|
| 301 | GAAGCTTGGG AAGAAAATAA AAAACGGACT GAGGAGGCGT ATATAGAACA |
|
| 351 | GCTTGGACCA AAATTTAGTA TACTCAAACA GAAAAACCCC GATTTAATTA |
|
| 401 | ATAAATTGGT AGAAGATTCC GTACTCACTC CTCATAGTAA TACATCACAG |
|
| 451 | ACTAGTCTCA ACAACATCTT CAATAAAAAA TTACACGTCA AAATCGAAAA |
|
| 501 | CAAATCCCAC GTCGCCGGAC AGGTGTTGGA ACTGACCAAG ATGACGCTGA |
|
| 551 | AAGATTCCCT TTGGGAACCG CGCCGCCATT CCGACATCCA TATGCTGGAA |
|
| 601 | ACTTCCGATA ATGCCCGCAT CCGCCTGAAC ACGAAAGATG AAAAACTGAC |
|
| 651 | CGTCCATAAA GCGTATCAGG GCGGTGCGGA TTTCCTGTTC GGCTACGACG |
|
| 701 | TGCGGGAGTC GGACAAACCC GCCCTGACCT TTGAAGAAAA AGTCAGCGGA |
|
| 751 | CAATCCGGCG TGGTTTTGGA ACGCCGGCCG GAAAATCTGA AAACGCTCGA |
|
| 801 | CGGGCGCAAA CTGATTGCGG CGGAAAAGGC AGACTCTAAT TCGTTTGCGT |
|
| 851 | TTAAACAAAA TTACCGGCAG GGACTGTACG AATTATTGCT CAAGCAATGC |
|
| 901 | GAAGGCGGAT TTTGCTTGGG CGTGCAGCGT TTGGCTATCC CCGAGGCGGA |
|
| 951 | AGCGGTTTTA TATGCCCAAC AGGCTTATGC GGCAAATACT TTGTTCGGGC |
|
| 1001 | TGCGTGCCGC CGACAGGGGC GACGACGTGT ATGCCGCCGA TCCGTCCCGT |
|
| 1051 | CAAAAATTGT GGCTGCGCTT CATCGGCGGC CGGTCGCATC AAAATATACG |
|
| 1101 | GGGCGGCGCG GCTGCGGACG GGCGGCGCAA AGGCGTGCAA ATCGGCGGCG |
|
| 1151 | AGGTGTTTGT ACGGCAAAAT GAAGGCAGCC GGCTGGCAAT CGGCGTGATG |
|
| 1201 | GGCGGCAGGG CTGGCCAGCA CGCATCAGTC AACGGCAAAG GCGGTGCGGC |
|
| 1251 | AGGCAGTTAT TTGCATGGTT ATGGCGGGGG TGTTTATGCT GCGTGGCATC |
|
| 1301 | AGTTGCGCGA TAAACAAACG GGTGCGTATT TGGACGGCTG GTTGCAATAC |
|
| 1351 | CAACGTTTCA AACACCGCAT CAATGATGAA AACCGTGCGG AACGCTACAA |
|
| 1401 | AACCAAAGGT TGGACGGCTT CTGTCGAAGG CGGCTACAAC GCGCTTGTGG |
|
| 1451 | CGGAAGGCGT TGTCGGAAAA GGCAATAATG TGCGGTTTTA CCTGCAACCG |
|
| 1501 | CAGGCGCAGT TTACCTACTT GGGCGTAAAC GGCGGCTTTA CCGACAGCGA |
|
| 1551 | GGGGACGGCG GTCGGACTGC TCGGCAGCGG TCAGTGGCAA AGCCGCGCCG |
|
| 1601 | GCATTCGGGC AAAAACCCGT TTTGCTTTGC GTAACGGTGT CAATCTTCAG |
|
| 1651 | CCTTTTGCCG CTTTTAATGT TTTGCACAGG TCAAAATCTT TCGGCGTGGA |
|
| 1701 | AATGGACGGC GAAAAACAGA CGCTGGCAGG CAGGACGGCG CTCGAAGGGC |
|
| 1751 | GGTTCGGCAT TGAAGCCGGT TGGAAAGGCC ATATGTCCGC ACGCATCGGA |
|
| 1801 | TACGGCAAAA GGACGGACGG CGACAAAGAA GCCGCATTGT CGCTCAAATG |
|
| 1851 | GCTGTTTTGA |
[1052]This encodes a protein having amino acid sequence <SEQ ID 454>:
[0000]| 1 | MFRAQLGSNT RSTKIGDDAD FSFSDKPKPG TSHYFSSGKT DQNSSEYGYD | |
|
| 51 | EINIQGKNYN SGILAVDNMP VVKKYITDTY GDNLKDAVKK QLQDLYKTRP |
|
| 101 | EAWEENKKRT EEAYIEQLGP KFSILKQKNP DLINKLVEDS VLTPHSNTSQ |
|
| 151 | TSLNNIFNKK LHVKIENKSH VAGQVLELTK MTLKDSLWEP RRHSDIHMLE |
|
| 201 | TSDNARIRLN TKDEKLTVHK AYQGGADFLF GYDVRESDKP ALTFEEKVSG |
|
| 251 | QSGVVLERRP ENLKTLDGRK LIAAEKADSN SFAFKQNYRQ GLYELLLKQC |
|
| 301 | EGGFCLGVQR LAIPEAEAVL YAQQAYAANT LFGLRAADRG DDVYAADPSR |
|
| 351 | QKLWLRFIGG RSHQNIRGGA AADGRRKGVQ IGGEVFVRQN EGSRLAIGVM |
|
| 401 | GGRAGQHASV NGKGGAAGSY LHGYGGGVYA AWHQLRDKQT GAYLDGWLQY |
|
| 451 | QRFKHRINDE NRAERYKTKG WTASVEGGYN ALVAEGVVGK GNNVRFYLQP |
|
| 501 | QAQFTYLGVN GGFTDSEGTA VGLLGSGQWQ SRAGIRAKTR FALRNGVNLQ |
|
| 551 | PFAAFNVLHR SKSFGVEMDG EKQTLAGRTA LEGRFGIEAG WKGHMSARIG |
|
| 601 | YGKRTDGDKE AALSLKWLF* |
Homology with a Predicted ORF from
N. gonorrhoeae [1053]ORF35 shows 51.7% identity over a 261aa overlap with a predicted ORF (ORF35ngh) from N. gonorrhoeae.
[0000]
[1054]A partial ORF35ngh nucleotide sequence <SEQ ID 455> is predicted to encode a protein having partial amino acid sequence <SEQ ID 456>:
[0000]| 1 | ..KKLRDRNSEY WKEETYHIKS NGRTYPNIPA LFPKHPFDPF ENINNSKKIS | |
|
| 51 | FYDKEYTEDY LVGFARGFGV EKRNGEEEKP LRQYFKDCVN TENSNNDNCK |
|
| 101 | ISSFGNYGPI LIKSDIFALA SQIKNSHINS EILSVGNYIE WLRPTLNKLT |
|
| 151 | GWQEHLYAGL DPFHYIEVTD NSHVIGQTID LGALELTNSL WKPRWNSNID |
|
| 201 | YLITKNAEIR FNTKNESLLV KEDYAGGARF RFAYDLKDKV PEIPVLTFEK |
|
| 251 | NITGTSDIIF EGKALDNLKH LDGHQIVKVN DTADKDAFRL SSKYRKGIYT |
|
| 301 | LSLQQRPEGF FTKVQERDDI AIYAQQAQAA NTLFALRLND KNSDIFDRTL |
|
| 351 | PRKGLWLRVI DGHSNQWVQG KTAPVEGYRK GVQLGGEVFT WQNESNQLSI |
|
| 401 | GLMGGQAEQR STFRNPDTDN LTTGNVKGFG AGVYATWHQL QDKQTGAYVD |
|
| 451 | SWMQYQRFRH RINTEYATER FTSKGITASI EAGYNALLAE HFTKKGNSLR |
|
| 501 | VYLQPQAQLT YLGVNGKFSD SENAQVNLLG SRQLQSRVGV QAKAQFAFTN |
|
| 551 | GVTFQPFVAV NSIYQQKPFG VEIDGDRRVI NNKTVIETQL GVAAKIKSHL |
|
| 601 | TLQASFNRQT SKHHHAKQGA LNLQWTF* |
[1055]Based on this prediction, these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 55
[1056]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 457>:
[0000]| 1 | ..GCGGAATATG TTCAGTTCTC TATAGATTTG TTCAGTGTGG GTAAATCGGG | |
|
| 51 | GGGCGGTATA CCTAAGGCTA AGCCTGTGTT TGATGCGAAA CCGAGATGGG |
|
| 101 | AGGTTGATAG GAAGCTTAAT AAATTGACAA CTCGTGAGCA GGTGGAGAAA |
|
| 151 | AATGTTCAGG AAACGAGAAG AAGGAGTCAG AGTAGTCAGT TTAAAGCCCA |
|
| 201 | TGCGCAACGA GAATGGGAAA ATAAAACAGG GTTAGATTTT AATCATTTTA |
|
| 251 | TAGGTGGTGA TATCAATAAA AAAGGCACAG TAACAGGAGG GCATAGTCTA |
|
| 301 | ACCCGTGGTG ATGTACGGGT GATACAACAA ACCTCGGCAC CTGATAAACA |
|
| 351 | TGGGGT.TTA TCAAGCGACA GTGGAAATTN A |
[1057]This corresponds to the amino acid sequence <SEQ ID 458; ORF46>:
[0000]| 1 | ..AEYVQFSIDL FSVGKSGGGI PKAKPVFDAK PRWEVDRKLN KLTTREQVEK | |
|
| 51 | NVQETRRRSQ SSQFKAHAQR EWENKTGLDF NHFIGGDINK KGTVTGGHSL |
|
| 101 | TRGDVRVIQQ TSAPDKHGXL SSDSGNX |
[1058]Further work revealed further partial nucleotide sequence <SEQ ID 459>:
[0000]| 1 | ..GCAGTGTGCC TnCCGATGCA TGCACACGCC TCAnATTTGG CAAACGATTC | |
|
| 51 | TTTTATCCGG CAGGTTCTCG ACCGTCAGCA TTTCGAACCC GACGGGAAAT |
|
| 101 | ACCACCTATT CGGCAGCAGG GGGGAACTTG CCGAGCGCCA GTCTCATATC |
|
| 151 | GGATTGGGAA AAATACAAAG CCATCAGTTG GGCAACCTGA TGATTCAACA |
|
| 201 | GGCGGCCATT AAAGGAAATA TCGGCTACAT TGTCCGCTTT TCCGATCACG |
|
| 251 | GGCACGAAGT CCATTCCCCs TTCGACAACC ATGCCTCACA TTCCGATTCT |
|
| 301 | GATGAAGCCG GTAGTCCCGT TGACGGATTT AGCCTTTACC GCATCCATTG |
|
| 351 | GGACGGATAC GAACACCATC CCGCCGACGG CTATGACGGG CCACAGGGCG |
|
| 401 | GCGGCTATCC CGCTCCCAAA GGCGCGAGGG ATATATACAG TTACGACATA |
|
| 451 | AAAGGCGTTG CCCAAAATAT CCGCCTCAAC CTGACCGACA ACCGCAGCAC |
|
| 501 | CGGACAACGG CTTGCCGACC GTTTCCACAA TGCCGGTAGT ATGCTGACGC |
|
| 551 | AAGGAGTAGG CGACGGATTC AAACGCGCCA CCCGATACAG CCCCGAGCTG |
|
| 601 | GACAGATCGG GCAATGCCGC CGAAGCCTTC AACGGCACTG CAGATATCGT |
|
| 651 | TAAAAACATC ATCGGCGCTG CAGGAGAAAT TGT |
[1059]This corresponds to the amino acid sequence <SEQ ID 460; ORF46-1>:
[0000]| 1 | ..AVCLPMHAHA SXLANDSFIR QVLDRQHFEP DGKYHLFGSR GELAERQSHI | |
|
| 51 | GLGKIQSHQL GNLMIQQAAI KGNIGYIVRF SDHGHEVHSP FDNHASHSDS |
|
| 101 | DEAGSPVDGF SLYRIHWDGY EHHPADGYDG PQGGGYPAPK GARDIYSYDI |
|
| 151 | KGVAQNIRLN LTDNRSTGQR LADRFHNAGS MLTQGVGDGF KRATRYSPEL |
|
| 201 | DRSGNAAEAF NGTADIVKNI IGAAGEI |
[1060]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1061]ORF46 shows 98.2% identity over a 111 aa overlap with a predicted ORF (ORF46ng) from N. gonorrhoeae:
[0000]
[1062]A partial ORF46ng nucleotide sequence <SEQ ID 461> is predicted to encode a protein having partial amino acid sequence <SEQ ID 462>:
[0000]| 1 | ..RRLKHCCHAR LGSAFHRKQD GAHQRFGRYG ATQRLCRSSH PRLGSPKPQC | |
|
| 51 | RTRHRSRQQY LYGSHPHQRD WSCPGKIQLG RHHGTSCRAV ADXRDRICER |
|
| 101 | EIRRQRQXCR CRLGKIPSLS IPKYPLKLEQ RYGKENITSS TVPPSNGKNV |
|
| 151 | KLADQRHPKT GVPFDGKGFP NFEKHVKYDT KLDIQELSGG GIPKAKPVFD |
|
| 201 | AKPRWEVDRK LNKLTTREQV EKNVQETRRR SQSSQFKAHA QREWENKTGL |
|
| 251 | DFNHFIGGDI NKKGAVTGGH SLTRGDVRVI QQTSAPDKHG VLSSDSGN* |
[1063]Further work revealed the complete gonococcal DNA sequence <SEQ ID 463>:
[0000]| 1 | TTGGGCATTT CCCGCAAAAT ATCCCTTATT CTGTCCATAC TGGCAGTGTG | |
|
| 51 | CCTGCCGATG CATGCACACG CCTCAGATTT GGcaAACGAT CCCTTTATCC |
|
| 101 | GgCaggttcT CGaccGTCAG CATTTCGaac ccgacggGAa ATACCaCCTA |
|
| 151 | TTcggCaGCA GGGGGGAGCT TgccnagcGC aacggccATa tcggattggG |
|
| 201 | aaacaTAcaa Agccatcagt tGggccacct gatgattcaa caggcggccg |
|
| 251 | ttgaaggaaA TAtcgGctac attgtccgct tttccgatca cgggcacaaa |
|
| 301 | ttccattcgc ccttcGAcaa ccaTGCCTCA CATTCCGATT CTGACGAAGC |
|
| 351 | CGGTAGTCCC GTTGACGGAT TCAGCCTTTA CCGCATCCAT TGGGACGGAT |
|
| 401 | ACGAACACCA TCCCGCCGAC GGCTATGACG GGCCACAGGG CGGCGGCTAT |
|
| 451 | CCCGCTCCCA AAGGCGCGAG GGATATATAC AGCTACGACA TAAAAGGCGT |
|
| 501 | TGCCCAAAAT ATCCGCCTCA ACCTGACCGA CAACCGCAGC ACCGGACAAC |
|
| 551 | GGCTTGCCGA CCGTTTCCAC AATGCCGGCG CTATGCTGAC GCAAGGAGTA |
|
| 601 | GGCGACGGAT TCAAACGCGC CACCCGATAC AGCCCCGAGC TGGACAGATC |
|
| 651 | GGGCAATGCc gccGAAGCCT TCAACGGCAC TGCAGATATC GTCAAAAACA |
|
| 701 | TCATCGGCGC GGCAGGAGAA ATTGTCGGCG CAGGCGATGC CGTGCagGGT |
|
| 751 | ATAAGCGAAG GCTCAAACAT TGCTGTCATG CACGGCTTGG GTCTGCTTTC |
|
| 801 | CACCGAAAAC AAGATGGCGC GCATCAACGA TTTGGCAGAT ATGGCGCAAC |
|
| 851 | TCAAAGACTA TGCCGCAGCA GCCATCCGCG ATTGGGCAGT CCAAAACCCC |
|
| 901 | AATGCCGCAC AAGGCATAGA AGCCGTCAGC AATATCTTTA TGGCAGCCAT |
|
| 951 | CCCCATCAAA GGGATTGGAG CTGTCCGGGG AAAATACGGC TTGGGCGGCA |
|
| 1001 | TCACGGCACA TCCTGTCAAG CGGTCGCAGA TGGGCGCGAT CGCATTGCCG |
|
| 1051 | AAAGGGAAAT CCGCCGTCAG CGACAATTTT GCCGATGCGG CATACGCCAA |
|
| 1101 | ATACCCGTCC CCTTACCATT CCCGAAATAT CCGTTCAAAC TTGGAGCAGC |
|
| 1151 | GTTACGGCAA AGAAAACATC ACCTCCTCAA CCGTGCCGCC GTCAAACGGC |
|
| 1201 | AAAAATGTCA AACTGGCAGA CCAACGCCAC CCGAAGACAG GCGTACCGTT |
|
| 1251 | TGACGGTAAA GGGTTTCCGA ATTTTGAGAA GCACGTGAAA TATGATACGA |
|
| 1301 | AGCTCGATAT TCAAGAATTA TCGGGGGGCG GTATACCTAA GGCTAAGCCT |
|
| 1351 | GTGTTTGATG CGAAACCGAG ATGGGAGGTT GATAGGAAGC TTAATAAATT |
|
| 1401 | GACAACTCGT GAGCAGGTGG AGAAAAATGT TCAGGAAACG AGAAGAAGGA |
|
| 1451 | GTCAGAGTAG TCAGTTTAAA GCCCATGCGC AACGAGAATG GGAAAATAAA |
|
| 1501 | ACAGGGTTAG ATTTTAATCA TTTTATAGGT GGTGATATCA ATAAGAAAGG |
|
| 1551 | CACAGTAACA GGAGGGCATA GTCTAACCCG TGGTGATGTA CGGGTGATAC |
|
| 1601 | AACAAACCTC GGCACCTGAT AAACATGGGG TTTATCAAGC GACAGTGGAA |
|
| 1651 | ATTAAAAAGC CTGATGGAAG TTGGGAGGTG AAAACGAAAA AAGGTGGGAA |
|
| 1701 | AGTGATGACC AAGCACACCA TGTTCCCAAA AGATTGGGAT GAGGCTAGAA |
|
| 1751 | TTAGGGCTGA AGTTACTTCG GCTTGGGAAA GTAGAATAAT GCTTAAGGAT |
|
| 1801 | AATAAATGGC AGGGTACAAG TAAATCGGGT ATTAAAATAG AAGGATTTAC |
|
| 1851 | CGAACCTAAT AGAACAGCAT ATCCCATTTA TGAATAG |
[1064]This corresponds to the amino acid sequence <SEQ ID 464; ORF46ng-1>:
[0000]| 1 | LGISRKISLI LSILAVCLPM HAHASDLAND PFIRQVLDRQ HFEPDGKYHL | |
|
| 51 | FGSRGELAXR NGHIGLGNIQ SHQLGHLMIQ QAAVEGNIGY IVRFSDHGHK |
|
| 101 | FHSPFDNHAS HSDSDEAGSP VDGFSLYRIH WDGYEHHPAD GYDGPQGGGY |
|
| 151 | PAPKGARDIY SYDIKGVAQN IRLNLTDNRS TGQRLADRFH NAGAMLTQGV |
|
| 201 | GDGFKRATRY SPELDRSGNA AEAFNGTADI VKNIIGAAGE IVGAGDAVQG |
|
| 251 | ISEGSNIAVM HGLGLLSTEN KMARINDLAD MAQLKDYAAA AIRDWAVQNP |
|
| 301 | NAAQGIEAVS NIFMAAIPIK GIGAVRGKYG LGGITAHPVK RSQMGAIALP |
|
| 351 | KGKSAVSDNF ADAAYAKYPS PYHSRNIRSN LEQRYGKENI TSSTVPPSNG |
|
| 401 | KNVKLADQRH PKTGVPFDGK GFPNFEKHVK YDTKLDIQEL SGGGIPKAKP |
|
| 451 | VFDAKPRWEV DRKLNKLTTR EQVEKNVQET RRRSQSSQFK AHAQREWENK |
|
| 501 | TGLDFNHFIG GDINKKGTVT GGHSLTRGDV RVIQQTSAPD KHGVYQATVE |
|
| 551 | IKKPDGSWEV KTKKGGKVMT KHTMFPKDWD EARIRAEVTS AWESRIMLKD |
|
| 601 | NKWQGTSKSG IKIEGFTEPN RTAYPIYE* |
[1065]ORF46ng-1 and ORF46-1 show 94.7% identity in 227 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1066]ORF46ng-1 shows 87.4% identity over a 486aa overlap with an ORF (ORF46a) from strain A of N. meningitidis:
[0000]
[1067]The complete length ORF46a DNA sequence <SEQ ID 465> is:
[0000]| 1 | TTGGGCATTT CCCGCAAAAT ATCCCTTATT CTGTCCATAC TGGCAGTGTG | |
|
| 51 | CCTGCCGATG CATGCACACG CCTCAGATTT GGCAAACGAT TCTTTTATCC |
|
| 101 | GGCAGGTTCT CGACCGTCAG CATTTCGAAC CCGACGGGAA ATACCACCTA |
|
| 151 | TTCGGCAGCA GGGGGGAACT TGCCGAGCGC AGCGGTCATA TCGGATTGGG |
|
| 201 | AAACATACAA AGCCATCAGT TGGGCAACCT GTTCATCCAG CAGGCGGCCA |
|
| 251 | TTAAAGGAAA TATCGGCTAC ATTGTCCGCT TTTCCGATCA CGGGCACGAA |
|
| 301 | GTCCATTCCC CCTTCGACAA CCATGCCTCA CATTCCGATT CTGATGAAGC |
|
| 351 | CGGTAGTCCC GTTGACGGAT TCAGCCTTTA CCGCATCCAT TGGGACGGAT |
|
| 401 | ACGAACACCA TCCCGCCGAC GGCTATGACG GGCCACAGGG CGGCGGCTAT |
|
| 451 | CCCGCTCCCA AAGGCGCGAG GGATATATAC AGCTACGACA TAAAAGGCGT |
|
| 501 | TGCCCAAAAT ATCCGCCTCA ACCTGACCGA CAACCGCAGC ACCGGACAAC |
|
| 551 | GGCTTGTCGA CCGTTTCCAC AATACCGGTA GTATGCTGAC GCAAGGAGTA |
|
| 601 | GGCGACGGAT TCAAACGCGC CACCCGATAC AGCCCCGAGC TGGACAGATC |
|
| 651 | GGGCAATGCC GCCGAAGCTT TCAACGGCAC TGCAGATATC GTCAAAAACA |
|
| 701 | TCATCGGCGC GGCAGGAGAA ATTGTCGGCG CAGGCGATGC CGTGCAGGGT |
|
| 751 | ATAAGCGAAG GCTCAAACAT TGCTGTTATG CACGGCTTGG GTCTGCTTTC |
|
| 801 | CACCGAAAAC AAGATGGCGC GCATCAACGA TTTGGCAGAT ATGGCGCAAC |
|
| 851 | TCAAAGACTA TGCCGCAGCA GCCATCCGCG ATTGGGCAGT CCAAAACCCC |
|
| 901 | AATGCCGCAC AAGGCATAGA AGCCGTCAGC AATATCTTTA CGGCAGTCAT |
|
| 951 | CCCCGTCAAA GGGATTGGAG CTGTTCGGGG AAAATACGGC TTGGGCGGCA |
|
| 1001 | TCACGGCACA TCCTGTCAAG CGGTCGCAGA TGGGCGAGAT CGCATTGCCG |
|
| 1051 | AAAGGGAAAT CCGCCGTCAG CGACAATTTT GCCGATGCGG CATACGCCAA |
|
| 1101 | ATACCCGTCC CCTTACCATT CCCGAAATAT CCGTTCAAAC TTGGAGCAGC |
|
| 1151 | GTTACGGCAA AGAAAACATC ACCTCCTCAA CCGTGCCGCC GTCAAACGGA |
|
| 1201 | AAGAATGTGA AACTGGCAAA CAAACGCCAC CCGAAGACCA AAGTGCCGTT |
|
| 1251 | TGACGGTAAA GGGTTTCCGA ATTTTGAAAA AGACGTAAAA TACGATACGA |
|
| 1301 | GAATTAATAC CGCTGTACCA CAAGTGAATC CTATAGATGA ACCCGTCTTT |
|
| 1351 | AATCCTAAAG GTTCTGTCGG ATCGGCTCAT TCTTGGTCTA TAACTGCCAG |
|
| 1401 | AATTCAATAC GCAAAATTAC CAAGGCAAGG TAGAATCAGA TATATCCCAC |
|
| 1451 | CTAAAAATTA CTCTCCTTCA GCACCGCTAC CAAAAGGACC TAATAATGGA |
|
| 1501 | TATTTGGATA AATTTGGTAA TGAATGGACT AAAGGTCCAT CAAGAACTAA |
|
| 1551 | AGGTCAAGAA TTTGAATGGG ATGTTCAATT GTCTAAAACA GGAAGAGAGC |
|
| 1601 | AACTTGGATG GGCTAGTAGG GATGGTAAGC ATTTAAATAT ATCAATTGAT |
|
| 1651 | GGAAAGATTA CACACAAATG A |
[1068]This corresponds to the amino acid sequence <SEQ ID 466>:
[0000]| 1 | LGISRKISLI LSILAVCLPM HAHASDLAND SFIRQVLDRQ HFEPDGKYHL | |
|
| 51 | FGSRGELAER SGHIGLGNIQ SHQLGNLFIQ QAAIKGNIGY IVRFSDHGHE |
|
| 101 | VHSPFDNHAS HSDSDEAGSP VDGFSLYRIH WDGYEHHPAD GYDGPQGGGY |
|
| 151 | PAPKGARDIY SYDIKGVAQN IRLNLTDNRS TGQRLVDRFH NTGSMLTQGV |
|
| 201 | GDGFKRATRY SPELDRSGNA AEAFNGTADI VKNIIGAAGE IVGAGDAVQG |
|
| 251 | ISEGSNIAVM HGLGLLSTEN KMARINDLAD MAQLKDYAAA AIRDWAVQNP |
|
| 301 | NAAQGIEAVS NIFTAVIPVK GIGAVRGKYG LGGITAHPVK RSQMGEIALP |
|
| 351 | KGKSAVSDNF ADAAYAKYPS PYHSRNIRSN LEQRYGKENI TSSTVPPSNG |
|
| 401 | KNVKLANKRH PKTKVPFDGK GFPNFEKDVK YDTRINTAVP QVNPIDEPVF |
|
| 451 | NPKGSVGSAH SWSITARIQY AKLPRQGRIR YIPPKNYSPS APLPKGPNNG |
|
| 501 | YLDKFGNEWT KGPSRTKGQE FEWDVQLSKT GREQLGWASR DGKHLNISID |
|
| 551 | GKITHK* |
[1069]Based on this analysis, including the presence of a RGD sequence in the gonococcal protein, typical of adhesins, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 56
[1070]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 467>:
[0000]| 1 | ATGAATATTC ACACCCTGCT CTCCAAACAA TGGACGCTGC CGCCATTCCT | |
|
| 51 | GCCGAAACGG CTGCTGCTGT CCCTGCTGAT ACTGCTTGCC CCCAATGCGG |
|
| 101 | TGTTTTGGGT TTTGGCACTG CTGACCGCCA CCGCCCGCCC GATTGTCAAT |
|
| 151 | TTGGACTATC TTCCCGCCGC GCTGCTGATC GCCCTGCCTT GGCGTTTCGT |
|
| 201 | CAAAATTGCC GGCGTATTGG CGTTTTGGCT GGCGGTTTTG TTTGACGGGC |
|
| 251 | TGATGATGGT GATCCAACTC TTCCCTTTTA TGGATCTCAT CGGCGCCATC |
|
| 301 | AACCTCGTCC CCTTCATCCT GACCGCCCCC GCCCCTTATC AGATAATGAC |
|
| 351 | CGGGCTG... |
[1071]This corresponds to the amino acid sequence <SEQ ID 468; ORF48>:
[0000]| 1 | MNIHTLLSKQ WTLPPFLPKR LLLSLLILLA PNAVFWVLAL LTATARPIVN | |
|
| 51 | LDYLPAALLI ALPWRFVKIA GVLAFWLAVL FDGLMMVIQL FPFMDLIGAI |
|
| 101 | NLVPFILTAP APYQIMTGL... |
[1072]Further work revealed the complete nucleotide sequence <SEQ ID 469>:
[0000]| 1 | ATGAATATTC ACACCCTGCT CTCCAAACAA TGGACGCTGC CGCCATTCCT | |
|
| 51 | GCCGAAACGG CTGCTGCTGT CCCTGCTGAT ACTGCTTGCC CCCAATGCGG |
|
| 101 | TGTTTTGGGT TTTGGCACTG CTGACCGCCA CCGCCCGCCC GATTGTCAAT |
|
| 151 | TTGGACTATC TTCCCGCCGC GCTGCTGATC GCCCTGCCTT GGCGTTTCGT |
|
| 201 | CAAAATTGCC GGCGTATTGG CGTTTTGGCT GGCGGTTTTG TTTGACGGGC |
|
| 251 | TGATGATGGT GATCCAACTC TTCCCTTTTA TGGATCTCAT CGGCGCCATC |
|
| 301 | AACCTCGTCC CCTTCATCCT GACCGCCCCC GCCCCTTATC AGATAATGAC |
|
| 351 | CGGGCTGTTG CTGCTGTATA TGCTGGCGAT GCCGTTTGTG TTGCAGAAAG |
|
| 401 | CCGCCGCCAA AACCGACTTC CGGCACATTG CCGTCTGCGC CGCCGTTGTG |
|
| 451 | GCGGCAGCCG GCTATTTCAC CGGCCATTTG AGTTACTACG ACCGGGGTCG |
|
| 501 | GATGGCCAAT ATCTTCGGCG CAAACAACTT CTACTACGCC AAAAGTCAGG |
|
| 551 | CGATGCTCTA CACCGTCAGC CAGAATGCCG ACTTTATTAC CGCCGGCCTG |
|
| 601 | GTCGATCCCG TCTTCCTCCC CTTGGGCAAT CAACAGCGTG CCGCCACGCA |
|
| 651 | TCTGAACGAG CCGAAATCTC AAAAAATCCT CTTTATCGTC GCCGAATCTT |
|
| 701 | GGGGGCTGCC GGCCAATCCC GAACTTCAAA ACGCCACTTT TGCCAAACTG |
|
| 751 | CTGGCGCAAA AAGACCGTTT TTCGGTTTGG GAAAGCGGCA GTTTTCCCTT |
|
| 801 | CATCGGCGCG ACGGTCGAAG GCGAAATGCG CGAACTGTGT GCCTACGGCG |
|
| 851 | GTTTGCGCGG GTTCGCACTG CGCCGCGCGC CCGACGAAAA ATTTGCCCGC |
|
| 901 | TGCCTCCCCA ACCGTTTGAA ACAAGAAGGT TACGCCACCT TTGCGATGCA |
|
| 951 | CGGCGCGGGC AGTTCGCTTT ACGACCGCTT CAGCTGGTAT CCGAGGGCGG |
|
| 1001 | GCTTTCAAGA AATCAAAACC GCCGAAAACC TGATCGGTAA AAAAACCTGC |
|
| 1051 | GCCATTTTCG GCGGCGTGTG CGACAGCGAG CTGTTCGGCG AAGTGTCGGC |
|
| 1101 | ATTTTTCAAA AAACACGACA AGGGACTGTT TTACTGGATG ACGCTGACCA |
|
| 1151 | GCCACGCCGA CTATCCCGAA TCCGACATTT TCAACCACAG GCTCAAATGC |
|
| 1201 | ACCGAATATG GCCTGCCCGC CGAAACCGAC CTCTGCCGCA ATTTCAGCCT |
|
| 1251 | GCACACCCAA TTCTTCGACC AACTGGCGGA TTTGATCCAA CGCCCCGAAA |
|
| 1301 | TGAAAGGCAC GGAAGTCATC ATCGTCGGCG ACCATCCGCC GCCCGTCGGC |
|
| 1351 | AACCTCAATG AAACCTTCCG CTACCTCAAA CAGGGGCACG TCGCCTGGCT |
|
| 1401 | GAACTTCAAA ATCAAATAA |
[1073]This corresponds to the amino acid sequence <SEQ ID 470; ORF48-1>:
[0000]| 1 | MNIHTLLSKQ WTLPPFLPKR LLLSLLILLA PNAVFWVLAL LTATARPIVN | |
|
| 51 | LDYLPAALLI ALPWRFVKIA GVLAFWLAVL FDGLMMVIQL FPFMDLIGAI |
|
| 101 | NLVPFILTAP APYQIMTGLL LLYMLAMPFV LQKAAAKTDF RHIAVCAAVV |
|
| 151 | AAAGYFTGHL SYYDRGRMAN IFGANNFYYA KSQAMLYTVS QNADFITAGL |
|
| 201 | VDPVFLPLGN QQRAATHLNE PKSQKILFIV AESWGLPANP ELQNATFAKL |
|
| 251 | LAQKDRFSVW ESGSFPFIGA TVEGEMRELC AYGGLRGFAL RRAPDEKFAR |
|
| 301 | CLPNRLKQEG YATFAMHGAG SSLYDRFSWY PRAGFQEIKT AENLIGKKTC |
|
| 351 | AIFGGVCDSE LFGEVSAFFK KHDKGLFYWM TLTSHADYPE SDIFNHRLKC |
|
| 401 | TEYGLPAETD LCRNFSLHTQ FFDQLADLIQ RPEMKGTEVI IVGDHPPPVG |
|
| 451 | NLNETFRYLK QGHVAWLNFK IK* |
[1074]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1075]ORF48 shows 94.1% identity over a 119aa overlap with an ORF (ORF48a) from strain A of N. meningitidis:
[0000]
[1076]The complete length ORF48a nucleotide sequence <SEQ ID 471> is:
[0000]| 1 | ATGAATATTC ACACCCTGCT CTCCAAACAA TGGACGCTGC CGCCATTCCT | |
|
| 51 | GCCGAAACGG CTGCTGCTGT CCCTGCTGAT ACTGCTNNCC CCCAATGCGG |
|
| 101 | TGTTTTGGGT TTTGGCACTG CTGACCGCCA CCGCCCGCCC GATTGTCAAT |
|
| 151 | TTGGANTACC TTCCCGCCGC GCTGCTGATC GCCCTGCCTT GGCGTNTCGT |
|
| 201 | CAAAATTGNC GGCGTATTGG CGTNTTGGCT GGCGGTTTTG TTTGACGGGC |
|
| 251 | TGATGATGGT GATCCAACTC TTCCCTTTTA TGGATCTCAT CGGCGCCATC |
|
| 301 | AACCTCGTCC CCTTCATCNT GACCGCCCCC GCCCTTTATC AGATAATGAC |
|
| 351 | CGGGCTGTTA CTGCTGTATA TGCTGGCGAT GCCGTTTGTG TTGCAGAAAG |
|
| 401 | CCGCCGCCAA AACCGACTTC CGACACATTG CCGCCTGTGC CGCCGTTGTG |
|
| 451 | GTGGCAGCCG GCTATTTTAC CGGCCATTTG AGTTANTACG ACCGGGGGCG |
|
| 501 | GATGGCCAAT ATCTTCGGCG CAAACAACTT CTATTACGCC AAAAGTCAGG |
|
| 551 | CGATGCTCTA CACCGTCAGC CAGAATGCCG ACTTTATTAC CGCCGGCCTG |
|
| 601 | GTCGATCCCG TCTTCCTCCC CTTGGGCAAT CAACAGCGTG CCGCCACGCA |
|
| 651 | TCTGAACGAG CCGAAATCTC AAAAAATCCT CTTTATCGTC GCCGAATCTT |
|
| 701 | GGGGGCTGCC GGCCAATCCC GAACTTCAAA ACGCCACTTT TGCCAAACTG |
|
| 751 | CTGGCGCAAA AAGANCGTTT TTCGGTTTGG GAAAGCGGCA GTTTTCCCTT |
|
| 801 | CATCGGCGCG ACGATCGAAG GCGAAATGCG CGAACTGTGT GCCTACGGCG |
|
| 851 | GTTTGCGCGG GTTCGCACTG CGCCGCGCGC CCGACGAAAA ATTTGCCCGC |
|
| 901 | TGCCTCCCCA ACCGTTTGAA ACAAGAAGGT TACGCCACCT TTGCGATGCA |
|
| 951 | CGGCGCGGGC AGTTCGCTTT ACGACCGCTT CAGCTGGTAT CCGAGGGCGG |
|
| 1001 | GCTTTCAAGA AATCAAAACC GCCGAAAACC TGATCGGTAA AAAAACCTGC |
|
| 1051 | GCCATTTTCG GCGGCGTGTG CGACAGCGAG CTGTTCGGCG AAGTGTCGGC |
|
| 1101 | ANTTTTCAAA AAACACGACA AGGGACTGTT TTACTGGATG ACGCTGACCA |
|
| 1151 | GCCACGCCGA CTATCCCGAA TCNGACATTT TCAACCACAG GCTCAAATGC |
|
| 1201 | ACCGAATATG GCCTGCCCGC CGAAACCGAC NTCTGCCGCA ATTTCAGCCT |
|
| 1251 | GCACACCCAA TTCTTCGACC AACTGGCGGA TTTGATCCAA CGCCCCGAAA |
|
| 1301 | TGAAAGGCAC GGAAGTCATC ATCGTCGGCG ACCATCCGCC GCCCGTCGGC |
|
| 1351 | AACCTCAATG AAACCTTCCG CTACCTCAAA CAGGGGCACG TCGNCTGGCT |
|
| 1401 | GAACTTCAAA ATCAAATAA |
[1077]This encodes a protein having amino acid sequence <SEQ ID 472>:
[0000]| 1 | MNIHTLLSKQ WTLPPFLPKR LLLSLLILLX PNAVFWVLAL LTATARPIVN | |
|
| 51 | LXYLPAALLI ALPWRXVKIX GVLAXWLAVL FDGLMMVIQL FPFMDLIGAI |
|
| 101 | NLVPFIXTAP ALYQIMTGLL LLYMLAMPFV LQKAAAKTDF RHIAACAAVV |
|
| 151 | VAAGYFTGHL SXYDRGRMAN IFGANNFYYA KSQAMLYTVS QNADFITAGL |
|
| 201 | VDPVFLPLGN QQRAATHLNE PKSQKILFIV AESWGLPANP ELQNATFAKL |
|
| 251 | LAQKXRFSVW ESGSFPFIGA TIEGEMRELC AYGGLRGFAL RRAPDEKFAR |
|
| 301 | CLPNRLKQEG YATFAMHGAG SSLYDRFSWY PRAGFQEIKT AENLIGKKTC |
|
| 351 | AIFGGVCDSE LFGEVSAXFK KHDKGLFYWM TLTSHADYPE SDIFNHRLKC |
|
| 401 | TEYGLPAETD XCRNFSLHTQ FFDQLADLIQ RPEMKGTEVI IVGDHPPPVG |
|
| 451 | NLNETFRYLK QGHVXWLNFK IK* |
[1078]ORF48a and ORF48-1 show 96.8% identity in 472 aa overlap:
[0000]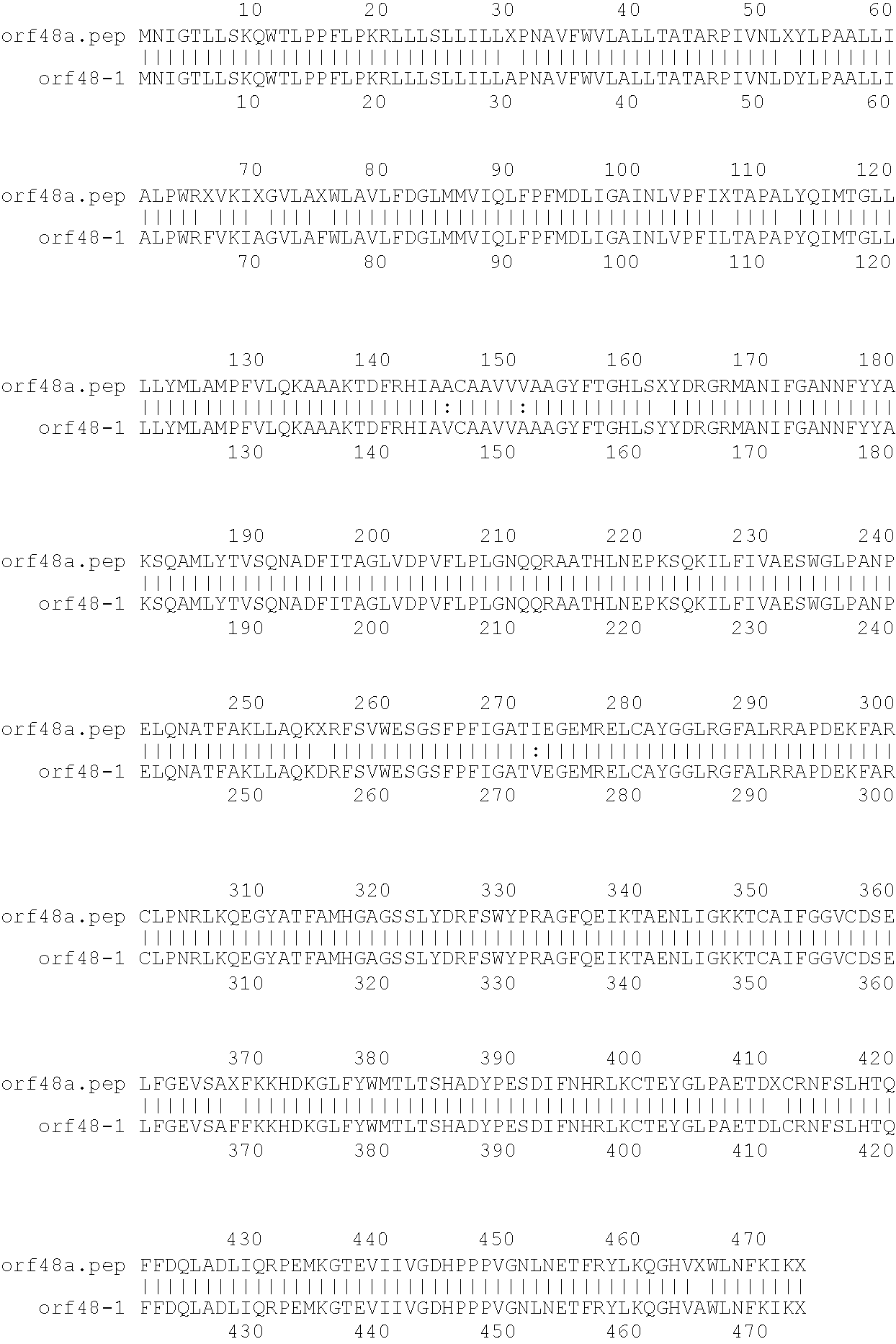
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1079]ORF48 shows 97.5% identity over a 119aa overlap with a predicted ORF (ORF48ng) from N. gonorrhoeae.
[0000]
[1080]The ORF48ng nucleotide sequence <SEQ ID 473> was predicted to encode a protein having amino acid sequence <SEQ ID 474>:
[0000]| 1 | MNIHALLSEQ WTLPPFLPKR LLLSLLILLA PNAVFWVLAL LTATARPIVN | |
|
| 51 | LDYLPAALLI ALPWRFVKIA GVLAFWPAVL FDGLMMVIQL FPFMDLIGAI |
|
| 101 | NLVPFILTAP APYQIMTGLL LLYMLAMPFV LQKAAVKTDF RHIAVCAAVV |
|
| 151 | AAARYFTGPF ELLRTGGRWQ YVQHRRLLLS GSRASFRRRQ KADVLRRLGN |
|
| 201 | PYASMGNGG.. |
[1081]Further work identified the complete gonococcal DNA sequence <SEQ ID 475>:
[0000]| 1 | ATGAATATTC ACGCCCTGCT CTCCGAACAA TGGACGCTGC CGCCATTCCT | |
|
| 51 | GCCGAAACGG CTGCTGCTGT CCCTGCTGAT ACTGCTGGCC CCCAATGCGG |
|
| 101 | TGTTTTGGGT TTTGGCACTG CTGACCGCCA CCGCCCGCCC GATTGTCAAT |
|
| 151 | TTGGACTACC TTCCCGCCGC GCTGCTGATC GCCCTGCCTT GGCGTTTCGT |
|
| 201 | CAAAATTGCC GGCGTATTGG CGTTTTGGCC GGCGGTTTTG TTTGACGGGC |
|
| 251 | TGATGATGGT GATCCAACTC TTCCCTTTTA TGGACCTCAT CGGCGCCATC |
|
| 301 | AACCTCGTCC CCTTCATCCT GACCGCCCCC GCCCCTTATC AGATAATGAC |
|
| 351 | CGGGCTGTTG CTGCTGTATA TGCTGGCGAT GCCGTTTGTG TTGCAAAAAG |
|
| 401 | CCGCCGTCAA AACCGACTTC CGACACATTG CCGTCTGTGC CGCCGTTGTG |
|
| 451 | GCGGCAGCCG GCTATTTCAC CGGCCATTTG AGTTACTACG ACCGGGGGCG |
|
| 501 | GATGGCCAAT ATCTTCGGCG CAAACAACTT CTATTACGCc aAAAGTCAGG |
|
| 551 | CGATGCTCTA CACCGTCAGC CAGAATGCCG ACTTTATTAC CGCCGgcctG |
|
| 601 | GTCGACCCCG TCTTCCTCCC CTTGGGCAAT CAGCAGCGTG CCGCCACGCG |
|
| 651 | GCTGAGTGAG CCGAAATCTC AAAAAATCCT CTTTATCGTC GCCGAATCTT |
|
| 701 | GGGGGCTGCC GGGCAATCCC GAGCTTCAAA ACGCCACTTT TGCCAAACTG |
|
| 751 | CTGGCGCAAA AAGACCGTTT TTCGGTTTGG GAAAGCGGCA GTTTTCCCTT |
|
| 801 | CATCGGCGCG ACGGTCGAAG GCGAAATGCG CGAATTGTGC GCCTACGGCG |
|
| 851 | GTTTGCGCGG GTTCGCACTG CGCCGCGCGC CCGACGAAAA ATTTGCCCGC |
|
| 901 | TGCCTCCCCA ACCGTTTGAA ACAAGAAGGT TACGCCACCT TTGCGATGCA |
|
| 951 | CGGCGCGGGT AGTTCGCTTT ACGACCGCTT CAGCTGGTAT CCGAGGGCGG |
|
| 1001 | GCTTTCAAAA AATCAAAACC GCCGAAAACC TGATCGGTAA AAAAACCTGC |
|
| 1051 | GCCATTTTCG GCGGCGTGTG CGACAGCGAG CTGTTCGGCG AAGTGTCGGC |
|
| 1101 | ATTTTTCAAA AAACACGACA AGGGACTGTT TTACTGGATG ACGCTGACCA |
|
| 1151 | GCCACGCCGA CTATCCCGAA TCCGACATTT TCAACCACAG GCTCAAATGC |
|
| 1201 | ACCGAATACG GCCTGCCCGC CGAAACCGAC CTCTGCCGCA ATTTCAGCCT |
|
| 1251 | GCACACCCAA TtcttcgACC AACTGGCGGA TTTGATCCGA CGCCCCGAAA |
|
| 1301 | TGAAAGGCAC GGAAGTCATC ATCGTCGGCG ACCATCCGCC GCCCGTCGGC |
|
| 1351 | AACCTCAATG AAACCTTCCG CTACCTCAAA CAGGGACACG TCGCCTGGCT |
|
| 1401 | GCACTTCAAA ATCAAATAA |
[1082]This encodes a protein having amino acid sequence <SEQ ID 476; ORF48ng-1>:
[0000]| 1 | MNIHALLSEQ WTLPPFLPKR LLLSLLILLA PNAVFWVLAL LTATARPIVN | |
|
| 51 | LDYLPAALLI ALPWRFVKIA GVLAFWPAVL FDGLMMVIQL FPFMDLIGAI |
|
| 101 | NLVPFILTAP APYQIMTGLL LLYMLAMPFV LQKAAVKTDF RHIAVCAAVV |
|
| 151 | AAAGYFTGHL SYYDRGRMAN IFGANNFYYA KSQAMLYTVS QNADFITAGL |
|
| 201 | VDPVFLPLGN QQRAATRLSE PKSQKILFIV AESWGLPGNP ELQNATFAKL |
|
| 251 | LAQKDRFSVW ESGSFPFIGA TVEGEMRELC AYGGLRGFAL RRAPDEKFAR |
|
| 301 | CLPNRLKQEG YATFAMHGAG SSLYDRFSWY PRAGFQKIKT AENLIGKKTC |
|
| 351 | AIFGGVCDSE LFGEVSAFFK KHDKGLFYWM TLTSHADYPE SDIFNHRLKC |
|
| 401 | TEYGLPAETD LCRNFSLHTQ FFDQLADLIR RPEMKGTEVI IVGDHPPPVG |
|
| 451 | NLNETFRYLK QGHVAWLHFK IK* |
[1083]ORG48ng-1 and ORF48-1 show 97.9% identity in 472 aa overlap:
[0000]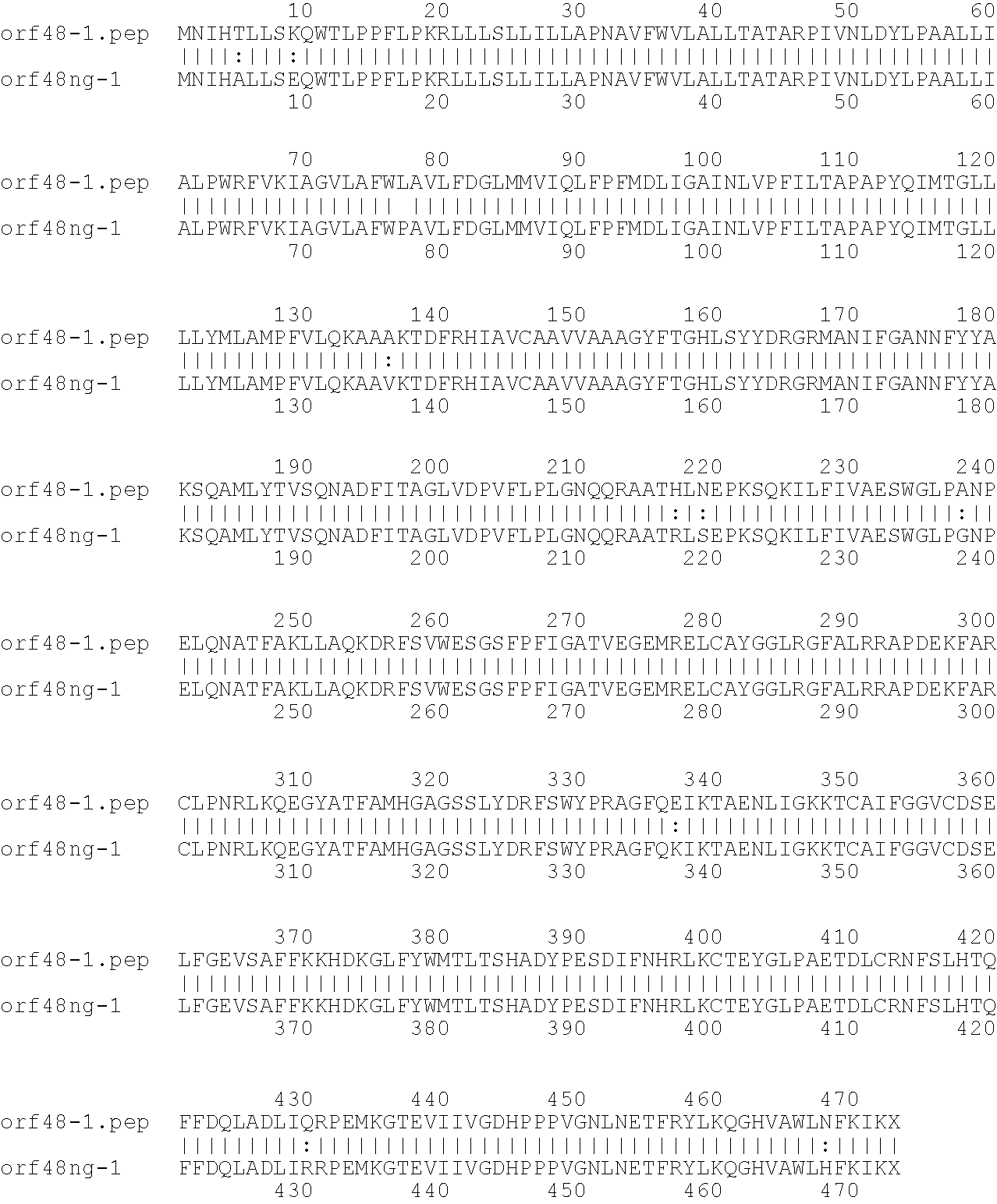
[1084]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and two putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 57
[1085]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 477>:
[0000]| 1 | ..GTGAGCGGAC GTTACCGCGC TTTGGATCGC GTTTCCAAAA TCATCATCGT | |
|
| 51 | TACTTTGAGT ATCGCCACGC TTGCCGCCGC CGGCATCGCT ATGTCGCGCG |
|
| 101 | GTATGCAGAT GCAGTCCGAT TTTATCGAGC CGACACCGTG GACGCTTGCC |
|
| 151 | GGTTTGGGCT TCCTGATCGC GCTGATGGGC TGGATGCCCG CGCCGATTGA |
|
| 201 | AATTTCCGCC ATCAATTCTT TGTGGGTAAC CGAAAAACAA CGCATCAATC |
|
| 251 | CTTCCGAATA CCGCGACGGG ATTTTTGAAT TCAACGTCGG TTATATCGCC |
|
| 301 | AGTGCGGTTT TGGCTTTGGT TTTCCTTGCA CTGGGCGC.G TAGCGCCGAA |
|
| 351 | CGGCAACGGC GA.ACAGTGC AGATGGCGGG CGGCAAATAT AACGGGCAAT |
|
| 401 | TGATCAATAT GTACGCC.. |
[1086]This corresponds to the amino acid sequence <SEQ ID 478; ORF53>:
[0000]| 1 | ..VSGRYRALDR VSKIIIVTLS IATLAAAGIA MSRGMQMQSD FIEPTPWTLA | |
|
| 51 | GLGFLIALMG WMPAPIEISA INSLWVTEKQ RINPSEYRDG IFEFNVGYIA |
|
| 101 | SAVLALVFLA LGXVAPNGNG XTVQMAGGKY NGQLINMYA.. |
[1087]Further work revealed the complete nucleotide sequence <SEQ ID 479>:
[0000]| 1 | ATGTCCGAAC AACATATTTC GACTTGGAAA AGTAAAATCA ACGCATTGGG | |
|
| 51 | TCCGGGGATC ATGATGGCTT CGGCGGCGGT CGGCGGTTCG CACCTGATTG |
|
| 101 | CCTCGACGCA GGCGGGCGCG CTTTACGGCT GGCAGATCGC GCTCATCATC |
|
| 151 | ATCCTGACCA ACCTCTTCAA ATACCCGTTT TTCCGCTTCA GCGCGCATTA |
|
| 201 | CACGCTGGAC ACGGGCAAGA GCCTGATTGA AGGTTATGCC GAGAAAAGCC |
|
| 251 | GCGTTTATTT GTGGGTATTC CTGATTTTGT GCATCCTCTC CGCCACGATT |
|
| 301 | AACGCGGGCG CGGTCGCCAT TGTAACCGCC GCCATCGTCA AAATGGCGAT |
|
| 351 | TCCCTCGCTG ATGTTTGATG CCGGCACGGT TGCCGCCTTG ATTATGGCAT |
|
| 401 | CCTGCCTGAT TATTTTGGTG AGCGGACGTT ACCGCGCTTT GGATCGCGTT |
|
| 451 | TCCAAAATCA TCATCGTTAC TTTGAGTATC GCCACGCTTG CCGCCGCCGG |
|
| 501 | CATCGCTATG TCGCGCGGTA TGCAGATGCA GTCCGATTTT ATCGAGCCGA |
|
| 551 | CACCGTGGAC GCTTGCCGGT TTGGGCTTCC TGATCGCGCT GATGGGCTGG |
|
| 601 | ATGCCCGCGC CGATTGAAAT TTCCGCCATC AATTCTTTGT GGGTAACCGA |
|
| 651 | AAAACAACGC ATCAATCCTT CCGAATACCG CGACGGGATT TTTGATTTCA |
|
| 701 | ACGTCGGTTA TATCGCCAGT GCGGTTTTGG CTTTGGTTTT CCTTGCACTG |
|
| 751 | GGCGCGTTTG TGCAATACGG CAACGGCGAA GCAGTGCAGA TGGCGGGCGG |
|
| 801 | CAAATATATC GGGCAATTGA TCAATATGTA CGCCGTTACC ATCGGCGGCT |
|
| 851 | GGTCGCGCCC GCTGGTGGCG TTTATCGCGT TTGCCTGTAT GTACGGCACG |
|
| 901 | ACGATTACCG TCGTGGACGG CTATGCCCGT GCCATTGCCG AACCCGTGCG |
|
| 951 | CCTGCTGCGC GGAAAAGACA AAACGGGCAA CGCCGAATTC TTTGCCTGGA |
|
| 1001 | ATATTTGGGT GGCGGGCAGC GGTTTGGCGG TGATTTTCTG GTTTGACGGC |
|
| 1051 | GTAATGGCGA ATCTGCTCAA ATTTGCGATG ATTGCCGCTT TTGTGTCCGC |
|
| 1101 | CCCTGTGTTT GCCTGGCTGA ATTACCGTTT GGTTAAAGGT GATGAAAAAC |
|
| 1151 | ACAAACTCAC ATCAGGTATG AATGCCCTTG CATTGGCAGG CTTGATTTAT |
|
| 1201 | CTGACCGGTT TTACCGTTTT GTTCTTATTG AATTTGGCGG GAATGTTCAA |
|
| 1251 | ATGA |
[1088]This corresponds to the amino acid sequence <SEQ ID 480; ORF53-1>:
[0000]| 1 | MSEQHISTWK SKINALGPGI MMASAAVGGS HLIASTQAGA LYGWQIALII | |
|
| 51 | ILTNLFKYPF FRFSAHYTLD TGKSLIEGYA EKSRVYLWVF LILCILSATI |
|
| 101 | NAGAVAIVTA AIVKMAIPSL MFDAGTVAAL IMASCLIILV SGRYRALDRV |
|
| 151 | SKIIIVTLSI ATLAAAGIAM SRGMQMQSDF IEPTPWTLAG LGFLIALMGW |
|
| 201 | MPAPIEISAI NSLWVTEKQR INPSEYRDGI FDFNVGYIAS AVLALVFLAL |
|
| 251 | GAFVQYGNGE AVQMAGGKYI GQLINMYAVT IGGWSRPLVA FIAFACMYGT |
|
| 301 | TITVVDGYAR AIAEPVRLLR GKDKTGNAEF FAWNIWVAGS GLAVIFWFDG |
|
| 351 | VMANLLKFAM IAAFVSAPVF AWLNYRLVKG DEKHKLTSGM NALALAGLIY |
|
| 401 | LTGFTVLFLL NLAGMFK* |
[1089]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1090]ORF53 shows 93.5% identity over a 139aa overlap with an ORF (ORF53a) from strain A of N. meningitidis:
[0000]
[1091]The complete length ORF53a nucleotide sequence <SEQ ID 481> is:
[0000]| 1 | ATGTCCGAAC AACATATTTC GACTTGGAAA AGTAAAATCA ACGCATTGGG | |
|
| 51 | ACCGGGGATT ATGATGGCTT CGGCGGCGGT CGGCGGTTCG CACCTGATTG |
|
| 101 | CCTCGACGCA GGCGGGCGCG CTTTACGGCT GGCAGATCGC GCTCATCATC |
|
| 151 | ATCCTGACCA ACCTCTTCAA ATACCCGTTT TTCCGCTTCA GCGCGCATTA |
|
| 201 | CACGCTGGAC ACGGGCAAGA GCCTGATTGA AGGTTATGCC GAGAAAAGCC |
|
| 251 | GCGTTTATTT GTGGGTATTC CTGATTTTGT GCATCCTCTC CGCCACGATT |
|
| 301 | AACGCGGGCG CGGTCGCCAT TGTAACCGCC GCCATCGTCA AAATGGCGAT |
|
| 351 | TCCCTCGCTG ATGTTTGATG CCGGCACGGT TGCCGCCTTG ATTATGGCAT |
|
| 401 | CCTGCCTGAT TATTTTGGTG AGCGGACGTT ACCGCGCTTT GGATCGCGTT |
|
| 451 | TCCAAAATCA TCATCGTTAC TTTGAGTATC GCCACGCTTG CCGCCGCCGG |
|
| 501 | CATCGCTATG TCGCGCGGTA TGCAGATGCA GTCCGATTTT ATCGAGCCGA |
|
| 551 | CACCGTGGAC GCTTGCCGGT TTGGGCTTCC TGATCGCGCT GATGGGCTGG |
|
| 601 | ATGCCCGCGC CGATTGAAAT TTCCGCCATC AATTCTTTGT GGGTAACCGA |
|
| 651 | AAAACAACGC ATCAATCCTT CCGAATACCG CGACGGGATT TTTGATTTCA |
|
| 701 | ACGTCGGTTA TATCGCCAGT GCGGTTTTGG CTTTGGTTTT CCTTGCACTG |
|
| 751 | GGCGCGTTTG TGCAATACGG CAACGGCGAA GCAGTGCAGA TGGCGGGCGG |
|
| 801 | CAAATATATC GGGCAATTGA TCAATATGTA CGCCGTTACC ATCGGCGGCT |
|
| 851 | GGTCGCGCCC GCTGGTGGCG TTTATCGCGT TTGCCTGTAT GTACGGCACG |
|
| 901 | ACGATTACCG TTGTGGACGG CTATGCCCGT GCCATTGCCG AACCCGTGCG |
|
| 951 | CCTGCTGCGC GGAAAAGACA AAACGGGCAA CGCCGAATTC TTTGCCTGGA |
|
| 1001 | ATATTTGGGT GGCGGGCAGC GGTTTGGCGG TGATTTTCTG GTTTGACGGC |
|
| 1051 | GTAATGGCGA ATCTGCTCAA ATTTGCGATG ATTGCCGCTT TTGTGTCCGC |
|
| 1101 | CCCTGTGTTT GCCTGGCTGA ATTACCGTTT GGTCAAAGGT GATGAAAAAC |
|
| 1151 | ACAAACTCAC ATCAGGTATG AATGCCCTTG CATTGGCAGG CTTGATTTAT |
|
| 1201 | CTGACCGGTT TTACCGTTTT GTTCTTATTG AATTTGGCGG GAATGTTCAA |
|
| 1251 | ATGA |
[1092]This encodes a protein having amino acid sequence <SEQ ID 482>:
[0000]| 1 | MSEQHISTWK SKINALGPGI MMASAAVGGS HLIASTQAGA LYGWQIALII | |
|
| 51 | ILTNLFKYPF FRFSAHYTLD TGKSLIEGYA EKSRVYLWVF LILCILSATI |
|
| 101 | NAGAVAIVTA AIVKMAIPSL MFDAGTVAAL IMASCLIILV SGRYRALDRV |
|
| 151 | SKIIIVTLSI ATLAAAGIAM SRGMQMQSDF IEPTPWTLAG LGFLIALMGW |
|
| 201 | MPAPIEISAI NSLWVTEKQR INPSEYRDGI FDFNVGYIAS AVLALVFLAL |
|
| 251 | GAFVQYGNGE AVQMAGGKYI GQLINMYAVT IGGWSRPLVA FIAFACMYGT |
|
| 301 | TITVVDGYAR AIAEPVRLLR GKDKTGNAEF FAWNIWVAGS GLAVIFWFDG |
|
| 351 | VMANLLKFAM IAAFVSAPVF AWLNYRLVKG DEKHKLTSGM NALALAGLIY |
|
| 401 | LTGFTVLFLL NLAGMFK* |
[1093]ORF 53a shows 100.0% identity in 417 aa overlap with ORF53-1:
[0000]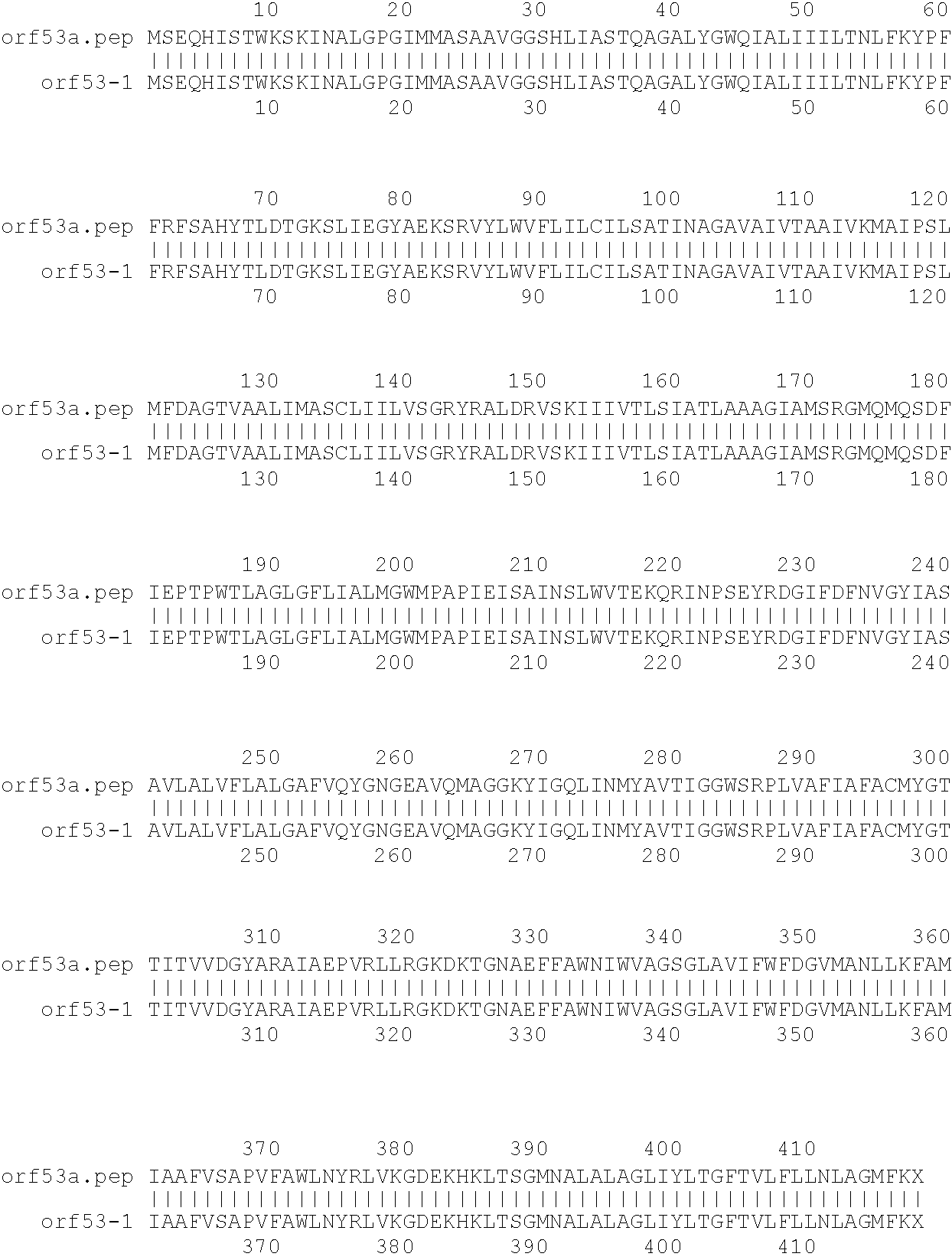
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1094]ORF53 shows 92.1% identity over a 139aa overlap with a predicted ORF (ORF53ng) from N. gonorrhoeae:
[0000]
[1095]An ORF53ng nucleotide sequence <SEQ ID 483> was predicted to encode a protein having amino acid sequence <SEQ ID 484>:
[0000]| 1 | MPKKSCVYLW VFLILCIASA TINAGAVAIV TAAIVKMAIP SLMFDAGTVA | |
|
| 51 | ALIMASCLII LVSGRYRALD RVSKIIIVTL SIATLAAAGI AMSRGMQMQP |
|
| 101 | DFIEPTPWTL AGLGFLIALM GWMPAPIEIS AINSLWVTEK QRINPSEYRD |
|
| 151 | GIFDFNVGYI ASAVLALVFL ALGAFVQYGN GEAVQMGGGK YIGQLINMYA |
|
| 201 | VTIGGGSRPL VAFIAFACMY GAASTVVDGY ARAIAEPVRL LRGKDKTARP |
|
| 251 | IVLLEKLGGR HRFGRDFLV* |
[1096]Further analysis revealed further partial DNA gonococcal sequence <SEQ ID 485>:
[0000]| 1 | ..aagaAAAGCT GCGTTTATTT GTGGGTTTTT TTGATTTTGT GTATCGCCTC | |
|
| 51 | CGCCACGATT AACGCGGGCG CGGTCGCCAT TGTAACCGCC GCCATCGTCA |
|
| 101 | AAATGGCGAT TCCCTCGCTG ATGTTTGATG CCGGCACGGT TGCCGCCTTG |
|
| 151 | ATTATGGCAT CCTGCCTGAT TATTTTGGTG AGCGGACGTT ACCGCGCTTT |
|
| 201 | GGATCGTGTT TCCAAAATCA TCATTGTTAC TTTGAGCATC GCCACGCTTG |
|
| 251 | CCGCCGCCGG CATCGCTATG TCGCGCGGTA TGCAGATGCA GCCCGATTTT |
|
| 301 | ATCGAGCCGA CACCGTGGAC GCTTGCCGGT TTGGGCTTCC TGATCGCGCT |
|
| 351 | GATGGGCTGG ATGCCCGCGC CGATCGAAAT TTCCGCCATC AATTCTTTGT |
|
| 401 | GGGTAACCGA AAAACAACGC ATCAATCCTT CTGAATACCG CGACGGGATT |
|
| 451 | TTCGATTTCA ACGTCGGTTA TATCGCcagT GCGGTTTTGG CTTTGGTTTT |
|
| 501 | CCTTGCACTG GGCGCGTTTG TGCAATACGG CAACGGCGAA GCAGTGCAGA |
|
| 551 | TGGCGGGCGG CAAATATATC GGGCAATTGA TTAATATGTA TGCCGTAACC |
|
| 601 | ATCGGCGGCT GGTCTCGTCC GCTGGTGGCG TTTATCGCGT TTGCCTGTAT |
|
| 651 | GTACGGCACG ACGATTACCG TTGTGGACGG TTATGCGCGT GCCATTGCCG |
|
| 701 | AACCCGTGCG CCTGCTGCGC GGCAGGGATA AAACCGGCAA CGCCGAGTTG |
|
| 751 | TTtgccTGGA ATATTTGGGT GGCGGGCAGC GGTTTGGCGG TGATTTTCTG |
|
| 801 | GTTTGACggc gcaaTGGCgG AACtgcTCAA ATTTGCGATG ATtgccgcCT |
|
| 851 | TTGTGTCCGC CCCTGTGTTC GCCTGGCTCA ACTACCGCCT CGTCAAAGGG |
|
| 901 | GACAAACGCC ACAGGCTTAC CGCCGGTATG AACGCCCTTG CCATTGTCGG |
|
| 951 | CCTGCTCTAC CTGGCCGGGT TTGCCGTTTT GTTCCTGTTG AACCTTACCG |
|
| 1001 | GACTTTTGGC ATAG |
[1097]This corresponds to the amino acid sequence <SEQ ID 486; ORF53ng-1>:
[0000]| 1 | ..KKSCVYLWVF LILCIASATI NAGAVAIVTA AIVKMAIPSL MFDAGTVAAL | |
|
| 51 | IMASCLIILV SGRYRALDRV SKIIIVTLSI ATLAAAGIAM SRGMQMQPDF |
|
| 101 | IEPTPWTLAG LGFLIALMGW MPAPIEISAI NSLWVTEKQR INPSEYRDGI |
|
| 151 | FDFNVGYIAS AVLALVFLAL GAFVQYGNGE AVQMAGGKYI GQLINMYAVT |
|
| 201 | IGGWSRPLVA FIAFACMYGT TITVVDGYAR AIAEPVRLLR GRDKTGNAEL |
|
| 251 | FAWNIWVAGS GLAVIFWFDG AMAELLKFAM IAAFVSAPVF AWLNYRLVKG |
|
| 301 | DKRHRLTAGM NALAIVGLLY LAGFAVLFLL NLTGLLA* |
[1098]ORF53ng-1 and ORF53-1 show 94.0% identity in 336 aa overlap:
[0000]
[1099]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 58
[1100]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 487>:
[0000]| 1 | ..TTGCGGGAAA CGGCATATGT TTTGGATAGT TTTGATCGTT ATTTTGTTGT | |
|
| 51 | TGCGCTTGCC GGCTTGTTTT TTGTCCGCGC ACAATCCGAA CGCGAGTGGA |
|
| 101 | TGCGCGAGGT TTCTGCGTGG CAGGAAAAGA AAGGGGAAAA ACAGGCGGAG |
|
| 151 | CTGCCTGAAA TCAAAGACGG TATGCCCGAT TTTCCCGAAC TTGCCCTGAT |
|
| 201 | GCTTTTCCAC GCCGTCAAAA CGGCAGTGTA TTGGCTGTTT GTCGGTGTCG |
|
| 251 | TCCGTTTCTG CCGAAACTAT CTGGCGCACG AATCCGAACC GGACAGGCCC |
|
| 301 | GTTCCGCCT.. |
[1101]This corresponds to the amino acid sequence <SEQ ID 488; ORF58>:
[0000]| 1 | ..LRETAYVLDS FDRYFVVALA GLFFVRAQSE REWMREVSAW QEKKGEKQAE | |
|
| 51 | LPEIKDGMPD FPELALMLFH AVKTAVYWLF VGVVRFCRNY LAHESEPDRP |
|
| 101 | VPP.. |
[1102]Further work revealed the complete nucleotide sequence <SEQ ID 489>:
[0000]| 1 | ATGTTTTGGA TAGTTTTGAT CGTTATTTTG TTGCTTGCGC TTGCCGGCTT | |
|
| 51 | GTTTTTTGTC CGCGCACAAT CCGAACGCGA GTGGATGCGC GAGGTTTCTG |
|
| 101 | CGTGGCAGGA AAAGAAAGGG GAAAAACAGG CGGAGCTGCC TGAAATCAAA |
|
| 151 | GACGGTATGC CCGATTTTCC CGAACTTGCC CTGATGCTTT TCCATGCCGT |
|
| 201 | CAAAACGGCA GTGTATTGGC TGTTTGTCGG TGTCGTCCGT TTCTGCCGAA |
|
| 251 | ACTATCTGGC GCACGAATCC GAACCGGACA GGCCCGTTCC GCCTGCTTCT |
|
| 301 | GCAAACCGTG CGGATGTTCC GACCGCATCC GACGGATATT CAGACAGTGG |
|
| 351 | AAACGGGACG GAAGAAGCGG AAACGGAAGA AGCAGAAGCT GCGGAGGAAG |
|
| 401 | AGGCTGCCGA TACGGAAGAC ATTGCAACTG CCGTAATCGA CAACCGCCGC |
|
| 451 | ATCCCATTCG ACCGGAGTAT TGCTGAAGGG TTGATGCCGT CTGAAAGCGA |
|
| 501 | AATTTCGCCC GTCCGTCCGG TTTTTAAAGA AATCACTTTG GAAGAAGCAA |
|
| 551 | CGCGTGCTTT AAACAGCGCG GCTTTAAGGG AAACGAAAAA ACGCTATATC |
|
| 601 | GATGCATTTG AGAAAAACGA AACAGCGGTC CCCAAAGTCC GCGTGTCCGA |
|
| 651 | TACCCCGATG GAAGGGCTGC AGATTATCGG TTTGGACGAC CCTGTGCTTC |
|
| 701 | AACGCACGTA TTCCCATATG TTCGATGCGG ACAAAGAAGC GTTTTCCGAG |
|
| 751 | TCTGCGGATT ACGGATTTGA GCCGTATTTT GAGAAGCAGC ATCCGTCTGC |
|
| 801 | CTTTTCTGCA GTCAAAGCCG AAAATGCACG GAATGCGCCG TTCCACCGTC |
|
| 851 | ATGCAGGGCA GGGGAAAGGG CAGGCGGAGG CAAAATCCCC GGATGTTTCC |
|
| 901 | CAAGGGCAGT CCGTTTCAGA CGGCACGGCC GTCCGCGATG CCCGCCGCCG |
|
| 951 | CGTTTCCGTC AATTTGAAAG AACCGAACAA GGCAACGGTT TCTGCGGAGG |
|
| 1001 | CGCGAATTTC TCGCCTGATT CCGGAAAGTC AGACGGTTGT CGGGAAACGG |
|
| 1051 | GATGTCGAAA TGCCGTCTGA AACCGAAAAT GTTTTCACGG AAACCGTTTC |
|
| 1101 | GTCTGTGGGA TACGGCGGTC CGGTTTATGA TGAAACTGCC GATATCCATA |
|
| 1151 | TTGAAGAACC TGCCGCGCCC GATGCTTGGG TGGTCGAACC ACCCGAAGTG |
|
| 1201 | CCGAAAGTTC CCATGACCGC AATCGATATT CAGCCGCCGC CTCCCGTATC |
|
| 1251 | GGAAATCTAC AACCGTACCT ATGAACCGCC GTCAGGATTC GAGCAGGTGC |
|
| 1301 | AACGCAGCCG CATTGCCGAG ACCGACCATC TTGCCGATGA TGTTTTGAAT |
|
| 1351 | GGAGGTTGGC AGGAGGAAAC CGCCGCTATT GCGGATGACG GCAGTGAAGG |
|
| 1401 | TGCGGCAGAG CGGTCAAGCG GGCAATATCT GTCGGAAACC GAAGCGTTCG |
|
| 1451 | GGCATGACAG TCAGGCGGTT TGTCCGTTTG AAAATGTGCC GTCTGAACGC |
|
| 1501 | CCGTCCTGCC GGGTATCGGA TACGGAAGCG GATGAAGGGG CGTTCCCATC |
|
| 1551 | TGAAGAAACC GGTGCGGTAT CCGAACACCT GCCGACAACC GACCTGCTTC |
|
| 1601 | TGCCTCCGCT GTTCAATCCC GAGGCGACGC AAACCGAAGA AGAACTGTTG |
|
| 1651 | GAAAACAGCA TCACCATCGA AGAAAAATTG GCGGAGTTCA AAGTCAAGGT |
|
| 1701 | CAAGGTTGTC GATTCTTATT CCGGCCCCGT AATTACGCGT TATGAAATCG |
|
| 1751 | AACCCGATGT CGGCGTGCGC GGCAATTCCG TTCTGAATCT GGAAAAAGAT |
|
| 1801 | TTGGCGCGTT CGCTCGGCGT GGCTTCCATC CGCGTTGTCG AAACCATCCC |
|
| 1851 | CGGCAAAACC TGCATGGGTT TGGAACTTCC GAACCCGAAA CGCCAAATGA |
|
| 1901 | TACGCCTGAG CGAAATCTTC AATTCGCCCG AGTTTGCCGA ATCCAAATCC |
|
| 1951 | AAGCTGACGC TCGCGCTCGG TCAGGACATC ACCGGACAGC CCGTCGTAAC |
|
| 2001 | CGACTTGGGA AAAGCACCGC ATTTGTTGGT TGCCGGCACG ACCGGTTCGG |
|
| 2051 | GCAAATCGGT GGGTGTCAAC GCGATGATTC TGTCTATGCT TTTCAAAGCC |
|
| 2101 | GCGCCGGAAG ACGTGCGTAT GATTATGATC GATCCGAAAA TGCTGGAATT |
|
| 2151 | GAGCATTTAC GAAGGCATCC CGCACCTGCT CGCCCCTGTC GTTACCGATA |
|
| 2201 | TGAAGCTGGC GGCAAACGCG CTGAACTGGT GTGTTAACGA AATGGAAAAA |
|
| 2251 | CGCTACCGCC TGATGAGCTT TATGGGCGTG CGTAATCTTG CGGGCTTCAA |
|
| 2301 | TCAAAAAATC GCCGAAGCCG CAGCAAGGGG AGAAAAAATC GGCAATCCGT |
|
| 2351 | TCAGCCTCAC GCCCGACGAT CCCGAACCTT TGGAAAAACT GCCGTTTATC |
|
| 2401 | GTGGTCGTGG TCGATGAGTT TGCCGACCTG ATGATGACGG CAGGCAAGAA |
|
| 2451 | AATCGAAGAA CTGATTGCCC GCCTCGCCCA AAAAGCCCGC GCGGCAGGCA |
|
| 2501 | TCCATTTGAT TCTTGCCACA CAACGCCCCA GCGTCGATGT CATCACGGGT |
|
| 2551 | CTGATTAAGG CGAACATCCC GACGCGTATC GCGTTCCAAG TGTCCAGCAA |
|
| 2601 | AATCGACAGC CGCACGATTC TCGACCAAAT GGGCGCGGAA AACCTGCTCG |
|
| 2651 | GTCAGGGCGA TATGCTGTTC CTGCTGCCGG GTACTGCCTA TCCGCAGCGC |
|
| 2701 | GTTCACGGCG CGTTTGCCTC GGATGAAGAG GTGCACCGCG TGGTCGAATA |
|
| 2751 | TTTGAAACAG TTTGGCGAAC CGGACTATGT TGACGATATT TTGAGCGGCG |
|
| 2801 | GCGGCAGCGA AGAGCTGCCC GGCATCGGGC GCAGCGGCGA CGACGAAACC |
|
| 2851 | GATCCGATGT ACGACGAGGC CGTATCCGTT GTCCTGAAAA CGCGCAAAGC |
|
| 2901 | CAGCATTTCG GGCGTACAGC GCGCCTTGCG TATCGGCTAC AACCGCGCCG |
|
| 2951 | CGCGTCTGAT TGACCAGATG GAGGCGGAAG GCATTGTGTC CGCACCGGAA |
|
| 3001 | CACAACGGCA ACCGTACGAT TCTCGTCCCC TTGGACAATG CTTGA |
[1103]This corresponds to the amino acid sequence <SEQ ID 490; ORF58-1>:
[0000]| 1 | MFWIVLIVIL LLALAGLFFV RAQSEREWMR EVSAWQEKKG EKQAELPEIK | |
|
| 51 | DGMPDFPELA LMLFHAVKTA VYWLFVGVVR FCRNYLAHES EPDRPVPPAS |
|
| 101 | ANRADVPTAS DGYSDSGNGT EEAETEEAEA AEEEAADTED IATAVIDNRR |
|
| 151 | IPFDRSIAEG LMPSESEISP VRPVFKEITL EEATRALNSA ALRETKKRYI |
|
| 201 | DAFEKNETAV PKVRVSDTPM EGLQIIGLDD PVLQRTYSHM FDADKEAFSE |
|
| 251 | SADYGFEPYF EKQHPSAFSA VKAENARNAP FHRHAGQGKG QAEAKSPDVS |
|
| 301 | QGQSVSDGTA VRDARRRVSV NLKEPNKATV SAEARISRLI PESQTVVGKR |
|
| 351 | DVEMPSETEN VFTETVSSVG YGGPVYDETA DIHIEEPAAP DAWVVEPPEV |
|
| 401 | PKVPMTAIDI QPPPPVSEIY NRTYEPPSGF EQVQRSRIAE TDHLADDVLN |
|
| 451 | GGWQEETAAI ADDGSEGAAE RSSGQYLSET EAFGHDSQAV CPFENVPSER |
|
| 501 | PSCRVSDTEA DEGAFPSEET GAVSEHLPTT DLLLPPLFNP EATQTEEELL |
|
| 551 | ENSITIEEKL AEFKVKVKVV DSYSGPVITR YEIEPDVGVR GNSVLNLEKD |
|
| 601 | LARSLGVASI RVVETIPGKT CMGLELPNPK RQMIRLSEIF NSPEFAESKS |
|
| 651 | KLTLALGQDI TGQPVVTDLG KAPHLLVAGT TGSGKSVGVN AMILSMLFKA |
|
| 701 | APEDVRMIMI DPKMLELSIY EGIPHLLAPV VTDMKLAANA LNWCVNEMEK |
|
| 751 | RYRLMSFMGV RNLAGFNQKI AEAAARGEKI GNPFSLTPDD PEPLEKLPFI |
|
| 801 | VVVVDEFADL MMTAGKKIEE LIARLAQKAR AAGIHLILAT QRPSVDVITG |
|
| 851 | LIKANIPTRI AFQVSSKIDS RTILDQMGAE NLLGQGDMLF LLPGTAYPQR |
|
| 901 | VHGAFASDEE VHRVVEYLKQ FGEPDYVDDI LSGGGSEELP GIGRSGDDET |
|
| 951 | DPMYDEAVSV VLKTRKASIS GVQRALRIGY NRAARLIDQM EAEGIVSAPE |
|
| 1001 | HNGNRTILVP LDNA* |
[1104]Computer analysis of this amino acid sequence predicts the indicated transmembrane region, and also gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1105]ORF58 shows 96.6% identity over a 89aa overlap with an ORF (ORF58a) from strain A of N. meningitidis:
[0000]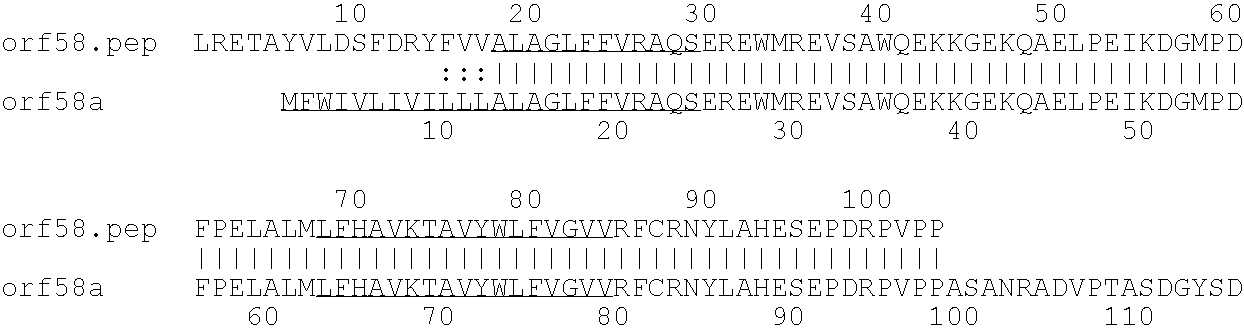
[1106]The complete length ORF58a nucleotide sequence <SEQ ID 491> is:
[0000]| 1 | ATGTTTTGGA TAGTTTTGAT CGTTATTTTG TTGCTTGCGC TTGCCGGCTT | |
|
| 51 | GTTTTTTGTC CGCGCACAAT CCGAACGCGA GTGGATGCGC GAGGTTTCTG |
|
| 101 | CGTGGCAGGA AAAGAAAGGG GAAAAACAGG CGGAGCTGCC TGAAATCAAA |
|
| 151 | GACGGTATGC CCGATTTTCC CGAACTTGCC CTGATGCTTT TCCATGCCGT |
|
| 201 | CAAAACGGCA GTGTATTGGC TGTTTGTCGG TGTCGTCCGT TTCTGCCGAA |
|
| 251 | ACTATCTGGC GCACGAATCC GAACCGGACA GGCCCGTTCC GCCTGCTTCT |
|
| 301 | GCAAATCGTG CGGATGTTCC GACCGCATCC GACGGATATT CAGACAGTGG |
|
| 351 | AAACGGGACG GAAGAAGCGG AAACGGAAGA AGCAGAAGCT GCGGAGGAAG |
|
| 401 | AGGCTGCCGA TACGGAAGAC ATTGCAACTG CCGTAATCGA CAACCGCCGC |
|
| 451 | ATCCCATTCG ACCGGAGTAT TGCTGAAGGG TTGATGCCGT CTGAAAGCGA |
|
| 501 | AATTTCGCCC GTCCGTCCGG TTTTTAAGGA AATCACTTTG GAAGAAGCAA |
|
| 551 | CGCGTGCTTT AAACAGCGCG GCTTTAAGGG AAACGAAAAA ACGCTATATC |
|
| 601 | GATGCATTTG AGAAAAACGA AACAGCGGTC CCCAAAGTCC GCGTGTCCGA |
|
| 651 | TACCCCGATG GAAGGGCTGC AGATTATCGG TTTGGACGAC CCTGTGCTTC |
|
| 701 | AACGCACGTA TTCCCGTATG TTCGATGCGG ACAAAGAAGC GTTTTCCGAG |
|
| 751 | TCTGCGGATT ACGGATTTGA GCCGTATTTT GAGAAGCAGC ATCCGTCTGC |
|
| 801 | CTTTTCTGCA GTCAAAGCCG AAAATGCACG GAATGCGCCG TTCCGCCGTC |
|
| 851 | ATGCAGGGCA GGGNAAAGGG CAGGCGGAGG CNAAATCCCC GGATGTTTCC |
|
| 901 | CAAGGGCAGT CCGTTTCAGA CGGCACAGCC GTCCGCGATG CCNGCCGCCG |
|
| 951 | CGTTTCCGTC AATTTGAAAG AACCGAACAA GGCAACGGTT TCTGCGGAGG |
|
| 1001 | CGCGGATTTC GCGCCTGATT CCGGAAAGTC GGACGGTTGT CGGGAAACGG |
|
| 1051 | GATGTCGAAA TGCCGTCTGA AACCGAAAAT GTTTTCACGG AAANTGTTTC |
|
| 1101 | GTCTGTGGGA TACGGCGNTC CGGTTTATGA TGAAACTGCC GATATCCATA |
|
| 1151 | TTGAAGAACC TGCCGCGCCC GATGCTTGGG TGGTCGAACC ACCCGAAGTG |
|
| 1201 | CCGAAAGTTC CCATGCCCGC AATNGATATT CCGCCGCCGC CTCCCGTATC |
|
| 1251 | GGAAATCTAC AACCGTACCT ATGAACCGCC GGCAGGATTC GAGCAGGTGC |
|
| 1301 | AACGCAGCCG CATTGCCGAA ACCGATCATC TTGCCGATGA TGTTTTGAAT |
|
| 1351 | GGAGGTTGGC AGGAGGAAAC CGCCGCTATT GCGAATGACG GCAGTGAGGG |
|
| 1401 | TGTGGCAGAG CGGTCAAGCG GGCAATATTT GTCGGAAACC GAAGCGTTCG |
|
| 1451 | GGCATGACAG TCAGGCGGTT TGTCCGTTTG AAAATGTGCC GTCTGAACGC |
|
| 1501 | CCGTCCCGCC GGGCATNGGA TACGGAAGCG GATGAAGGGG CGTTCCAATC |
|
| 1551 | TGAAGAAACC GGTGCGGTAT CCGAACACCT GCCGACAACC GACCTGCTTC |
|
| 1601 | TGCCGCCGCT GTTCAATCCC GGGGCGACGC AAACCGAAGA AGANCTGTTG |
|
| 1651 | GANAACAGCA TCACCATCGA AGAAAAATNG GCGGAGTTCA AAGTCAAGGT |
|
| 1701 | CAAGGTTGTC GATTCTTATT CCGGCCCCGT GATTACGCGT TATGAAATCG |
|
| 1751 | AACCCGATGT CGGCGTGCGC GGCAATTCCG TTCTAAATCT GGAAAAAGAN |
|
| 1801 | TTGGCGCGTT CGCTCGGCGT GGCTTCCATC CGCGTTGTCG AAACCATCCT |
|
| 1851 | CGGCAAAACC TGTATGGGTT TGGAACTTCC GAACCCGAAA CGCCAAATGA |
|
| 1901 | TACGCCTGAG CGAAATCTTC AATTCGCCCG AGTTTGCCGA ATCCAAATCC |
|
| 1951 | AAGCTGACGC TCGCGCTCGG TCAGGACATC ACCGGACAGC CCGTCGTAAC |
|
| 2001 | CGACTTGGGC AAAGCACCGC ATTTGTTGGT TGCCGGCACG ACCGGTTCGG |
|
| 2051 | GCAAATCGGT GGGTGTCAAC GCGATGATTC TGTCTATGCT TTTCAAAGCC |
|
| 2101 | GCGCCGGAAG ACGTGCGTAT GATTATGATC GATCCGAAAA TGCTGGAATT |
|
| 2151 | GAGCATTTAC GAAGGCATCC CGCACCTGCT CGCCCCTGTC GTTACCGATA |
|
| 2201 | TGAAGCTGGC GGCAAACGCG CTGAACTGGT GTGTTAACGA AATGGAAAAA |
|
| 2251 | CGCTACCGCC TGATGAGCTT TATGGGCGTG CGCAATCTTG CGGGTNTCAA |
|
| 2301 | TCAAAAAATC GCCGAAGCCG CAGCAAGGGG GGAGAAAATC GGCAACCCGT |
|
| 2351 | TCAGCCTCAC GCCCGACAAT CCCGAACCTT TGGANAAATT GCCGTTTATC |
|
| 2401 | GTGGTCGTGG TTGATGAGTT TGCCGACCTG ATGATGACGG CAGGCAAGAA |
|
| 2451 | AATCGAAGAA CTGATTGCCC GCCTCGCCCA AAAAGCCCGC GCGGCAGGCA |
|
| 2501 | TCCATCTTAT CCTTGCCACA CAACGCCCCA GTGTCGATGT CATCACGGGT |
|
| 2551 | CTGATTAAGG CGAACATCCC GACGCGTATC GCGTTCCAAG TGTCCAGCAA |
|
| 2601 | AATCGACAGC CGCACGATTC TTGACCAAAT GGGTGCGGAA AACCTGCTCG |
|
| 2651 | GGCAGGGCGA TATGCTGTTC CTGCCGCCGG GTACGGCCTA TCCGCAGCGC |
|
| 2701 | GTTCACGGCG CGTTTGCCTC GGATGAAGAG GTGCACCGCG TGGTCGAATA |
|
| 2751 | TCTGAAACAG TTTGGCGAAC CGGACTATGT TGACGATATN TTGAGCGGCG |
|
| 2801 | GTATGTCCGA CGATTTGCTG GGAATCAGCC GGAGCGGCGA CGGCGAAACC |
|
| 2851 | GATCCGATGT ACGACGAGGC CGTGTCNGTT GTTTTGAAAA CGCGCAAAGC |
|
| 2901 | CAGCATTTCT GGCGTGCAGC GCGCATTGCG TATCGGCTAT AATCGCGCCG |
|
| 2951 | CGCGTCTGAT TGACCAGATG GAGGCGGAAG GCATTGTGTC CGCACCGGAA |
|
| 3001 | CACAACGGCA ACCGTACGAT TCTCGTCCCC TTNGACAATG CTTGA |
[1107]This encodes a protein having amino acid sequence <SEQ ID 492>:
[0000]| 1 | MFWIVLIVIL LLALAGLFFV RAQSEREWMR EVSAWQEKKG EKQAELPEIK | |
|
| 51 | DGMPDFPELA LMLFHAVKTA VYWLFVGVVR FCRNYLAHES EPDRPVPPAS |
|
| 101 | ANRADVPTAS DGYSDSGNGT EEAETEEAEA AEEEAADTED IATAVIDNRR |
|
| 151 | IPFDRSIAEG LMPSESEISP VRPVFKEITL EEATRALNSA ALRETKKRYI |
|
| 201 | DAFEKNETAV PKVRVSDTPM EGLQIIGLDD PVLQRTYSRM FDADKEAFSE |
|
| 251 | SADYGFEPYF EKQHPSAFSA VKAENARNAP FRRHAGQGKG QAEAKSPDVS |
|
| 301 | QGQSVSDGTA VRDAXRRVSV NLKEPNKATV SAEARISRLI PESRTVVGKR |
|
| 351 | DVEMPSETEN VFTEXVSSVG YGXPVYDETA DIHIEEPAAP wDAWVVEPPEV |
|
| 401 | PKVPMPAXDI PPPPPVSEIY NRTYEPPAGF EQVQRSRIAE TDHLADDVLN |
|
| 451 | GGWQEETAAI ANDGSEGVAE RSSGQYLSET EAFGHDSQAV CPFENVPSER |
|
| 501 | PSRRAXDTEA DEGAFQSEET GAVSEHLPTT DLLLPPLFNP GATQTEEXLL |
|
| 551 | XNSITIEEKX AEFKVKVKVV DSYSGPVITR YEIEPDVGVR GNSVLNLEKX |
|
| 601 | LARSLGVASI RVVETILGKT CMGLELPNPK RQMIRLSEIF NSPEFAESKS |
|
| 651 | KLTLALGQDI TGQPVVTDLG KAPHLLVAGT TGSGKSVGVN AMILSMLFKA |
|
| 701 | APEDVRMIMI DPKMLELSIY EGIPHLLAPV VTDMKLAANA LNWCVNEMEK |
|
| 751 | RYRLMSFMGV RNLAGXNQKI AEAAARGEKI GNPFSLTPDN PEPLXKLPFI |
|
| 801 | VVVVDEFADL MMTAGKKIEE LIARLAQKAR AAGIHLILAT QRPSVDVITG |
|
| 851 | LIKANIPTRI AFQVSSKIDS RTILDQMGAE NLLGQGDMLF LPPGTAYPQR |
|
| 901 | VHGAFASDEE VHRVVEYLKQ FGEPDYVDDX LSGGMSDDLL GISRSGDGET |
|
| 951 | DPMYDEAVSV VLKTRKASIS GVQRALRIGY NRAARLIDQM EAEGIVSAPE |
|
| 1001 | HNGNRTILVP XDNA* |
[1108]ORF58a and ORF58-1 show 96.6% identity in 1014 aa overlap:
[0000]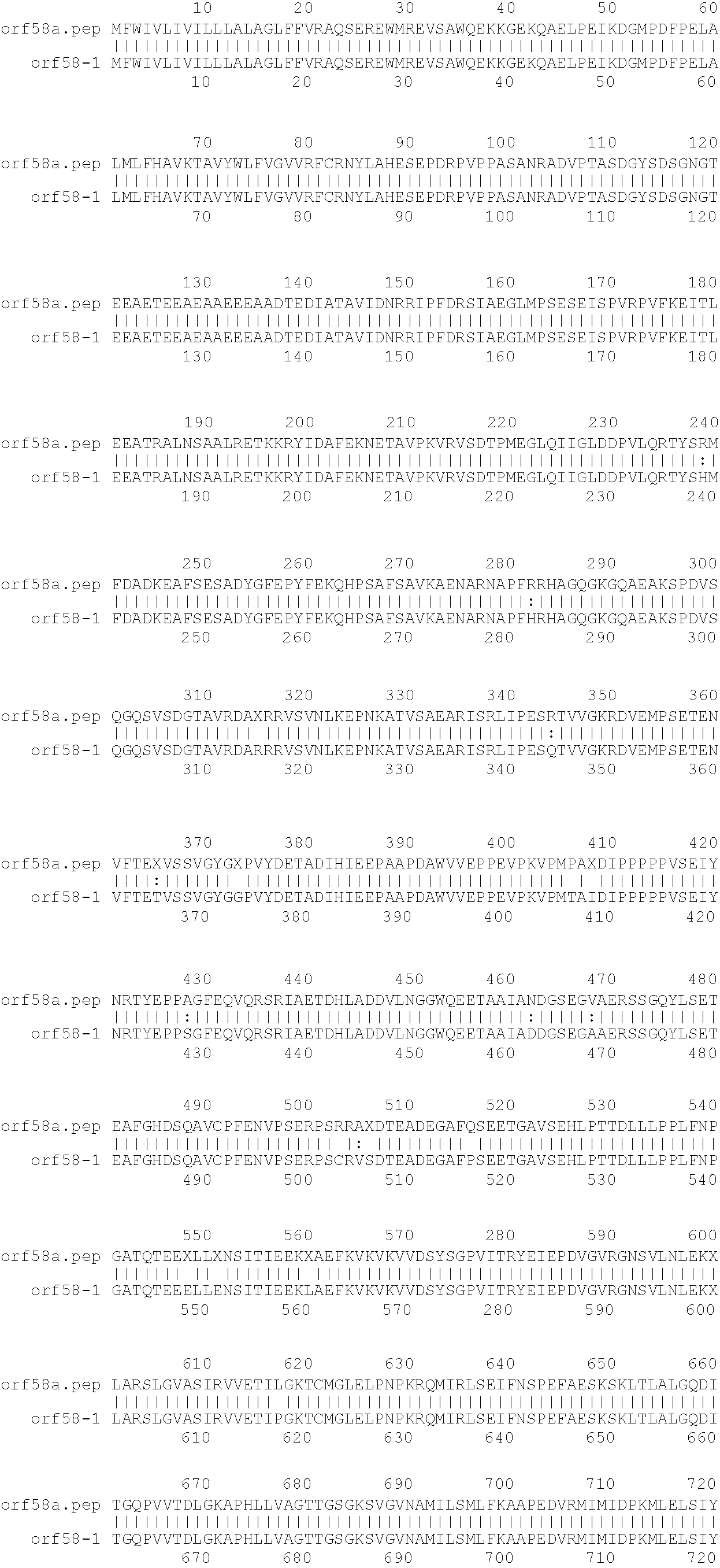

[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1109]ORF58 shows complete identity over a 9aa overlap with a predicted ORF (ORF58ng) from N. gonorrhoeae.
[0000]
[1110]The ORF58ng nucleotide sequence <SEQ ID 493> is predicted to encode a protein having partial amino acid sequence <SEQ ID 494>:
[0000]| 1 | ..SEPDRPVPPA SANRADVPTA SDGYSDSGNG TEEAETEAAE AAEEEAADTE | |
|
| 51 | DIATAVIDNR RIPFDRSIAE GLMQSESKTS PVRPVFKEIT LEEATRALSS |
|
| 101 | AALRETKKRY IDAFEKNGTA VPKVRVSDTP MEGLQIIGLD DPVLQRTYSR |
|
| 151 | MFDADKEAFS ESADYGFEPY FEKQHPSAFS AVKAENARNA PFRRHAGQEK |
|
| 201 | GQAEAKSPDV SQGQSVSDGT AVRDARRRVS VNLKEPNKAT VSAEARISRL |
|
| 251 | IPESRTVVGK RDVEMPSETE NVFTETVSSV GYGGPVYDEA ADIHIEEPAA |
|
| 301 | PDAWVVEPPE VPEVAVPEID ILPPPPVSEI YNRTYEPPAG FEQAQRSRIA |
|
| 351 | ETDHLAADVL NGGWQEETAA IADDGSEGAA ERSSGQYLSE TEAFGHDSQA |
|
| 401 | VCPFEDVPSE RPSCRVSDTE ADEGAFQSEE TGAVSEHLPT TDLLLPPLFN |
|
| 451 | PEATQTEEEL LENSITIEEK LAEFKVKVKV VDSYSGPVIT RYEIEPDVGV |
|
| 501 | RGNSVLNLEK DLARSLGVAS IRVVETIPGK TCMGLELPNP KRQMIRLSEI |
|
| 551 | FNSPEFAESK SKLTLALGQD ITGQPVVTDL GKAPHLLVAG TTGSGKSVGV |
|
| 601 | NAMILSMLFK AAPEDVRMIM IDPKMLELSI YEGITHLLAP VVTDMKLAAN |
|
| 651 | ALNWCVNEME KRYRLMSFMG VRNLAGFNQK IAEAAARGEK IGNPFSLTPD |
|
| 701 | DPEPLEKLPF IVVVVDEFAD LMMTAGKKIE ELIARLAQKA RAAGIHLILA |
|
| 751 | TQRPSVDVIT GLIKANIPTR IAFQVSSKID SRTILDQMGA ENLLGQGDML |
|
| 801 | FLPPGTAYPQ RVHGAFASDE EVHRVVEYLK QFGEPDYVDD ILSGGGSEEL |
|
| 851 | PGIGRSGDGE TDPMYDEAVS VVLKTRKASI SGVQRALRIG YNRAARLIDQ |
|
| 901 | MEAEGIVSAP EHNGNRTILV PLDNA* |
[1111]This partial gonococcal sequence contains a predicted transmembrane region and a predicted ATP/GTP-binding site motif A (P-loop; double underlined). Furthermore, it has a domain homologous to the FTSK cell division protein of E. coli. Alignment of ORF58ng and FtsK (accession number p46889) show a 65% amino acid identity in 459 overlap:
[0000]| ORF58ng: | 467 | IEEKLAEFKVKVKVVDSYSGPVITRYEIEPDVGVRGNSVLNLEKDLARSLGVASIRVVET | 526 | |
| | +E +LA+F++K VV+ GPVITR+E+ GV+ + NL +DLARSL ++RVVE |
| FtsK: | 868 | VEARLADFRIKADVVNYSPGPVITRFELNLAPGVKAARISNLSRDLARSLSTVAVRVVEV | 927 |
|
| ORF58ng: | 527 | IPGKTCMGLELPNPKRQMIRLSEIFNSPEFAESKSKLTLALGQDITGQPVVTDLGKAPHL | 586 |
| | IPGK +GLELPN KRQ + L E+ ++ +F ++ S LT+ LG+DI G+PVV DL K PHL |
| FtsK: | 928 | IPGKPYVGLELPNKKRQTVYLREVLDNAKFRDNPSPLTVVLGKDIAGEPVVADLAKMPHL | 987 |
|
| ORF58ng: | 587 | LVAGTTGSGKSVGVNAMILSMLFKAAPEDVRMIMIDPKMLELSIYEGITHLLAPVVTDMK | 646 |
| | LVAGTTGSGKSVGVNAMILSML+KA PEDVR IMIDPKMLELS+YEGI HLL VVTDMK |
| FtsK: | 988 | LVAGTTGSGKSVGVNAMILSMLYKAQPEDVRFIMIDPKMLELSVYEGIPHLLTEVVTDMK | 1047 |
|
| ORF58ng: | 647 | LAANALNWCVNEMEKRYRLMSFMGVRNLAGFNQKIAEAAARGEKIGNPFSLTPDDPEP-- | 704 |
| | AANAL WCVNEME+RY+LMS +GVRNLAG+N+KIAEA I +P+ D + |
| FtsK: | 1048 | DAANALRWCVNEMERRYKLMSALGVRNLAGYNEKIAEADRMMRPIPDPYWKPGDSMDAQH | 1107 |
|
| ORF58ng: | 705 | --LEKLPFIVVVVDEFADLMMTAGKKIEELIARLAQKARAAGIHLILATQRPSVDVITGL | 762 |
| | L+K P+IVV+VDEFADLMMT GKK+EELIARLAQKARAAGIHL+LATQRPSVDVITGL |
| FtsK: | 1108 | PVLKKEPYIVVLVDEFADLMMTVGKKVEELIARLAQKARAAGIHLVLATQRPSVDVITGL | 1167 |
|
| ORF58ng: | 763 | IKANIPTRIAFQVSSKIDSRTILDQMGAENLLGQGDMLFLPPGTAYPQRVHGAFASDEEV | 822 |
| | IKANIPTRIAF VSSKIDSRTILDQ GAE+LLG GDML+ P + P RVHGAF D+EV |
| FtsK: | 1168 | IKANIPTRIAFTVSSKIDSRTILDQAGAESLLGMGDMLYSGPNSTLPVRVHGAFVRDQEV | 1227 |
|
| ORF58ng: | 823 | HRVVEYLKQFGEPDYVDDILSGGGSEELPGIGRSGDGETDPMYDEAVSVVLKTRKASISG | 882 |
| | H VV+ K G P YVD I S SE G G G E DP++D+AV V + RKASISG |
| FtsK: | 1228 | HAVVQDWKARGRPQYVDGITSDSESEGGAG-GFDGAEELDPLFDQAVQFVTEKRKASISG | 1286 |
|
| ORF58ng: | 883 | VQRALRIGYNRAARLIDQMEAEGIVSAPEHNGNRTILVP | 921 |
| | VQR RIGYNRAAR+I+QMEA+GIVS HNGNR +L P |
| FtsK: | 1287 | VQRQFRIGYNRAARIIEQMEAQGIVSEQGHNGNREVLAP | 1325 |
[1112]Further work on ORF58ng revealed the complete gonococcal DNA sequence to be <SEQ ID 495>:
[0000]| 1 | ATGTTTTGGA TAGTTTTGAT CGTTATtgtg TTGCTTGCGC TTGCCGGCCT | |
|
| 51 | GTTTTTTGTC CGCGCACAAT CCGAACGCGA GTGGATGCGC GAGGTTTCTG |
|
| 101 | CGTGGCAGGA AAAGAAAGGG GAAAAACAGG CGGAGCTGCC TGAAATCAAA |
|
| 151 | GACGGTATGC CCGATTTTCC CGAGTTTTCC CTGATGCTTT TCCATGCCGT |
|
| 201 | CAAAACGGCA GTGTATTGGC TGTTTGTCGG TGTCGTCCGT TTCTGCCGAA |
|
| 251 | ACTATCTGGC GCACGAATCC GAACCGGACA GGCCCGTTCC GCCTGCTTCT |
|
| 301 | GCAAACCGTG CGGATGTTCC GACCGCATCC GACGGGTATT CAGACAGTGG |
|
| 351 | AAACGGGACG GAAGAAGCGG AAACGGAAGC AGCAGAAGCT GCGGAGGAAG |
|
| 401 | AGGCTGCCgA TACgGAAGAC ATTGCAACTG CCGTAATCGA CAACCGCCGC |
|
| 451 | ATCCcatTCG ACCGGAGTAT TGCTGAAGGG TTGATGCAGT CTGAAAGCAA |
|
| 501 | AACTTCGCCC GTCCGTCCGG TTTTTAAGGA AATCACTTTG GAAGAAGCAA |
|
| 551 | CGCGTGCTTT AAGCAGCGCG GCTTTAAGGG AAACGAAAAA ACGCTATATC |
|
| 601 | GATGCATTTG AGAAAAACGG AACAGCCGTC CCCAAAGTAC GCGTGTCCGA |
|
| 651 | TACCCCGATG GAAGGGCTGC AGATTATCGG TTTGGACGAC CCTGTGCTTC |
|
| 701 | AACGCACGTA TTCCCGTATG TTTGATGCGG ACAAAGAAGC GTTTTCCGAG |
|
| 751 | TCTGCGGATT ACGGATTTGA GCCGTATTTT GAGAAGCAGC ATCCGTCTGC |
|
| 801 | CTTTTCTGCA GTCAAAGCCG AAAATGCACG GAATGCGCCG TTCCGCCGTC |
|
| 851 | ATGCAGGGCA GGAGAAAGGG CAGGCGGAGG CAAAATCCCC GGATGTTTCC |
|
| 901 | CAAGGGCAGT CCGTTTCAGA CGGCACAGCC GTCCGCGATG CCCGCCGCCG |
|
| 951 | CGTTTCCGTC AATTTGAAAG AACCGAACAA GGCAACGGTT TCTGCGGAGG |
|
| 1001 | CGCGGATTTC GCGCCTGATT CCGGAAAGTC GGACGGTTGT CGGGAAACGG |
|
| 1051 | GATGTCGAAA TGCCGTCTGA AACCGAAAAT GTTTTCACGG AAACCGTTTC |
|
| 1101 | GTCTGTGGGA TACGGCGGTC CGGTTTATGA TGAAGCTGCC GATATCCATA |
|
| 1151 | TTGAAGAGCC TGCCGCGCCC GATGCTTGGG TGGTCGAACC ACCCGAAGTG |
|
| 1201 | CCGGAGGTAG CCGTACCCGA AATCGATATT CTGCCGCCGC CTCCCGTATC |
|
| 1251 | GGAAATCTAC AACCGTACCT ATGAGCCGCC GGCAGGATTC GAGCAGGCGC |
|
| 1301 | AACGCAGCCG CATTGCCGAA ACCGACCATC TTGCCGCTGA TGTTTTGAAT |
|
| 1351 | GGAGGTTGGC AGGAGGAAAC CGCCGCTATT GCAGATGACG GCAGTGAGGG |
|
| 1401 | TGCGGCAGAG CGGTCAAGCG GGCAATATCT GTCGGAAACC GAAGCGTTCG |
|
| 1451 | GGCATGACAG TCAGGCGGTT TGTCCGTTTG AAGATGTGCC GTCTGAACGC |
|
| 1501 | CCGTCCTGCC GGGTATCGGA TACGGAAGCG GATGAAGGGG CGTTCCAATC |
|
| 1551 | GGAAGAGACC GGTGCGGTAT CCGAACACCT GCCGACAACC GACCTGCTTC |
|
| 1601 | TGCCTCCGCT GTTCAATCCC GAGGCGACGC AAACCGAAGA AGAACTGTTG |
|
| 1651 | GAAAACAGCA TCACCATCGA AGAAAAATTG GCGGAGTTCA AAGTCAAGGT |
|
| 1701 | CAAGGTTGTC GATTCTTATT CCGGCCCCGT GATTACGCGT TATGAAATCG |
|
| 1751 | AACCCGATGT CGGCGTGCGC GGCAATTCCG TTCTGAATTT GGAAAAAGAC |
|
| 1801 | TTGGCGCGTT CGCTCGGCGT GGCTTCCATC CGCGTTGTCG AAACCATCCC |
|
| 1851 | CGGCAAAACC TGCATGGGTT TGGAACTTCC GAACCCGAAA CGCCAAATGA |
|
| 1901 | TACGCCTGAG CGAAATTTTC AATTCGCCCG AGTTTGCCGA ATCCAAATCC |
|
| 1951 | AAGCTGACGC TCGCGCTCGG TCAGGACATT ACCGGACAGC CCGTCGTAAC |
|
| 2001 | CGACTTGGGC AAAGCACCGC ATTTGCTGGT TGCCGGCACG ACCGGTTCGG |
|
| 2051 | GCAAATCGGT GGGTGTCAAC GCGATGATTC TGTCTATGCT TTTCAAAGCC |
|
| 2101 | GCGCCGGAAG ACGTGCGTAT GATTATGATC GATCCGAAAA TGCTGGAATT |
|
| 2151 | GAGCATTTAC GAAGGCATCA CGCACCTGCT CGCCCCTGTC GTTACCGATA |
|
| 2201 | TGAAGCTGGC GGCAAACGCG CTGAACTGGT GTGTTAACGA AATGGAAAAA |
|
| 2251 | CGCTACCGCC TGATGAGCTT TATGGGCGTG CGCAATCTTG CGGGCTTCAA |
|
| 2301 | CCAAAAAATC GCCGAAGCCG CAGCAAGGGG AGAAAAAATC GGCAATCCGT |
|
| 2351 | TCAGCCTCAC GCCCGACGAT CCCGAACCTT TGGAAAAACT GCCGTTTATC |
|
| 2401 | GTGGTCGTGG TCGATGAGTT TGCCGATTTG ATGATGACGG CAGGCAAGAA |
|
| 2451 | AATCGAAGAA CTGATTGCGC GCCTCGCCCA AAAAGCCCGC GCGGCAGGCA |
|
| 2501 | TCCACCTTAT CCTTGCCACA CAACGCCCCA GCGTCGATGT CATCACGGGT |
|
| 2551 | CTGATTAAGG CGAACATCCC GACGCGTATC GCGTTCCAAG TGTCCAGCAA |
|
| 2601 | AATCGACAGC CGCACGATTC TCGACCAAAT GGGCGCGGAA AACCTGCTCG |
|
| 2651 | GTCAGGGCGA TATGCTGTTC CTGCCGCCGG GTACTGCCTA TCCGCAGCGC |
|
| 2701 | GTTCACGGCG CGTTTGCCTC GGATGAAGAG GTGCACCGCG TGGTCGAATA |
|
| 2751 | TCTGAAGCAG TTTGGCGAGC CGGACTATGT TGACGATATT TTGAGCGGCG |
|
| 2801 | GCGGCAGCGA AGAGCTGCCC GGCATCGGGC GCAGCGGCGA CGGCGAAACC |
|
| 2851 | GATCCGATGT ACGACGAGGC CGTATCCGTT GTCCTGAAAA CGCGCAAAGC |
|
| 2901 | CAGCATTTCG GGCGTACAGC GCGCCTTGCG CATCGGCTAC AACCGCGCCG |
|
| 2951 | CGCGTCTGAT TGACCAAATG GAAGCGGAAG GCATTGTGTC CGCACCGGAA |
|
| 3001 | CACAACGGCA ACCGTACGAT TCTCGTCCCC TTGGACAATG CTTGA |
[1113]This corresponds to the amino acid sequence <SEQ ID 496; ORF58ng-1>:
[0000]| 1 | MFWIVLIVIV LLALAGLFFV RAQSEREWMR EVSAWQEKKG EKQAELPEIK | |
|
| 51 | DGMPDFPEFS LMLFHAVKTA VYWLFVGVVR FCRNYLAHES EPDRPVPPAS |
|
| 101 | ANRADVPTAS DGYSDSGNGT EEAETEAAEA AEEEAADTED IATAVIDNRR |
|
| 151 | IPFDRSIAEG LMQSESKTSP VRPVFKEITL EEATRALSSA ALRETKKRYI |
|
| 201 | DAFEKNGTAV PKVRVSDTPM EGLQIIGLDD PVLQRTYSRM FDADKEAFSE |
|
| 251 | SADYGFEPYF EKQHPSAFSA VKAENARNAP FRRHAGQEKG QAEAKSPDVS |
|
| 301 | QGQSVSDGTA VRDARRRVSV NLKEPNKATV SAEARISRLI PESRTVVGKR |
|
| 351 | DVEMPSETEN VFTETVSSVG YGGPVYDEAA DIHIEEPAAP DAWVVEPPEV |
|
| 401 | PEVAVPEIDI LPPPPVSEIY NRTYEPPAGF EQAQRSRIAE TDHLAADVLN |
|
| 451 | GGWQEETAAI ADDGSEGAAE RSSGQYLSET EAFGHDSQAV CPFEDVPSER |
|
| 501 | PSCRVSDTEA DEGAFQSEET GAVSEHLPTT DLLLPPLFNP EATQTEEELL |
|
| 551 | ENSITIEEKL AEFKVKVKVV DSYSGPVITR YEIEPDVGVR GNSVLNLEKD |
|
| 601 | LARSLGVASI RVVETIPGKT CMGLELPNPK RQMIRLSEIF NSPEFAESKS |
|
| 651 | KLTLALGQDI TGQPVVTDLG KAPHLLVAGT TGSGKSVGVN AMILSMLFKA |
|
| 701 | APEDVRMIMI DPKMLELSIY EGITHLLAPV VTDMKLAANA LNWCVNEMEK |
|
| 751 | RYRLMSFMGV RNLAGFNQKI AEAAARGEKI GNPFSLTPDD PEPLEKLPFI |
|
| 801 | VVVVDEFADL MMTAGKKIEE LIARLAQKAR AAGIHLILAT QRPSVDVITG |
|
| 851 | LIKANIPTRI AFQVSSKIDS RTILDQMGAE NLLGQGDMLF LPPGTAYPQR |
|
| 901 | VHGAFASDEE VHRVVEYLKQ FGEPDYVDDI LSGGGSEELP GIGRSGDGET |
|
| 951 | DPMYDEAVSV VLKTRKASIS GVQRALRIGY NRAARLIDQM EAEGIVSAPE |
|
| 1001 | HNGNRTILVP LDNA* |
[1114]ORF58ng-1 and ORF58-1 show 97.2% identity in 1014 aa overlap:
[0000]

[1115]Furthermore, ORF58ng-1 shows significant homology to the E. coli protein FtsK:
[0000]| sp|P46889|FTSK_ECOLI CELL DIVISION PROTEIN FTSK >gi|1651412|gnl|PID|d1015290 (Dl | |
| division protein FtsK [Escherichia coli] >gi|1651418|gnl|PID|d1015296 (D90727) Cell |
| division protein FtsK [Escherichia coli] >gi|1787117 (AE000191) cell division |
| protein FtsK [Escherichia coli] Length = 1329 |
| Score = 576 bits (1469), Expect = e−163 |
| Identities = 301/459 (65%), Positives = 353/459 (76%), Gaps = 5/459 (1%) |
|
| Query: | 556 | IEEKLAEFKVKVKVVDSYSGPVITRYEIEPDVGVAGNSVLNLEKDLARSLGVASIRVVET | 615 | |
| | +E +LA+F++K VV+ GPVITR+E+ GV+ + NL +DLARSL ++RVVE |
| Sbjct: | 868 | VEARLADFRIKADVVNYSPGPVITRFELNLAPGVKAARISNLSRDLARSLSTVAVRVVEV | 927 |
|
| Query: | 616 | IPGKTCMGLELPNPKRQMIRLSEIFNSPEFAESKSKLTLALGQDITGQPVVTDLGKAPHL | 675 |
| | IPGK +GLELPN KRQ + L E+ ++ +F ++ S LT+ LG+DI G+PVV DL K PHL |
| Sbjct: | 928 | IPGKPYVGLELPNKKRQTVYLREVLDNAKFRDNPSPLTVVLGKDIAGEPVVADLAKMPHL | 987 |
|
| Query: | 676 | LVAGTTGSGKSVGVNAMILSMLFKAAPEDVRMIMIDPKMLELSIYEGITHLLAPVVTDMK | 735 |
| | LVAGTTGSGKSVGVNAMILSML+KA PEDVR IMIDPKMLELS+YEGI HLL VVTDMK |
| Sbjct: | 988 | LVAGTTGSGKSVGVNAMILSMLYKAQPEDVRFIMIDPKMLELSVYEGIPHLLTEVVTDMK | 1047 |
|
| Query: | 736 | LAANALNWCVNEMEKRYRLMSFMGVRNLAGFNQKIAEAAARGEKIGNPFSLTPDDPEP-- | 793 |
| | AANAL WCVNEME+RY+LMS +GVRNLAG+N+KIAEA I +P+ D + |
| Sbjct: | 1048 | DAANALRWCVNEMERRYKLMSALGVRNLAGYNEKIAEADRMMRPIPDPYWKPGDSMDAQH | 1107 |
|
| Query: | 794 | --LEKLPFIVVVVDEFADLMMTAGKKIEELIARLAQKARAAGIHLILATQRPSVDVITGL | 851 |
| | L+K P+IVV+VDEFADLMMT GKK+EELIARLAQKARAAGIHL+LATQRPSVDVITGL |
| Sbjct: | 1108 | PVLKKEPYIVVLVDEFADLMMTVGKKVEELIARLAQKARAAGIHLVLATQRPSVDVITGL | 1167 |
|
| Query: | 852 | IKANIPTRIAFQVSSKIDSRTILDQMGAENLLGQGDMLFLPPGTAYPQRVHGAFASDEEV | 911 |
| | IKANIPTRIAF VSSKIDSRTILDQ GAE+LLG GDML+ P + P RVHGAF D+EV |
| Sbjct: | 1168 | IKANIPTRIAFTVSSKIDSRTILDQAGAESLLGMGDMLYSGPNSTLPVRVHGAFVRDQEV | 1227 |
|
| Query: | 912 | HRVVEYLKQFGEPDYVDDILSGGGSEELPGIGRSGDGETDPMYDEAVSVVLKTRKASISG | 971 |
| | H VV+ K G P YVD I S SE G G G E DP++D+AV V + RKASISG |
| Sbjct: | 1228 | HAVVQDWKARGRPQYVDGITSDSESEGGAG-GFDGAEELDPLFDQAVQFVTEKRKASISG | 1286 |
|
| Query: | 972 | VQRALRIGYNRAARLIDQMEAEGIVSAPEHNGNRTILVP | 1010 |
| | VQR RIGYNRAAR+I+QMEA+GIVS HNGNR +L P |
| Sbjct: | 1287 | VQRQFRIGYNRAARIIEQMEAQGIVSEQGHNGNREVLAP | 1325 |
[1116]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 59
[1117]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 497>:
[0000]
[1118]This corresponds to the amino acid sequence <SEQ ID 498; ORF101>:
[0000]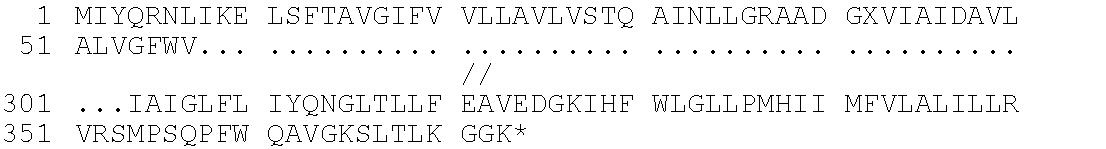
[1119]Further work revealed the complete nucleotide sequence <SEQ ID 499>:
[0000]| 1 | ATGATTTATC AAAGAAACCT CATCAAAGAA CTCTCTTTTA CCGCCGTCGG | |
|
| 51 | CATTTTCGTC GTCCTCTTGG CGGTATTGGT CTCCACGCAG GCAATCAACC |
|
| 101 | TGCTCGGCCG TGCCGCCGAC GGGCGTGTCG CCATCGATGC CGTGTTGGCA |
|
| 151 | TTGGTCGGCT TCTGGGTCAT CGGTATGACG CCGCTTTTGC TGGTGTTGAC |
|
| 201 | CGCATTTATC AGTACGTTGA CCGTGTTGAC CCGCTACTGG CGCGACAGCG |
|
| 251 | AAATGTCGGT CTGGCTATCC TGCGGATTGG CATTGAAACA ATGGATACGC |
|
| 301 | CCGGTGATGC AGTTTGCCGT GCCGTTTGCC GTTTTGGTTG CCGTCATGCA |
|
| 351 | GCTTTGGGTG ATACCGTGGG CAGAGCTACG CAGCCGCGAA TACGCTGAAA |
|
| 401 | TCCTGAAGCA GAAGCAGGAA TTGTCTTTGG TGGAGGCAGG CGAGTTCAAC |
|
| 451 | AGTTTGGGCA AGCGCAACGG CAGGGTTTAT TTTGTCGAAA CCTTCGATAC |
|
| 501 | CGAATCCGGC ATCATGAAAA ACCTGTTCCT GCGCGAACAG GACAAAAACG |
|
| 551 | GCGGCGACAA CATCATCTTC GCCAAAGAAG GTAACTTCTC GCTGAACGAC |
|
| 601 | AACAAACGCA CGCTCGAATT GCGCCACGGC TACCGTTACA GCGGCACGCC |
|
| 651 | CGGACGCGCC GACTACAATC AGGTTTCCTT CCAAAAACTC AACCTGATTA |
|
| 701 | TCAGCACCAC GCCCAAACTC ATCGACCCCG TTTCCCACCG CCGTACCATT |
|
| 751 | CCGACCGCCC AACTGATTGG CAGCAGCAAC CCGCAACATC AGGCGGAATT |
|
| 801 | GATGTGGCGC ATCTCGCTGA CCGTCAGCGT CCTCCTACTC TGCCTGCTTG |
|
| 851 | CCGTGCCGCT TTCCTATTTC AACCCGCGCA GCGGACATAC CTACAATATC |
|
| 901 | TTGATTGCCA TCGGTTTGTT TTTAATTTAC CAAAACGGGC TGACCCTGCT |
|
| 951 | TTTTGAAGCC GTGGAAGACG GCAAAATCCA TTTTTGGCTC GGACTGCTGC |
|
| 1001 | CTATGCACAT TATCATGTTT GCCGTTGCAC TCATCCTGTT GCGCGTCCGC |
|
| 1051 | AGTATGCCCA GCCAGCCCTT CTGGCAGGCG GTTGGCAAAA GTCTGACATT |
|
| 1101 | GAAAGGCGGA AAATGA |
[1120]This corresponds to the amino acid sequence <SEQ ID 500; ORF101-1>:
[0000]| 1 | MIYQRNLIKE LSFTAVGIFV VLLAVLVSTQ AINLLGRAAD GRVAIDAVLA | |
|
| 51 | LVGFWVIGMT PLLLVLTAFI STLTVLTRYW RDSEMSVWLS CGLALKQWIR |
|
| 101 | PVMQFAVPFA VLVAVMQLWV IPWAELRSRE YAEILKQKQE LSLVEAGEFN |
|
| 151 | SLGKRNGRVY FVETFDTESG IMKNLFLREQ DKNGGDNIIF AKEGNFSLND |
|
| 201 | NKRTLELRHG YRYSGTPGRA DYNQVSFQKL NLIISTTPKL IDPVSHRRTI |
|
| 251 | PTAQLIGSSN PQHQAELMWR ISLTVSVLLL CLLAVPLSYF NPRSGHTYNI |
|
| 301 | LIAIGLFLIY QNGLTLLFEA VEDGKIHFWL GLLPMHIIMF AVALILLRVR |
|
| 351 | SMPSQPFWQA VGKSLTLKGG K* |
[1121]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1122]ORF101 shows 91.2% identity over a 57aa overlap and 95.7% identity over a 69aa overlap with an ORF (ORF101a) from strain A of N. meningitidis:
[0000]
[1123]The complete length ORF101a nucleotide sequence <SEQ ID 501> is:
[0000]| 1 | ATGATTTATC AAAGAAACCT CATCAAAGAA CTCTCTTTTA CCGCCGTCGG | |
|
| 51 | CATTTTCGTC GTCCTCTTGG CGGTATTGGT CTCCACGCAG GCAATCAACC |
|
| 101 | TGCTCGGCCN TGCCGCCGAC NGGCGTNTCG CCATCGATGC CGTGTTGGCA |
|
| 151 | TTGGTCGGCT TCTGGGTCNN NNGNATGACG CCGCTTTTGC TNGTGTTGAC |
|
| 201 | CGCATTTATC AGTACGTTGA CCGTGTTGAC CCGCTACTGG CGNGACAGCG |
|
| 251 | AAATGTCGGT CTGGNTATCC TGCGGATTGG CATTGAAACA ATGGATACGC |
|
| 301 | CCGGTGATGC AGTTTGCCGT GCCGTTTGCC GTTTTGGTTG CCGTCATGCA |
|
| 351 | GCTTTGGGTG ATACCGTGGG CAGAGCTACG CAGCCGCGAA TACGCTGAAA |
|
| 401 | TCCTGAAGCA GAAGCAGGAA TTGTCTTTGG TGGAGGCAGG CGGGTTCAAC |
|
| 451 | AGTTTGGGCA AGCGCAACGG CAGGGTTTAT TTTGTCGAAA CCTTCGATAC |
|
| 501 | CGAATCCGGC ATCATGAAAA ACCTGTTCCT GCGCGAACAG GACAAAAACG |
|
| 551 | GCGGCGACAA CATCATCTTC NCCAAAGAAA GTAACTTCTC GCTGAACGAC |
|
| 601 | AACAAACGCA CGCTCGAATT GCGCCACGGC TACCGTTACA GCGGCACGCC |
|
| 651 | CGGACGCGCC GACTACAATC AGGTTTCCTT CCNAAAACTC AACCTGATTA |
|
| 701 | TCAGCACCAC GCCCAAACTC ATCGACCCCG TTTCCCACCG CCGTACNATN |
|
| 751 | CCNACNGCCC AACTGATTGG CAGCAGCAAC CCGCAACATC ANGCGGAATT |
|
| 801 | GATGTGGCGC ATCTCGCTGA CCGTCAGCGT CCTCCTACTC TGCCTGCTTG |
|
| 851 | CCGTGCCGCT TTCCTATTTC AACCCGCGCA GCGGACATAC CTACAATATC |
|
| 901 | TTGANTGCCA TCGGTTTGTT TTTAATTTAC CAAAACGGGC TGACCCTGCT |
|
| 951 | TTTTGAAGCC GTGGAAGACG GCAAAATCCA TTTTTGGCTC GGACTGCTGC |
|
| 1001 | CTATGCACAT CATCATGTTC GTCATCGCAA TCGTACTTCT GCGCGTCCGC |
|
| 1051 | AGCATGCCCA GCCAGCCCTT CTGGCAGGCG GTTGGCAAAA GTCTGACATT |
|
| 1101 | GAAAGGCGGA AAATGA |
[1124]This encodes a protein having amino acid sequence <SEQ ID 502>:
[0000]| 1 | MIYQRNLIKE LSFTAVGIFV VLLAVLVSTQ AINLLGXAAD XRXAIDAVLA | |
|
| 51 | LVGFWVXXMT PLLLVLTAFI STLTVLTRYW RDSEMSVWXS CGLALKQWIR |
|
| 101 | PVMQFAVPFA VLVAVMQLWV IPWAELRSRE YAEILKQKQE LSLVEAGGFN |
|
| 151 | SLGKRNGRVY FVETFDTESG IMKNLFLREQ DKNGGDNIIF XKESNFSLND |
|
| 201 | NKRTLELRHG YRYSGTPGRA DYNQVSFXKL NLIISTTPKL IDPVSHRRTX |
|
| 251 | PTAQLIGSSN PQHXAELMWR ISLTVSVLLL CLLAVPLSYF NPRSGHTYNI |
|
| 301 | LXAIGLFLIY QNGLTLLFEA VEDGKIHFWL GLLPMHIIMF VIAIVLLRVR |
|
| 351 | SMPSQPFWQA VGKSLTLKGG K* |
[1125]ORF101a and ORF101-1 show 95.4% identity in 371 aa overlap:
[0000]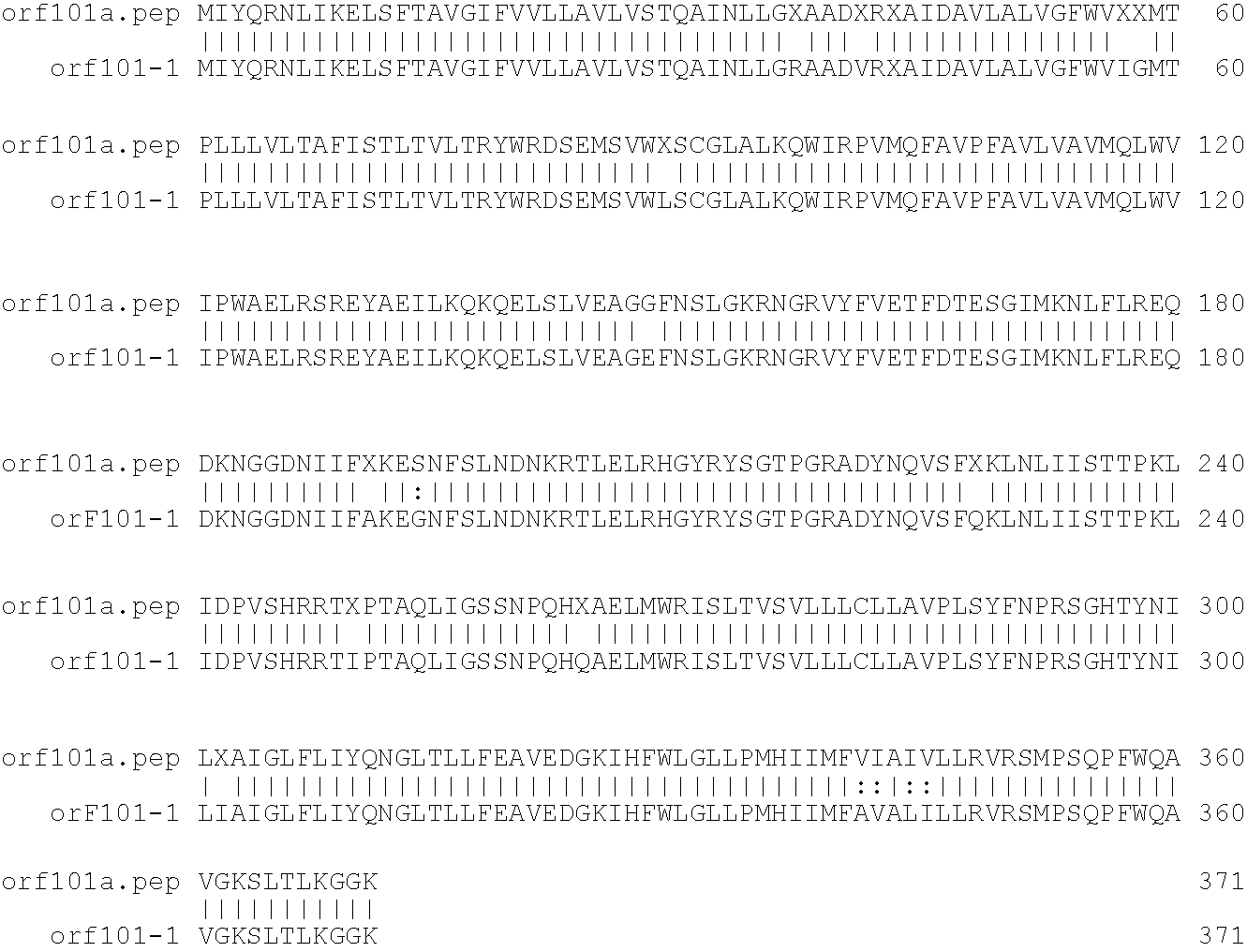
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1126]ORF101 shows 96.5% identity in 57aa overlap at the N-terminal domain and 95.1% identity in 61 aa overlap at the C-terminal domain, respectively, with a predicted ORF (ORF101ng) from N. gonorrhoeae:
[0000]
[1127]The ORF101ng nucleotide sequence <SEQ ID 503> is predicted to encode a protein having partial amino acid sequence <SEQ ID 504>:
[0000]| 1 | MIYQRNLIKE LSFTAVGIFV VLLAVLVSTQ AINLLGRAAD GRVAIDAVLA | |
|
| 51 | LVGFWVIGMT PLLLVLTAFI STLTVLTRYW RDSEMSVWLS CGLALKQWIR |
|
| 101 | PVMQFAVPFA ILIAVMQLWV IPWAELRSRE YAEILKQKQE LSLVEAGEFN |
|
| 151 | NLGKRNGRVY FVETFDTESG IMKNLFLREQ DKNGGDNIIF AKEGNFSLKD |
|
| 201 | NKRTLELRHG YRYSGTPGRA DYNQVSFQKL NLIISTTPKL IDPVSHRRTI |
|
| 251 | STAQLIGSSN PQHQAELMWR ISLTVSVLLL CLLAVPLSYF NPRSGHTYNI |
|
| 301 | LIAIGLFLIY QNGLTLLFEA VEDGKIHFWL GLLPMHIIMF VIAIVLLRVR |
|
| 351 | SMPSQPFWQA VG... |
[1128]Further work revealed the complete nucleotide sequence <SEQ ID 505>:
[0000]| 1 | ATGATTTATC AAAGAAACCT CATCAAAGAA CTCTCTTTTA CCGCCGTCGG | |
|
| 51 | CATTTTCGTC GTCCTCTTGG CGGTGTTGGT GTCCACGCAG GCGATCAACC |
|
| 101 | TGCTTGGCCG CGCAGCTGAC GGGCGTGTCG CCATCGATGC CGTGTTGGCC |
|
| 151 | TTAGTCGGCT TCTGGGTCAT CGGTATGACC CCGCTTTTGC TGGTGTTGAC |
|
| 201 | CGCATTCATC AGCACGCTGA CCGTATTGAC CCGCTACTGG CGCGACAGCG |
|
| 251 | AAATGTCGGT CTGGCTATCC TGCGGATTGG CGTTGAAACA GTGGATACGC |
|
| 301 | CCCGTCATGC AGTTTGCCGT GCCGTTTGCC ATCCTGATTG CCGTCATGCA |
|
| 351 | GCTTTGGGTG ATACCGTGGG CAGAGCTGCG CAGCCGCGAA TATGCCGAAA |
|
| 401 | TTTTGAAGCA GAAGCAGGAA TTGTCTTTGG TGGAAGCCGG CGAGTTCAAT |
|
| 451 | AACTTGGGCA AGCGCAACGG CAgggtttaT TtcgtcgaaA CCTTTGACAC |
|
| 501 | CGaatccgGC ATCATGAAAA ACCTGTtcct GcGCGAACAG GACAAAAACG |
|
| 551 | gcggcgacaA CATCATCTTC GCcaaaGAag gtaactTctc gctgaaggaC |
|
| 601 | AACAAAcgca cgctcgaATT GCGCCACGGC TACCGTTACA GCGGcacgcC |
|
| 651 | CGGacGCGCc gactaCAATC AGGTTtcctt cCAAAAacTc aacctgATta |
|
| 701 | TCAGCACCAC GCCCAAacTT ATCGaccCCG TTTCCCACCG CCGCACCATT |
|
| 751 | tcgacCGCCC AAcTGATTGG CAGCAGCAAT CCGCAACATC AGGCAGAATT |
|
| 801 | GATGTGGCGC ATCTCGCTGA CCGTCAGCGT CCTCCTGCTC TGCCTACTCG |
|
| 851 | CCGTGCCGCT TTCCTATTTC AACCCGCGCA GCGGACATAC CTACAATATC |
|
| 901 | TTGATTGCCA TCGGTTTGTT TTTAATTTAC CAAAACGGGC TGACCCTGCT |
|
| 951 | TTTTGAAGCC GTGGAAGACG GCAAAATCCA TTTTTGGCTC GGACTGCTGC |
|
| 1001 | CTATGCACAT CATCATGTTC GTCATCGCAA TCGTACTTCT GCGCGTCCGC |
|
| 1051 | AGTATGCCCA GCCAGCCCTT CTGGCAGGCG GTTGGCAAAA GTCTGACATT |
|
| 1101 | GAAAGgcgGA AAATGA |
[1129]This corresponds to the amino acid sequence <SEQ ID 506; ORF101ng-1>:
[0000]| 1 | MIYQRNLIKE LSFTAVGIFV VLLAVLVSTQ AINLLGRAAD GRVAIDAVLA | |
|
| 51 | LVGFWVIGMT PLLLVLTAFI STLTVLTRYW RDSEMSVWLS CGLALKQWIR |
|
| 101 | PVMQFAVPFA ILIAVMQLWV IPWAELRSRE YAEILKQKQE LSLVEAGEFN |
|
| 151 | NLGKRNGRVY FVETFDTESG IMKNLFLREQ DKNGGDNIIF AKEGNFSLKD |
|
| 201 | NKRTLELRHG YRYSGTPGRA DYNQVSFQKL NLIISTTPKL IDPVSHRRTI |
|
| 251 | STAQLIGSSN PQHQAELMWR ISLTVSVLLL CLLAVPLSYF NPRSGHTYNI |
|
| 301 | LIAIGLFLIY QNGLTLLFEA VEDGKIHFWL GLLPMHIIMF VIAIVLLRVR |
|
| 351 | SMPSQPFWQA VGKSLTLKGG K* |
[1130]ORF101ng-1 and ORF101-1 show 97.6% identity in 371 aa overlap:
[0000]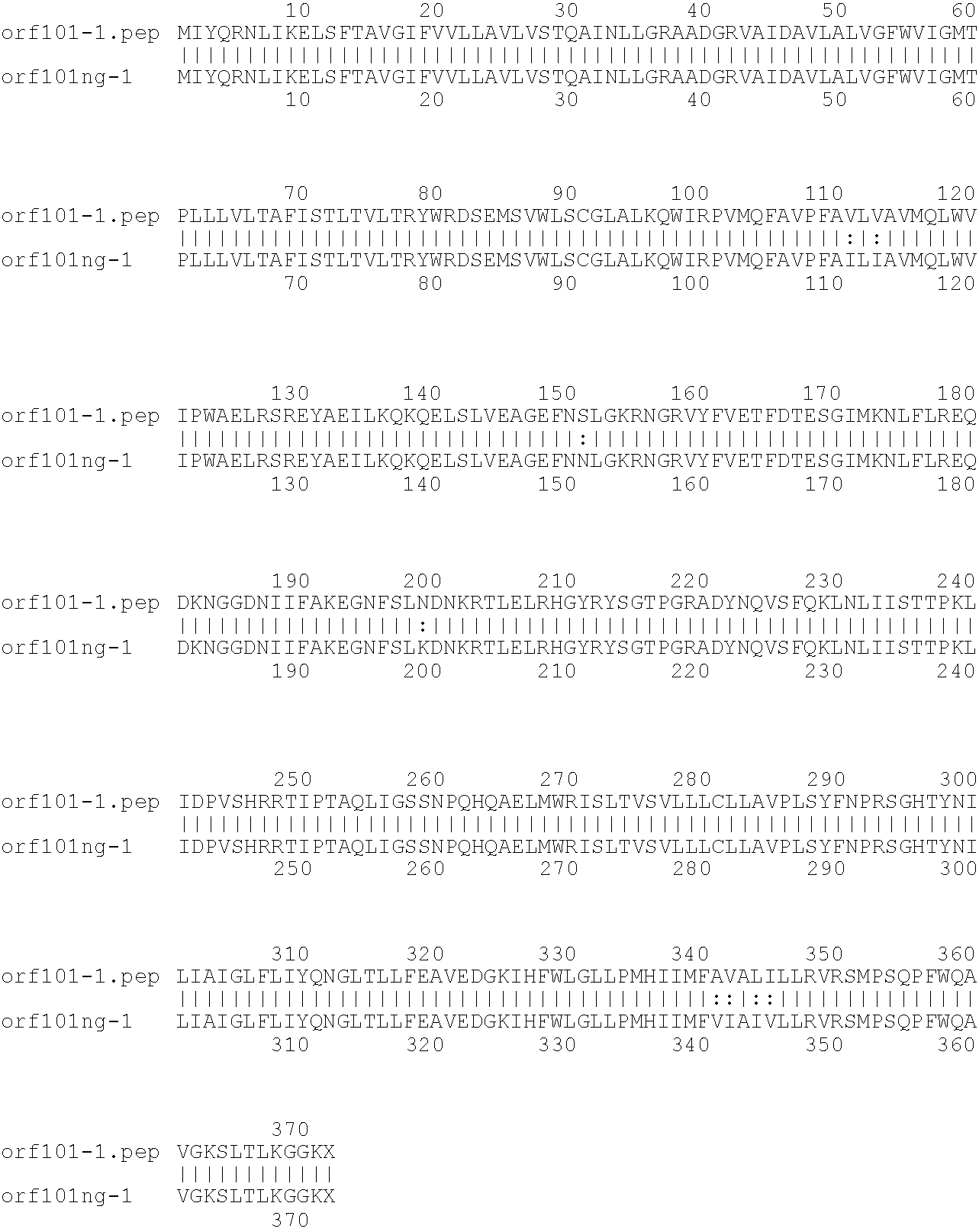
[1131]Based on this analysis, including the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 60
[1132]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 507>:
[0000]| 1 | ..GGTGGTGGTT TTATCAATGC TTCCTGTGCC ACTTTGACGA CAGCCAAACC | |
|
| 51 | GCAATATCAA GCAGGAGACC TTAGCGCTTT TAAGATAAGG CAAGGCAATG |
|
| 101 | TTGTAATCGC CGGACACGGT TTGGATGCAC GTGATACCGA TTACACACGT |
|
| 151 | ATTCTCAGTT ATCATTCCAA AATCGATGCA CCCGTATGGG GACAAGATGT |
|
| 201 | TCGTGTCGTC GCGGGACAAA ACGATGTGGC CGCAACAGGT GATGCACATT |
|
| 251 | CGCCTATTCT CAATAATGCT GCTGCCAATA CGTCAAACAA TACAGCCAAC |
|
| 301 | AACGGCACAC ATATCCCTTT ATTTGCGATT GATACAGGCA AATTAGGAGG |
|
| 351 | TAT.GTATGC CAACAAAATC ACCTTGATCA GTACGGTCGA GCAAGCAGGC |
|
| 401 | ATTCGTAA |
[1133]This corresponds to the amino acid sequence <SEQ ID 508; ORF113>:
[0000]| 1 | ..GGGFINASCA TLTTAKPQYQ AGDLSAFKIR QGNVVIAGHG LDARDTDYTR | |
|
| 51 | ILSYHSKIDA PVWGQDVRVV AGQNDVAATG DAHSPILNNA AANTSNNTAN |
|
| 101 | NGTHIPLFAI DTGKLGGXVC QQNHLDQYGR ASRHS* |
[1134]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with pspA Putative Secreted Protein of N. meningitidis (Accession AF030941)
[1135]ORF and pspA show 44% aa identity in 179aa overlap:
[0000]| orf113 | GGGFINASCATLTTAKPQYQAGDLSAFKIRQGNVVIAGHGLDARDTDYTRILSYHSKIDA | 60 | |
| GGG INA+ TLT+ P G+L+ F + G VVI G GLD D DYTRILS ++I+A |
| pspa | GGGLINAASVTLTSGVPVLNNGNLTGFDVSSGKVVIGGKGLDTSDADYTRILSRAAEINA | 256 |
|
| orf113 | PVWGQDVRVVAGQNDVAATGDAHSPILXXXXXXXXXXXXXXGTHIPLFAIDTGKLGGMYA | 120 |
| VWG+DV+VV+G+N + G + P AIDT LGGMYA |
| pspa | GVWGKDVKVVSGKNKLDFDG---------SLAKTASAPSSSDSVTPTVAIDTATLGGMYA | 307 |
|
| orf113 | NKITLISTVEQAGIRNQGQWFASAGNVAVNAEGKLVNTGMIAATGENHAVSLHARNVHN | 179 |
| +KITLIST A IRN+G+ FA+ G V ++A+GKL N+G I A +++ A+ V N |
| pspa | DKITLISTDNGAVIRNKGRIFAATGGVTLSADGKLSNSGSIDAA----EITISAQTVDN | 362 |
Homology with a Predicted ORF from
N. gonorrhoeae [1136]ORF113 shows 86.5% identity in 52aa overlap at the N-terminal part and 94.1% identity in 17aa overlap at the C-terminal part with a predicted ORF (ORF113ng) from N. gonorrhoeae:
[0000]
[1137]The complete length ORF113ng nucleotide sequence <SEQ ID 509> is predicted to encode a protein having amino acid sequence <SEQ ID 510>:
[0000]| 1 | MNKTLYRVIF NRKRGAVVAV AETTKREGKS CADSGSGSVY VKSVSFIPTH | |
|
| 51 | SKAFCFSALG FSLCLALGTV NIAFADGIIT DKAAPKTQQA TILQTGNGIP |
|
| 101 | QVNIQTPTSA GVSVNQYAQF DVGNRGAILN NSRSNTQTQL GGWIQGNPWL |
|
| 151 | TRGEARVVVN QINSSHPSQL NGYIEVGGRR AEVVIANPAG IAVNGGGFIN |
|
| 201 | ASRATLTTGQ PQYQAGDFSG FKIRQGNAVI AGHGLDARDT DFTRILVCQQ |
|
| 251 | NHLDQYGRTS RHS* |
[1138]Based on this analysis, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 61
[1139]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 511>:
[0000]| 1 | ..TCAACGGGAC ATAGCGAACA AAATTACACT TTGCCGCGAG AAATCACACG | |
|
| 51 | CAACATTTCA CTGGGTTCAT TTGCCTATGA ATCGCATCGC AAAGCATTAA |
|
| 101 | GCCATCATGC GCCCAGCCAA GGCACTGAGT TGCCGCAAAG CAACGGTATT |
|
| 151 | TCGCTACCCT ATACGTCCAA TTCTTTTACC CCATTACCCA GCAGCAGCTT |
|
| 201 | ATACATTATC AATCCTGTCA ATAAAGGCTA TCTTGTTGAA ACCGATCCAC |
|
| 251 | GCTTTGCCAA CTACCGTCAA TGGTTGGGTA GTGACTATAT GCtGGACAGC |
|
| 301 | CTCAAACTAG ACCCAAACAA TTTACATAAA CGTTTGGGTG ATGGTTATTA |
|
| 351 | CGAGCAACGT TTAATCAATG AACAAATCGC AGAGCTGACA GGGCATCGTC |
|
| 401 | GTTTAGAcGG TTATCAAAAC GACGAAGAAC AATTTAAAGC CTTAATGGAT |
|
| 451 | AATGGCGCGA CTGCGGCACG TTcGATGAAT CTCAGCGTTG GCATTGCATT |
|
| 501 | AAGTGCCGAG CAAGTAGCGC AACTGACCAG CGATATTGTT TGGTTGGTAC |
|
| 551 | AAAAAGAAGT TAAGCTTCCT GATGGCGGCA CACAAACCGT ATTGGTGCCA |
|
| 601 | CAGGTTTATG TACGCGTTAA AAATGGCGAC ATAGACGGTA AAGGTGCATT |
|
| 651 | GTTGTCAGGC AGCAATACAC AAATCAATGT TTCAGGCAGC CTGAAAAACT |
|
| 701 | CAGGCACGAT TGCAGGgCGC AATGCGCTTA TTATCAATAC CGATACGCTA |
|
| 751 | GACAATATCG GTGGGCGTAT TCATGCGCAA AAATCAGCGG TTACGGCCAC |
|
| 801 | ACAAGACATC AATAATATTG GCGGCATGCT TTCTGCCGAA CAGACATTAT |
|
| 851 | TGCTCAACGC AGGCAACAAC ATCAACAGCC AAAGCACCAC CGCCAGCAGT |
|
| 901 | CAAAATACAC AAGGCAGCAG CACCTACCTA GACCGAATGG CAGGTATTTA |
|
| 951 | TATCACAGGC AAAGAAAAAG GTGTTT.. |
[1140]This corresponds to the amino acid sequence <SEQ ID 512; ORF115>:
[0000]| 1 | ..STGHSEQNYT LPREITRNIS LGSFAYESHR KALSHHAPSQ GTELPQSNGI | |
|
| 51 | SLPYTSNSFT PLPSSSLYII NPVNKGYLVE TDPRFANYRQ WLGSDYMLDS |
|
| 101 | LKLDPNNLHK RLGDGYYEQR LINEQIAELT GHRRLDGYQN DEEQFKALMD |
|
| 151 | NGATAARSMN LSVGIALSAE QVAQLTSDIV WLVQKEVKLP DGGTQTVLVP |
|
| 201 | QVYVRVKNGD IDGKGALLSG SNTQINVSGS LKNSGTIAGR NALIINTDTL |
|
| 251 | DNIGGRIHAQ KSAVTATQDI NNIGGMLSAE QTLLLNAGNN INSQSTTASS |
|
| 301 | QNTQGSSTYL DRMAGIYITG KEKGV.. |
[1141]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the pspA Putative Secreted Protein of N. meningitidis (Accession Number AF030941)
[1142]ORF115 and pspA protein show 50% aa identity in 325aa overlap:
[0000]| Orf115: | 1 | STGHSEQNYTLPREITRNISLGSFAYESHRKALSHHAPSQGTELPQSNGISLPYTSNSFT | 60 | |
| | STG+S Y E++ +I +G AY+ + + P + NGI +T |
| pspA: | 778 | STGYSRSPYEPAPEVS-SIRMGISAYKGYAPQQASDIPGTVVPVVAENGIHPTFT----- | 831 |
|
| Orf115: | 61 | PLPSSSLYIINPVNKGYLVETDPRFANYRQWLGSDYMLDSLKLDPNNLHKRLGDGYYEQR | 120 |
| | LP+SSL+ I P NKGYL+ETDP F +YR+WLGS YML +L+ DPN++HKRLGDGYYEQ+ |
| pspA: | 832 | -LPNSSLFAIAPNNKGYLIETDPAFTDYRKWLGSGYMLAALQQDPNHIHKRLGDGYYEQK | 890 |
|
| Orf115: | 121 | LINEQIAELTGHRRLDGYQNDEEQFKALMDNGATAARSMNLSVGIALSAEQVAQLTSDIV | 180 |
| | L+NEQIA+LTG+RRLDGY NDEEQFKALMDNG T A+ + L+ GIALSAEQVA+LTSDIV |
| pspA: | 891 | LVNEQIAKLTGYRRLDGYTNDEEQFKALMDNGITIAKELQLTPGIALSAEQVARLTSDIV | 950 |
|
| Orf115: | 181 | WLVQKEVKLPDGGTQTVLVPQVYVRVKNGDIDGKGALLSGSNTQINVSGSLKN-SGTIAG | 239 |
| | WL + V LPDG TQTVL P+VYVR + D++G+GALLSGS I SG+++N G IAG |
| pspA: | 951 | WLENETVTLPDGTTQTVLKPKVYVRARPKDMNGQGALLSGSVVDIG-SGAIENRGGLIAG | 1009 |
|
| Orf115: | 240 | RNALIINTDTLDNIGGRIHAQKSAVTATQDINNIGGMLSAEQTLLLNAGXXXXXXXXXXX | 299 |
| | R ALI+N + N+ G + + A DI N G + AE LLL A |
| pspA: | 1010 | REALILNAQNIKNLQGDLQGKNIFAAAGSDITNTGS-IGAENALLLKASNNIESRSETRS | 1068 |
|
| Orf115: | 300 | XXXXXXXXXYLDRMAGIYITGKEKG | 324 |
| | + R+AGIY+TG++ G |
| pspA: | 1069 | NQNEQGSVRNIGRVAGIYLTGRQNG | 1093 |
Homology with a Predicted ORF from
N. gonorrhoeae [1143]ORF115 shows 91.9% identity over a 334aa overlap with a predicted ORF (ORF115ng) from N. gonorrhoeae:
[0000]
[1144]An ORF115ng nucleotide sequence <SEQ ID 513> was predicted to encode a protein having amino acid sequence <SEQ ID 514>:
[0000]| 1 | MLVQTEKDGL HNEQTFGEKK VFSENGKLHN YWRARRKGHD ETGHREQNYT | |
|
| 51 | LPEEITRDIS LGSFAYESHS KALSRHAPSQ GTELPQSNRD NIRTAKSNGI |
|
| 101 | SLPYTPNSFT PLPGSSLYII NPANKGYLVE TDPRFANYRQ WLGSDYMLGS |
|
| 151 | LKLDPNNLHK RLGDGYYEQR LINEQIAELT GHRRLDGYQN DEEQFKALMD |
|
| 201 | NGATAARSMN LSVGIALSAE QAAQLTSDIV WLVQKEVKLP DGGTQTVLMP |
|
| 251 | QVYVRVKNGG IDGKGALLSG SNTQINVSGS LKNSGTIAGR NALIINTDTL |
|
| 301 | DNIGGRIHAQ KSAVTATQDI NNIGGILSAE QTLLLNAGNN INNQSTAKSS |
|
| 351 | QNAQGSSTYL DRMAGIYITG KEKGVLAAQA GKDINIIAGQ ISNQSDQGQT |
|
| 401 | RLQAGRDINL DTVQTGKYQE IHFDADNHTI RGSTNEVGSS IQTKGDVTLL |
|
| 451 | SGNNLNAKAA EVGSAKGTLA VYAKNDITIS SGIHAGQVDD ASKHTGRSGG |
|
| 501 | GNKLVITDKA QSHHETAQSS TFEGKQVVLQ AGNDANILGS NVISDNGTRI |
|
| 551 | QAGNHVRIGT TQTQSQSETY HQTQKSGLMS AGIGFTIGSK TNTQENQSQS |
|
| 601 | NEHTGSTVGS LKGDTTIVAS KHYEQTGSNV SSPEGNNLIS TQSMDIGAAQ |
|
| 651 | NQLNSKTTQT YEQKGLTVAF SSPVTDLAQQ AIAVAHKAAK QFDKAKTTAL |
|
| 701 | MPWRLPMQVG RLFKQAKAPK K* |
[1145]Further work revealed the following partial gonococcal DNA sequence <SEQ ID 515>:
[0000]| 1 | TTGCTTGTGC AAACAGAAAA AGACGGTTTG CATAACGAGC AAACCTTTGG | |
|
| 51 | CGAGAAGAAA GTCTTCAGCG AAAATGGTAA GTTGCACAAC TACTGGCGTG |
|
| 101 | CGCGTCGTAA AGGACATGAT GAAACAGGGC ATCGTGAACA AAATTATACT |
|
| 151 | TTGCCGGAGG AAATCACACG CGACATTTCA CTGGGTTCAT TTGCCTATGA |
|
| 201 | ATCGCATAGC AAAGCATTAA GCCGTCATGC GCCCAGCCAA GGCACTGAGT |
|
| 251 | TGCCACAAAG TAACCGGGAT AATATCCGTA CTGCGAAAAG CAACGGTATT |
|
| 301 | TCGCTACCCT ATACGCCCAA TTCTTTTACC CCATTACCCG GCAGCAGCTT |
|
| 351 | ATACATTATC AATCCTGCCA ATAAAGGCTA TCTTGTTGAA ACCGATCCAC |
|
| 401 | GCTTTGCCAA CTACCGTCAA TGGTTGGGTA GTGACTATAT GCTGGGCAGC |
|
| 451 | CTCAAACTAG ACCCAAACAA TTTACATAAA CGTTTGGGTG ATGGTTATTA |
|
| 501 | CGAGCAACGT TTAATCAATG AACAAATCGC AGAGCTGACA GGGCATCGTC |
|
| 551 | GTTTAGACGG TTATCAAAAC GACGAAGAAC AATTTAAAGC CTTAATGGAT |
|
| 601 | AATGGCGCGA CTGCGGCACG TTCGATGAAT CTCAGCGTTG GCATTGCATT |
|
| 651 | AAGTGCCGAG CAAGCAGCGC AACTGACCAG CGATATTGTT TGGTTGGTAC |
|
| 701 | AAAAAGAAGT TAAACTTCCT GATGGCGGCA CACAAACCGT ATTGATGCCA |
|
| 751 | CAGGTTTATG TACGCGTTAA AAATGGCGGC ATAGACGGTA AAGGTGCATT |
|
| 801 | GTTGTCAGGC AGCAATACAC AAATCAATGT TTCAGGCAGC CTGAAAAACT |
|
| 851 | CAGGCACGAT TGCAGGGCGC AATGCGCTTA TTATCAATAC CGATACGCTA |
|
| 901 | GACAATATCG GTGGGCGTAT TCATGCGCAA AAATCAGCGG TTACGGCCAC |
|
| 951 | ACAAGACATC AATAATATTG GCGGCATTCT TTCTGCCGAA CAGACATTAT |
|
| 1001 | TGCTCAATGC GGGTAACAAC ATCAACAACC AAAGCACGGC CAAGAGCAGT |
|
| 1051 | CAAAATGCAC AAGGTAGCAG CACCTACCTA GACCGAATGG CAGGTATTTA |
|
| 1101 | TATCACAGGC AAAGAAAAAG GTGTTTTAGC AGCGCAGGCA GGCAAAGACA |
|
| 1151 | TCAACATCAT TGCCGGTCAA ATCAGCAATC AATCAGATCA AGGGCAAACC |
|
| 1201 | CGGCTGCAGG CAGGACGCGA CATTAACCTG GATACGGTAC AAACCGGCAA |
|
| 1251 | ATATCAAGAA ATCCATTTTG ATGCCGATAA CCATACCATC CGAGGTTCAA |
|
| 1301 | CGAACGAAGT CGGCAGCAGC ATTCAAACAA AAGGCGATGT TACCCtatTG |
|
| 1351 | TCAGGGAATA ATCTCAATGC CAAAGCTGCC GAAGTCGGCA GCGCAAAAGG |
|
| 1401 | CACACTTGCC GTGTATGCTA AAAATGACAT TACTATCAGC TCAGGCATCC |
|
| 1451 | ATGCCGGCCA AGTTGATGAT GCGTCCAAAC ATACAGGCAG AAGCGGCGGC |
|
| 1501 | GGTAATAAAT TAGTCATTAC CGATAAAGCC CAAAGTCATC ACGAAACTGC |
|
| 1551 | TCAAAGCAGC ACCTTTGAAG GCAAGCAAGT TGTATTGCAG GCAGGAAACG |
|
| 1601 | ATGCCAACAT CCTTGGCAGT AATGTTATTT CCGATAATGG CACCCGGATT |
|
| 1651 | CAAGCAGGCA ATCATGTTCG CATTGGTACA ACCCAAACTC AAAGCCAAAG |
|
| 1701 | CGAAACCTAT CATCAAACCC AAAAATCAGG ATTGATGAGT GCAGGTATCG |
|
| 1751 | GCTTCACTAT TGGCAGCAAG ACAAACACAC AAGAAAACCA ATCCCAAAGC |
|
| 1801 | AACGAACATA CAGGCAGTAC CGTAGGCAGC CTGAAAGGCG ATACCACCAT |
|
| 1851 | TGTTGCAAGC AAACACTACG AACAAACCGG CAGCAACGTT TCCAGCCCTG |
|
| 1901 | AGGGCAACAA CCTTATCAGC ACGCAAAGTA TGGATATTGG CGCAGCACAA |
|
| 1951 | AACCAATTAA ACAGCAAAAC CACCCAAACC TACGAACAAA AAGGCTTAAC |
|
| 2001 | GGTGGCATTC AGTTCGCCCG TTACCGATTT GGCACAACAA GCGATTGCCG |
|
| 2051 | TAGCACACAA AGCAGCAAAC AAGTCGGACA AAGCAAAAAC GACCGCGTTA |
|
| 2101 | ATGCCATGGC GGCTGCCAAT GCAGGTTGGC AGGCCTATCA AACAGGCAAA |
|
| 2151 | GGCGCACAAA ACTTAG |
[1146]This corresponds to the amino acid sequence <SEQ ID 516; ORF115ng-1>:
[0000]| 1 | LLVQTEKDGL HNEQTFGEKK VFSENGKLHN YWRARRKGHD ETGHREQNYT | |
|
| 51 | LPEEITRDIS LGSFAYESHS KALSRHAPSQ GTELPQSNRD NIRTAKSNGI |
|
| 101 | SLPYTPNSFT PLPGSSLYII NPANKGYLVE TDPRFANYRQ WLGSDYMLGS |
|
| 151 | LKLDPNNLHK RLGDGYYEQR LINEQIAELT GHRRLDGYQN DEEQFKALMD |
|
| 201 | NGATAARSMN LSVGIALSAE QAAQLTSDIV WLVQKEVKLP DGGTQTVLMP |
|
| 251 | QVYVRVKNGG IDGKGALLSG SNTQINVSGS LKNSGTIAGR NALIINTDTL |
|
| 301 | DNIGGRIHAQ KSAVTATQDI NNIGGILSAE QTLLLNAGNN INNQSTAKSS |
|
| 351 | QNAQGSSTYL DRMAGIYITG KEKGVLAAQA GKDINIIAGQ ISNQSDQGQT |
|
| 401 | RLQAGRDINL DTVQTGKYQE IHFDADNHTI RGSTNEVGSS IQTKGDVTLL |
|
| 451 | SGNNLNAKAA EVGSAKGTLA VYAKNDITIS SGIHAGQVDD ASKHTGRSGG |
|
| 501 | GNKLVITDKA QSHHETAQSS TFEGKQVVLQ AGNDANILGS NVISDNGTRI |
|
| 551 | QAGNHVRIGT TQTQSQSETY HQTQKSGLMS AGIGFTIGSK TNTQENQSQS |
|
| 601 | NEHTGSTVGS LKGDTTIVAS KHYEQTGSNV SSPEGNNLIS TQSMDIGAAQ |
|
| 651 | NQLNSKTTQT YEQKGLTVAF SSPVTDLAQQ AIAVAHKAAN KSDKAKTTAL |
|
| 701 | MPWRLPMQVG RPIKQAKAHK T* |
[1147]This gonococcal protein (ORF115ng-1) shows 91.9% identity with ORF 115 over 334aa:
[0000]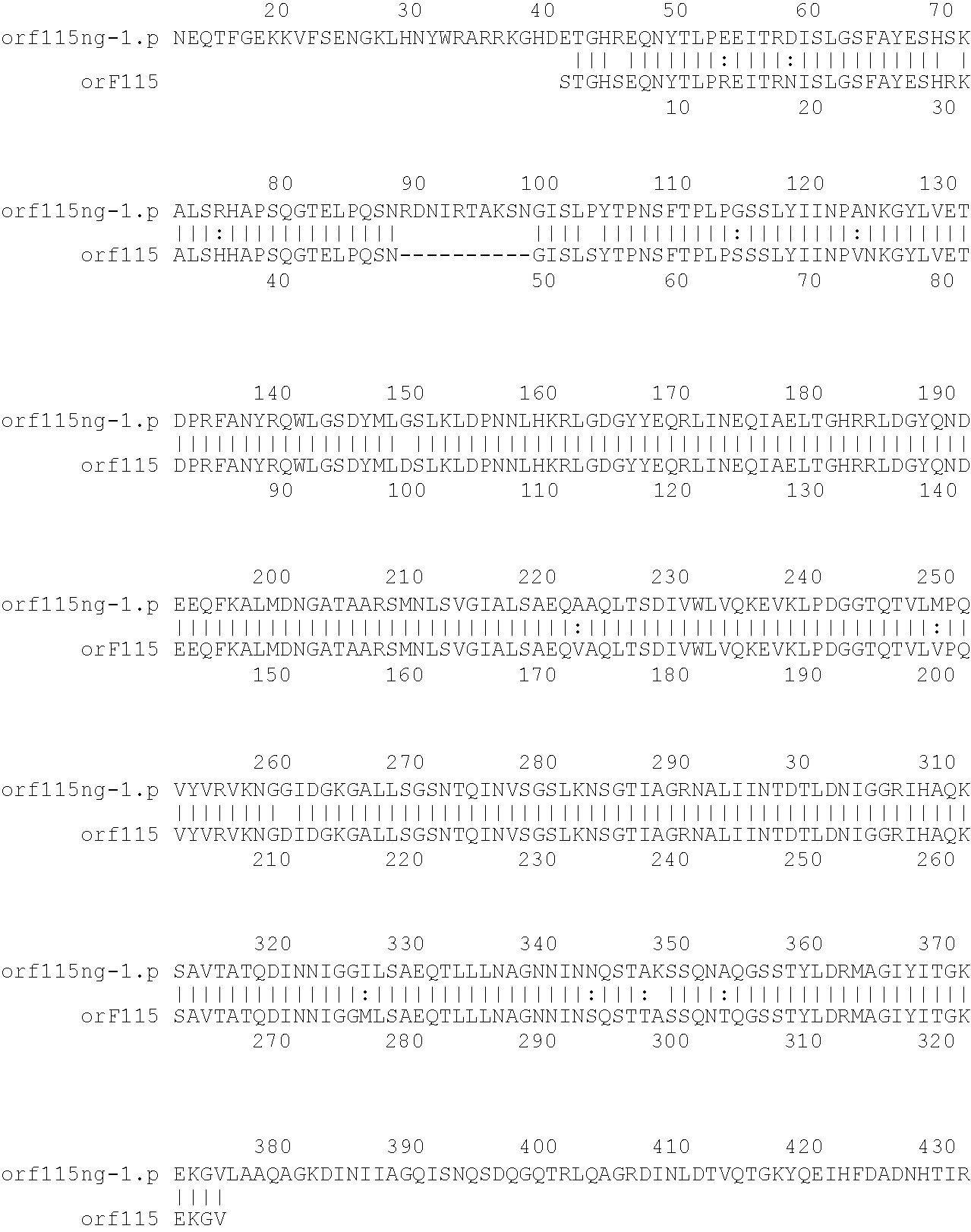
[1148]In addition, it shows homology with a secreted N. meningitidis protein in the database:
[0000]| gi|2623258 (AF030941) putative secreted protein | |
| [Neisseria meningitidis] Length = 2273 |
| Score = 604 bits (1541), Expect = e−172 |
| Identities = 325/678 (47%), Positives = 449/678 (65%), Gaps = 22/678 (3%) |
|
| Query: | 1 | LLVQTEKDGLHNEQTFGEKKVFSENGKLHNYWRARRKGHDETGHREQNYTLPEEITRDIS | 60 | |
| | L+V T + L N++T G K + ++ G LH Y R +KG D TG+ Y E++ I |
| Sbjct: | 739 | LIVGTPESALDNDETLGTKTI-TDKGDLHRYHRHHKKGRDSTGYSRSPYEPAPEVS-SIR | 796 |
|
| Query: | 61 | LGSFAYESHSKALSRHAPSQGTELPQSNRDNIRTAKSNGISLPYTPNSFTPLPGSSLYII | 120 |
| | +G AY+ + AP Q +++P + + NGI +T LP SSL+ I |
| Sbjct: | 797 | MGISAYKGY-------APQQASDIPGTV---VPVVAENGIHPTFT------LPNSSLFAI | 840 |
|
| Query: | 121 | NPANKGYLVETDPRFANYRQWLGSDYMLGSLKLDPNNLHKRLGDGYYEQRLINEQIAELT | 180 |
| | P NKGYL+ETDP F +YR+WLGS YML +L+ DPN++HKRLGDGYYEQ+L+NEQIA+LT |
| Sbjct: | 841 | APNNKGYLIETDPAFTDYRKWLGSGYMLAALQQDPNHIHKRLGDGYYEQKLVNEQIAKLT | 900 |
|
| Query: | 181 | GHRRLDGYQNDEEQFKALMDNGATAARSMNLSVGIALSAEQAAQLTSDIVWLVQKEVKLP | 240 |
| | G+RRLDGY NDEEQFKALMDNG T A+ + L+ GIALSAEQ A+LTSDIVWL + V LP |
| Sbjct: | 901 | GYRRLDGYTNDEEQFKALMDNGITIAKELQLTPGIALSAEQVARLTSDIVWLENETVTLP | 960 |
|
| Query: | 241 | DGGTQTVLMPQVYVRVKNGGIDGKGALLSGSNTQINVSGSLKN-SGTIAGRNALIINTDT | 299 |
| | DG TQTVL P+VYVR + ++G+GALLSGS I SG+++N G IAGR ALI+N |
| Sbjct: | 961 | DGTTQTVLKPKVYVRARPKDMNGQGALLSGSVVDIG-SGAIENRGGLIAGREALILNAQN | 1019 |
|
| Query: | 300 | LDNIGGRIHAQKSAVTATQDINNIGGILSAEQTLLLNAGNNINNQSTAKSSQNAQGSSTY | 359 |
| | + N+ G + + A DI N G I AE LLL A NNI ++S +S+QN QGS |
| Sbjct: | 1020 | IKNLQGDLQGKNIFAAAGSDITNTGSI-GAENALLLKASNNIESRSETRSNQNEQGSVRN | 1078 |
|
| Query: | 360 | LDRMAGIYITGKEKGVLAAQAGKDINIIAGQISNQSDQGQTRLQAGRDINLDTVQTGKYQ | 419 |
| | + R+AGIY+TG++ G + AG +I + A +++NQS+ GQT L AG DI DT + Q |
| Sbjct: | 1079 | IGRVAGIYLTGRQNGSVLLDAGNNIVLTASELTNQSEDGQTVLNAGGDIRSDTTGISRNQ | 1138 |
|
| Query: | 420 | EIHFDADNHTIRGSTNEVGSSIQTKGDVTLLSGNNLNAKAAEVGSAKGTLAVYAKNDITI | 479 |
| | FD+DN+ IR NEVGS+I+T+G+++L + ++ +AAEVGS +G L + A DI + |
| Sbjct: | 1139 | NTIFDSDNYVIRKEQNEVGSTIRTRGNLSLNAKGDIRIRAAEVGSEQGRLKLAAGRDIKV | 1198 |
|
| Query: | 480 | SSGIHAGQVDDASKHTGRSGGGNKLVITDKAQSHHETAQSSTFEGKQVVLQAGNDANILG | 539 |
| | +G + +DA K+TGRSGGG K +T ++ + A S T +GK+++L +G D + G |
| Sbjct: | 1199 | EAGKAHTETEDALKYTGRSGGGIKQKMTRHLKNQNGQAVSGTLDGKEIILVSGRDITVTG | 1258 |
|
| Query: | 540 | SNVISDNGTRIQAGNHVRIGTTQTQSQSETYHQTQKSGLM-SAGIGFTIGSKTNTQENQS | 598 |
| | SN+I+DN T + A N++ + +T+S+S ++ +KSGLM S GIGFT GSK +TQ N+S |
| Sbjct: | 1259 | SNIIADNHTILSAKNNIVLKAAETRSRSAEMNKKEKSGLMGSGGIGFTAGSKKDTQTNRS | 1318 |
|
| Query: | 599 | QSNEHTGSTVGSLKGDTTIVASKHYEQTGSNVSSPEGNNLISTQSMDIGAAQNQLNSKTT | 658 |
| | ++ HT S VGSL G+T I A KHY QTGS +SSP+G+ IS+ + I AAQN+ + ++ |
| Sbjct: | 1319 | ETVSHTESVVGSLNGNTLISAGKHYTQTGSTISSPQGDVGISSGKISIDAAQNRYSQESK | 1378 |
|
| Query: | 659 | QTYEQKGLTVAFSSPVTD | 676 |
| | Q YEQKG+TVA S PV + |
| Sbjct: | 1379 | QVYEQKGVTVAISVPVVN | 1396 |
[1149]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 62
[1150]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 517>:
[0000]| 1 | ..TCAGGGAATA ACCTCAATGC CAAAGCTGCC GAAGTCAGCA GCGCAAACGG | |
|
| 51 | TACACTCGCT GTGTCTGCCA ATAATGACAT CAACATCAGC GCAGGCATCA |
|
| 101 | ACACGACCCA TGTTGATGAT GCGTCCAAAC ACACAGGCAG AAGCGGTGGT |
|
| 151 | GGCAATAAAT TAGTCATTAC CGATAAAGCC CAAAGTCATC ACGAAACCGC |
|
| 201 | CCAAAGCAGC ACCTTTGAAG GCAAGCAAGT TGTATTGCAG GCAGGAAACG |
|
| 251 | ATGCCAACAT CCTTGGCAGC AATGTTATTT CCGATAATGG CACCCAGATT |
|
| 301 | CAAGCAGGCA ATCATGTTCG CATTGGTACA ACCCAAACTC AAAGCCAAAG |
|
| 351 | CGAAACCTAT CATCAAACCC AGAAATCAGG ATTGATGAGT GCAGGTATCG |
|
| 401 | GCTTCACTAT TGGCAGCAAG ACAAACACAC AAGAAAACCA ATCCCAAAGC |
|
| 451 | AACGAACATA CAGGCAGTAC CGTAGGCAGC TTGAAAGGCG ATACCACCAT |
|
| 501 | TGTTGCAGGC AAACACTACG AACAAATCGG CAGTACCGTT TCCAGCCCGG |
|
| 551 | AAGGCAACAA TACCATCTAT GCCCAAAGCA TAGACATTCA AGCGGCACAC |
|
| 601 | AACAAATTAA ACAGTAATAC CACCCAAACC TATGAACAAA AAGG.CTAAC |
|
| 651 | GGTGGCATTC AGTTCGCCCG TTACCGATTT GGCACAACAA ... |
[1151]This corresponds to the amino acid sequence <SEQ ID 518; ORF117>:
[0000]| 1 | ..SGNNLNAKAA EVSSANGTLA VSANNDINIS AGINTTHVDD ASKHTGRSGG | |
|
| 51 | GNKLVITDKA QSHHETAQSS TFEGKQVVLQ AGNDANILGS NVISDNGTQI |
|
| 101 | QAGNHVRIGT TQTQSQSETY HQTQKSGLMS AGIGFTIGSK TNTQENQSQS |
|
| 151 | NEHTGSTVGS LKGDTTIVAG KHYEQIGSTV SSPEGNNTIY AQSIDIQAAH |
|
| 201 | NKLNSNTTQT YEQKXLTVAF SSPVTDLAQQ ... |
[1152]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the pspA Putative Secreted Protein of N. meningitidis (Accession Number AF030941)
[1153]ORF117 and pspA protein show 45% aa identity in 224aa overlap:
[0000]| Orf117: | 4 | NLNAKAAEVSSANGTLAVSANNDINISAGINTTHVDDASKHTGRSGGGNKLVITDKAQSH | 63 | |
| | ++ +AAEV S G L ++A DI + AG T +DA K+TGRSGGG K +T ++ |
| pspA: | 1173 | DIRIRAAEVGSEQGRLKLAAGRDIKVEAGKAHTETEDALKYTGRSGGGIKQKMTRHLKNQ | 1232 |
|
| Orf117: | 64 | HETAQSSTFEGKQVVLQAGNDANILGSNVISDNGTQIQAGNHVRIGTTQTQSQSETYHQT | 123 |
| | + A S T +GK+++L +G D + GSN+I+DN T + A N++ + +T+S+S ++ |
| pspA: | 1233 | NGQAVSGTLDGKEIILVSGRDITVTGSNIIADNHTILSAKNNIVLKAAETRSRSAEMNKK | 1292 |
|
| Orf117: | 124 | QKSGLM-SAGIGFTIGSKTNTQENQSQSNEHTGSTVGSLKGDTTIVAGKHYEQIGSTVSS | 182 |
| | +KSGLM S GIGFT GSK +TQ N+S++ HT S VGSL G+T I AGKHY Q GST+SS |
| pspA: | 1293 | EKSGLMGSGGIGFTAGSKKDTQTNRSETVSHTESVVGSLNGNTLISAGKHYTQTGSTISS | 1352 |
|
| Orf117: | 183 | PEGNNTIYAQSIDIQAAHNKLNSNTTQTYEQKXLTVAFSSPVTD | 226 |
| | P+G+ I + I I AA N+ + + Q YEQK +TVA S PV + |
| pspA: | 1353 | PQGDVGISSGKISIDAAQNRYSQESKQVYEQKGVTVAISVPVVN | 1396 |
Homology with a Predicted ORF from
N. gonorrhoeae [1154]ORF117 shows 90% identity over a 230aa overlap with a predicted ORF (ORF117ng) from N. gonorrhoeae:
[0000]
[1155]An ORF117ng nucleotide sequence <SEQ ID 519> was predicted to encode a protein having amino acid sequence <SEQ ID 520>:
[0000]| 1 | ..LLVQTEKDGL HNEQTFGEKK VFSENGKLHN YWRARRKGHD ETGHREQNYT | |
|
| 51 | LPEEITRDIS LGSFAYESHS KALSRHAPSQ GTELPQSNRD NIRTAKSNGI |
|
| 101 | SLPYTPNSFT PLPGSSLYII NPANKGYLVE TDPRFANYRQ WLGSDYMLGS |
|
| 151 | LKLDPNNLHK RLGDGYYEQR LINEQIAELT GHRRLDGYQN DEEQFKALMD |
|
| 201 | NGATAARSMN LSVGIALSAE QAAQLTSDIV WLVQKEVKLP DGGTQTVLMP |
|
| 251 | QVYVRVKNGG IDGKGALLSG SNTQINVSGS LKNSGTIAGR NALIINTDTL |
|
| 301 | DNIGGRIHAQ KSAVTATQDI NNIGGILSAE QTLLLNAGNN INNQSTAKSS |
|
| 351 | QNAQGSSTYL DRMAGIYITG KEKGVLAAQA GKDINIIAGQ ISNQSDQGQT |
|
| 401 | RLQAGRDINL DTVQTGKYQE IHFDADNHTI RGSTNEVGSS IQTKGDVTLL |
|
| 451 | SGNNLNAKAA EVGSAKGTLA VYAKNDITIS SGIHAGQVDD ASKHTGRSGG |
|
| 501 | GNKLVITDKA QSHHETAQSS TFEGKQVVLQ AGNDANILGS NVISDNGTRI |
|
| 551 | QAGNHVRIGT TQTQSQSETY HQTQKSGLMS AGIGFTIGSK TNTQENQSQS |
|
| 601 | NEHTGSTVGS LKGDTTIVAS KHYEQTGSNV SSPEGNNLIS TQSMDIGAAQ |
|
| 651 | NQLNSKTTQT YEQKGLTVAF SSPVTDLAQQ AIAVAHKAAE QFDKAKTTAL |
|
| 701 | MPWRLPMQVG RLFKQAKAPK K* |
[1156]Further work revealed the following gonococcal partial DNA sequence <SEQ ID 521>:
[0000]| 1 | TTGCTTGTGC AAACAGAAAA AGACGGTTTG CATAACGAGC AAACCTTTGG | |
|
| 51 | CGAGAAGAAA GTCTTCAGCG AAAATGGTAA GTTGCACAAC TACTGGCGTG |
|
| 101 | CGCGTCGTAA AGGACATGAT GAAACAGGGC ATCGTGAACA AAATTATACT |
|
| 151 | TTGCCGGAGG AAATCACACG CGACATTTCA CTGGGTTCAT TTGCCTATGA |
|
| 201 | ATCGCATAGC AAAGCATTAA GCCGTCATGC GCCCAGCCAA GGCACTGAGT |
|
| 251 | TGCCACAAAG TAACCGGGAT AATATCCGTA CTGCGAAAAG CAACGGTATT |
|
| 301 | TCGCTACCCT ATACGCCCAA TTCTTTTACC CCATTACCCG GCAGCAGCTT |
|
| 351 | ATACATTATC AATCCTGCCA ATAAAGGCTA TCTTGTTGAA ACCGATCCAC |
|
| 401 | GCTTTGCCAA CTACCGTCAA TGGTTGGGTA GTGACTATAT GCTGGGCAGC |
|
| 451 | CTCAAACTAG ACCCAAACAA TTTACATAAA CGTTTGGGTG ATGGTTATTA |
|
| 501 | CGAGCAACGT TTAATCAATG AACAAATCGC AGAGCTGACA GGGCATCGTC |
|
| 551 | GTTTAGACGG TTATCAAAAC GACGAAGAAC AATTTAAAGC CTTAATGGAT |
|
| 601 | AATGGCGCGA CTGCGGCACG TTCGATGAAT CTCAGCGTTG GCATTGCATT |
|
| 651 | AAGTGCCGAG CAAGCAGCGC AACTGACCAG CGATATTGTT TGGTTGGTAC |
|
| 701 | AAAAAGAAGT TAAACTTCCT GATGGCGGCA CACAAACCGT ATTGATGCCA |
|
| 751 | CAGGTTTATG TACGCGTTAA AAATGGCGGC ATAGACGGTA AAGGTGCATT |
|
| 801 | GTTGTCAGGC AGCAATACAC AAATCAATGT TTCAGGCAGC CTGAAAAACT |
|
| 851 | CAGGCACGAT TGCAGGGCGC AATGCGCTTA TTATCAATAC CGATACGCTA |
|
| 901 | GACAATATCG GTGGGCGTAT TCATGCGCAA AAATCAGCGG TTACGGCCAC |
|
| 951 | ACAAGACATC AATAATATTG GCGGCATTCT TTCTGCCGAA CAGACATTAT |
|
| 1001 | TGCTCAATGC GGGTAACAAC ATCAACAACC AAAGCACGGC CAAGAGCAGT |
|
| 1051 | CAAAATGCAC AAGGTAGCAG CACCTACCTA GACCGAATGG CAGGTATTTA |
|
| 1101 | TATCACAGGC AAAGAAAAAG GTGTTTTAGC AGCGCAGGCA GGCAAAGACA |
|
| 1151 | TCAACATCAT TGCCGGTCAA ATCAGCAATC AATCAGATCA AGGGCAAACC |
|
| 1201 | CGGCTGCAGG CAGGACGCGA CATTAACCTG GATACGGTAC AAACCGGCAA |
|
| 1251 | ATATCAAGAA ATCCATTTTG ATGCCGATAA CCATACCATC CGAGGTTCAA |
|
| 1301 | CGAACGAAGT CGGCAGCAGC ATTCAAACAA AAGGCGATGT TACCCtatTG |
|
| 1351 | TCAGGGAATA ATCTCAATGC CAAAGCTGCC GAAGTCGGCA GCGCAAAAGG |
|
| 1401 | CACACTTGCC GTGTATGCTA AAAATGACAT TACTATCAGC TCAGGCATCC |
|
| 1451 | ATGCCGGCCA AGTTGATGAT GCGTCCAAAC ATACAGGCAG AAGCGGCGGC |
|
| 1501 | GGTAATAAAT TAGTCATTAC CGATAAAGCC CAAAGTCATC ACGAAACTGC |
|
| 1551 | TCAAAGCAGC ACCTTTGAAG GCAAGCAAGT TGTATTGCAG GCAGGAAACG |
|
| 1601 | ATGCCAACAT CCTTGGCAGT AATGTTATTT CCGATAATGG CACCCGGATT |
|
| 1651 | CAAGCAGGCA ATCATGTTCG CATTGGTACA ACCCAAACTC AAAGCCAAAG |
|
| 1701 | CGAAACCTAT CATCAAACCC AAAAATCAGG ATTGATGAGT GCAGGTATCG |
|
| 1751 | GCTTCACTAT TGGCAGCAAG ACAAACACAC AAGAAAACCA ATCCCAAAGC |
|
| 1801 | AACGAACATA CAGGCAGTAC CGTAGGCAGC CTGAAAGGCG ATACCACCAT |
|
| 1851 | TGTTGCAAGC AAACACTACG AACAAACCGG CAGCAACGTT TCCAGCCCTG |
|
| 1901 | AGGGCAACAA CCTTATCAGC ACGCAAAGTA TGGATATTGG CGCAGCACAA |
|
| 1951 | AACCAATTAA ACAGCAAAAC CACCCAAACC TACGAACAAA AAGGCTTAAC |
|
| 2001 | GGTGGCATTC AGTTCGCCCG TTACCGATTT GGCACAACAA GCGATTGCCG |
|
| 2051 | TAGCACACAA AGCAGCAAAC AAGTCGGACA AAGCAAAAAC GACCGCGTTA |
|
| 2101 | ATGCCATGGC GGCTGCCAAT GCAGGTTGGC AGGCCTATCA AACAGGCAAA |
|
| 2151 | GGCGCACAAA ACTTAG |
[1157]This corresponds to the amino acid sequence <SEQ ID 522; ORF117ng-1>:
[0000]| 1 | LLVQTEKDGL HNEQTFGEKK VFSENGKLHN YWRARRKGHD ETGHREQNYT | |
|
| 51 | LPEEITRDIS LGSFAYESHS KALSRHAPSQ GTELPQSNRD NIRTAKSNGI |
|
| 101 | SLPYTPNSFT PLPGSSLYII NPANKGYLVE TDPRFANYRQ WLGSDYMLGS |
|
| 151 | LKLDPNNLHK RLGDGYYEQR LINEQIAELT GHRRLDGYQN DEEQFKALMD |
|
| 201 | NGATAARSMN LSVGIALSAE QAAQLTSDIV WLVQKEVKLP DGGTQTVLMP |
|
| 251 | QVYVRVKNGG IDGKGALLSG SNTQINVSGS LKNSGTIAGR NALIINTDTL |
|
| 301 | DNTGGRIHAQ KSAVTATQDI NNIGGILSAE QTLLLNAGNN INNQSTAKSS |
|
| 351 | QNAQGSSTYL DRMAGIYITG KEKGVLAAQA GKDINIIAGQ ISNQSDQGQT |
|
| 401 | RLQAGRDINL DTVQTGKYQE IHFDADNHTI RGSTNEVGSS IQTKGDVTLL |
|
| 451 | SGNNLNAKAA EVGSAKGTLA VYAKNDITIS SGIHAGQVDD ASKHTGRSGG |
|
| 501 | GNKLVITDKA QSHHETAQSS TFEGKQVVLQ AGNDANILGS NVISDNGTRI |
|
| 551 | QAGNHVRIGT TQTQSQSETY HQTQKSGLMS AGIGFTIGSK TNTQENQSQS |
|
| 601 | NEHTGSTVGS LKGDTTIVAS KHYEQTGSNV SSPEGNNLIS TQSMDIGAAQ |
|
| 651 | NQLNSKTTQT YEQKGLTVAF SSPVTDLAQQ AIAVAHKAAN KSDKAKTTAL |
|
| 701 | MPWRLPMQVG RPIKQAKAHK T* |
[1158]ORF117ng-1 shows the same 90% identity over a 230aa overlap with ORF117. In addition, it shows homology with a secreted N. meningitidis protein in the database:
[0000]| gi|2623258 (AF030941) putative secreted protein [Neisseria meningitidis] | |
| Length = 2273 |
| Score = 604 bits (1541), Expect = e−172 |
| Identities = 325/678 (47%), Positives = 449/678 (65%), Gaps = 22/678 (3%) |
|
| Query: | 1 | LLVQTEKDGLHNEQTFGEKKVFSENGKLHNYWRARRKGHDETGHREQNYTLPEEITRDIS | 60 | |
| | L+V T + L N++T G K + ++ G LH Y R +KG D TG+ Y E++ I |
| Sbjct: | 739 | LIVGTPESALDNDETLGTKTI-TDKGDLHRYHRHHKKGRDSTGYSRSPYEPAPEVS-SIR | 796 |
|
| Query: | 61 | LGSFAYESHSKALSRHAPSQGTELPQSNRDNIRTAKSNGISLPYTPNSFTPLPGSSLYII | 120 |
| | +G AY+ + AP Q +++P + + NGI +T LP SSL+ I |
| Sbjct: | 797 | MGISAYKGY-------APQQASDIPGTV---VPVVAENGIHPTFT------LPNSSLFAI | 840 |
|
| Query: | 121 | NPANKGYLVETDPRFANYRQWLGSDYMLGSLKLDPNNLHKRLGDGYYEQRLINEQIAELT | 180 |
| | P NKGYL+ETDP F +YR+WLGS YML +L+ DPN++HKRLGDGYYEQ+L+NEQIA+LT |
| Sbjct: | 841 | APNNKGYLIETDPAFTDYRKWLGSGYMLAALQQDPNHIHKRLGDGYYEQKLVNEQIAKLT | 900 |
|
| Query: | 181 | GHARLDGYQNDEEQFKALMDNGATAARSMNLSVGIALSAEQAAQLTSDIVWLVQKEVKLP | 240 |
| | G+RRLDGY NDEEQFKALMDNG T A+ + L+ GIALSAEQ A+LTSDIVWL + V LP |
| Sbjct: | 901 | GYRRLDGYTNDEEQFKALMDNGITIAKELQLTPGIALSAEQVARLTSDIVWLENETVTLP | 960 |
|
| Query: | 241 | DGGTQTVLMPQVYVRVKNGGIDGKGALLSGSNTQINVSGSLKN-SGTIAGRNALIINTDT | 299 |
| | DG TQTVL P+VYVR + ++G+GALLSGS I SG+++N G IAGR ALI+N |
| Sbjct: | 961 | DGTTQTVLKPKVYVRARPKDMNGQGALLSGSVVDIG-SGAIENRGGLIAGREALILNAQN | 1019 |
|
| Query: | 300 | LDNIGGRIHAQKSAVTATQDINNIGGILSAEQTLLLNAGNNINNQSTAKSSONAQGSSTY | 359 |
| | + N+ G + + A DI N G I AE LLL A NNI ++S +S+QN QGS |
| Sbjct: | 1020 | IKNLQGDLQGKNIFAAAGSDITNTGSI-GAENALLLKASNNIESRSETRSNQNEQGSVRN | 1078 |
|
| Query: | 360 | LDRMAGIYITGKEKGVLAAQAGKDINIIAGQISNQSDQGQTRLQAGRDINLDTVQTGKYQ | 419 |
| | + R+AGIY+TG++ G + AG +I + A +++NQS+ GQT L AG DI DT + Q |
| Sbjct: | 1079 | IGRVAGIYLTGRQNGSVLLDAGNNIVLTASELTNQSEDGQTVLNAGGDIRSDTTGISRNQ | 1138 |
|
| Query: | 420 | EIHFDADNHTIRGSTNEVGSSIQTKGDVTLLSGNNLNAKAAEVGSAKGTLAVYAKNDITI | 479 |
| | FD+DN+ IR NEVGS+I+T+G+++L + ++ +AAEVGS +G L + A DI + |
| Sbjct: | 1139 | NTIFDSDNYVIRKEQNEVGSTIRTRGNLSLNAKGDIRIRAAEVGSEQGRLKLAAGRDIKV | 1198 |
|
| Query: | 480 | SSGIHAGQVDDASKHTGRSGGGNKLVITDKAQSHHETAQSSTFEGKQVVLQAGNDANILG | 539 |
| | +G + +DA K+TGRSGGG K +T ++ + A S T +GK+++L +G D + G |
| Sbjct: | 1199 | EAGKAHTETEDALKYTGRSGGGIKQKMTRHLKNQNGQAVSGTLDGKEIILVSGRDITVTG | 1258 |
|
| Query: | 540 | SNVISDNGTRIQAGNHVRIGTTQTQSQSETYHQTQKSGLM-SAGIGFTIGSKTNTQENQS | 598 |
| | SN+I+DN T + A N++ + +T+S+S ++ +KSGLM S GIGFT GSK +TQ N+S |
| Sbjct: | 1259 | SNIIADNHTILSAKNNIVLKAAETRSRSAEMNKKEKSGLMGSGGIGFTAGSKKDTQTNRS | 1318 |
|
| Query: | 599 | QSNEHTGSTVGSLKGDTTIVASKHYEQTGSNVSSPEGNNLISTQSMDIGAAQNQLNSKTT | 658 |
| | ++ HT S VGSL G+T I A KHY QTGS +SSP+G+ IS+ + I AAQN+ + ++ |
| Sbjct: | 1319 | ETVSHTESVVGSLNGNTLISAGKHYTQTGSTISSPQGDVGISSGKISIDAAQNRYSQESK | 1378 |
|
| Query: | 659 | QTYEQKGLTVAFSSPVTD | 676 |
| | Q YEQKG+TVA S PV + |
| Sbjct: | 1379 | QVYEQKGVTVAISVPVVN | 1396 |
[1159]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 63
[1160]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 523>:
[0000]| 1 | ATGATTTACA TCGTACTGTT TCTAGCTGTC GTCCTCGCCG TTGTCGCCTA | |
|
| 51 | CAACATGTAT CAGGAAAACC AATACCGCAA AAAAGTGCGC GACCAGTTCG |
|
| 101 | GACACTCCGA CAAAGATGCC CTGCTCAACA GCAwAACCAG CCATGTCCGC |
|
| 151 | GACGGCAAAC CGTCCGGCGG GTCAGTCATG ATGCCGAAAC CCCAACCGGC |
|
| 201 | GGTCAAAAAA ACGGCAAAAC CCCAAGACCC CGyCATGCGC AACCTGCAAG |
|
| 251 | AACAGGATGC CGTCTACATC GCCAAGCAGA AACAGGCAAA AGCCTCCCCG |
|
| 301 | TTCAAAACCG AAATCGAAAC CGCCTTGGAA GAAAGCGGCA TTATCGGCAA |
|
| 351 | CTCCGCCCAC ACCGTTTCCG AACCCCAAAC CGGACATTCC GCAACGAAAC |
|
| 401 | CTGCCGACGC GTCGGCAAAA CCTGCACCCG TTCCGCAAAC ACCTGCAAAA |
|
| 451 | CCGCTGATTA CGCTCAAAGA ACTGTCAAAA GTCGAATTAT CCTGGTTTGA |
|
| 501 | CGTGCGCATC GACTTCATCT CCTAT... |
[1161]This corresponds to the amino acid sequence <SEQ ID 524; ORF119>:
[0000]| 1 | MIYIVLFLAV VLAVVAYNMY QENQYRKKVR DQFGHSDKDA LLNSXTSHVR | |
|
| 51 | DGKPSGGSVM MPKPQPAVKK TAKPQDPXMR NLQEQDAVYI AKQKQAKASP |
|
| 101 | FKTEIETALE ESGIIGNSAH TVSEPQTGHS ATKPADASAK PAPVPQTPAK |
|
| 151 | PLITLKELSK VELSWFDVRI DFISY... |
[1162]Further work revealed the complete nucleotide sequence <SEQ ID 525>:
[0000]| 1 | ATGATTTACA TCGTACTGTT TCTAGCTGTC GTCCTCGCCG TTGTCGCCTA | |
|
| 51 | CAACATGTAT CAGGAAAACC AATACCGCAA AAAAGTGCGC GACCAGTTCG |
|
| 101 | GACACTCCGA CAAAGATGCC CTGCTCAACA GCAAAACCAG CCATGTCCGC |
|
| 151 | GACGGCAAAC CGTCCGGCGG GTCAGTCATG ATGCCGAAAC CCCAACCGGC |
|
| 201 | GGTCAAAAAA ACGGCAAAAC CCCAAGACCC CGCCATGCGC AACCTGCAAG |
|
| 251 | AACAGGATGC CGTCTACATC GCCAAGCAGA AACAGGCAAA AGCCTCCCCG |
|
| 301 | TTCAAAACCG AAATCGAAAC CGCCTTGGAA GAAAGCGGCA TTATCGGCAA |
|
| 351 | CTCCGCCCAC ACCGTTTCCG AACCCCAAAC CGGACATTCC GCACCGAAAC |
|
| 401 | CTGCCGACGC GCCGGCAAAA CCTGCACCCG TTCCGCAAAC ACCTGCAAAA |
|
| 451 | CCGCTGATTA CGCTCAAAGA ACTGTCAAAA GTCGAATTAC CCTGGTTTGA |
|
| 501 | CGTGCGCTTC GACTTCATCT CCTATATCGC GCTGACCGAA GCCAAAGAAC |
|
| 551 | TGCACGCACT GCCGCGCCTT TCCAACCGCT GCCGCTACCA GATTGTCGGC |
|
| 601 | TGCACCATGG ACGACCATTT CCAGATTGCC GAACCCATCC CGGGCATCCG |
|
| 651 | CTATCAGGCA TTTATCGTGG GTATTCAGGC AGTCAGCCGC AACGGACTTG |
|
| 701 | CCTCGCAGGA AGAACTCTCC GCATTCAACC GCCAGGTGGA CGCATTCGCA |
|
| 751 | CAAAGCATGG GCGGTCAGAC GCTGCACACC GACCTTGCCG CCTTTATCGA |
|
| 801 | AGTGGCTTCC GCACTGGACG CATTCTGCGC GCGCGTCGAC CAGACCATCG |
|
| 851 | CCATCCATTT GGTTTCCCCG ACCAGCATCA GCGGCGTAGA ACTGCGTTCC |
|
| 901 | GCCGTAACGG GCGTGGGTTT CGTTTTGGAA GACGACGGCG CGTTCCACTA |
|
| 951 | TACCGACACG TCGGGCTCGA CCATGTTCTC CATCTGCTCG CTCAACAACG |
|
| 1001 | AGCCGTTTAC CAACGCCCTT TTGGACAACC AGTCCTACAA AGGCTTCAGT |
|
| 1051 | ATGCTGCTCG ACATCCCGCA CTCTCCGGCA GGCGAAAAAA CCTTCGACGA |
|
| 1101 | TTTGTTTATG GATTTGGCGG TACGCCTGTC CGGCCAGTTG AACCTGAATC |
|
| 1151 | TGGTCAACGA CAAAATGGAA GAAGTTTCGA CCCAATGGCT CAAAGACGTG |
|
| 1201 | CGCACTTATG TATTGGCGCG TCAGTCCGAG ATGCTCAAAG TCGGTATCGA |
|
| 1251 | ACCGGGCGGC AAAACCGCAT TGCGCCTGTT CTCCTAA |
[1163]This corresponds to the amino acid sequence <SEQ ID 526; ORF119-1>:
[0000]| 1 | MIYIVLFLAV VLAVVAYNMY QENQYRKKVR DQFGHSDKDA LLNSKTSHVR | |
|
| 51 | DGKPSGGSVM MPKPQPAVKK TAKPQDPAMR NLQEQDAVYI AKQKQAKASP |
|
| 101 | FKTEIETALE ESGIIGNSAH TVSEPQTGHS APKPADAPAK PAPVPQTPAK |
|
| 151 | PLITLKELSK VELPWFDVRF DFISYIALTE AKELHALPRL SNRCRYQIVG |
|
| 201 | CTMDDHFQIA EPIPGIRYQA FIVGIQAVSR NGLASQEELS AFNRQVDAFA |
|
| 251 | QSMGGQTLHT DLAAFIEVAS ALDAFCARVD QTIAIHLVSP TSISGVELRS |
|
| 301 | AVTGVGFVLE DDGAFHYTDT SGSTMFSICS LNNEPFTNAL LDNQSYKGFS |
|
| 351 | MLLDIPHSPA GEKTFDDLFM DLAVRLSGQL NLNLVNDKME EVSTQWLKDV |
|
| 401 | RTYVLARQSE MLKVGIEPGG KTALRLFS* |
[1164]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1165]ORF119 shows 93.7% identity over a 175aa overlap with an ORF (ORF119a) from strain A of N. meningitidis:
[0000]
[1166]The complete length ORF119a nucleotide sequence <SEQ ID 527> is:
[0000]| 1 | ATGATTTACA TCGTACTGTT CCTCGCCGCC GTCCTCGCCG TTGTCGCCTA | |
|
| 51 | CAATATGTAT CAGGAAAACC AATACCGCAA AAAAGTGCGC GACCAGTTCG |
|
| 101 | GGCACTCCGA CAAAGATGCC CTGCTCAACA GCAAAACCAG CCATGTCCGC |
|
| 151 | GACGGCAAAC CGTCCGGCGG GCCAGTCATG ATGCCGAAAC CCCAACCGGC |
|
| 201 | GGTCAAAAAA ACGGCAAAAT CCCAAGACCC CGCCATGCGC AACCTGCAAG |
|
| 251 | AGCAGGATGC CGTCTACATC GCCAAGCAGA AACAGGCAAA AGCCTCCCCG |
|
| 301 | TTCAAAACCG AAATCGAAAC CGCCTTGGAA GAAAGCGGCA TTATCGGCAA |
|
| 351 | CTCCGCCCAC ACCGTTCCCG AACCCCAAAC CGGACATTCC GCACCAAAAC |
|
| 401 | CTGCCGACGC GCCGGCAAAA CCTGTTCCCG TTCCGCAAAC GCCGGCAAAA |
|
| 451 | CCGCTGATTA CGCTCAAAGA GCTGTCGAAG GTCGAGCTGC CCTGGTTTGA |
|
| 501 | CGTGCGCTTC GACTTCATCT CTTATATCGC GCTGACCGAA GCCAAAGAAC |
|
| 551 | TGCACGCACT GCCGCGCCTT TCCAACCGCT GCCGCTACCA GATTGTCGGC |
|
| 601 | TGCACCATGG ACGACCATTT CCAGATTGCC GAACCCATCC CGGGCATCCG |
|
| 651 | CTATCAGGCA TTTATCGTGG GTATTCAGGC AGTCAGCCGC AACGGACTTG |
|
| 701 | CCTCGCAGGA AGAACTCTCC GCATTCAACC GCCAGGTGGA TGCATTCGCA |
|
| 751 | CACAGCATGG GCGGTCAGAC GCTGCACACC GACCTTGCCG CCTTTATCGA |
|
| 801 | AGTGGCTTCC GCACTGGACG CATTCTGCGC GCGCGTCGAC CAGACTATCG |
|
| 851 | CCATCCATTT GGTTTCCCCG ACCAGCATCA GCGGCGTAGA ACTGCGTTCC |
|
| 901 | GCCGTAACGG GCGTGGGTTT CGTTTTGGAA GACGACGGCG CGTTCCACTA |
|
| 951 | TACCGACACG TCGGGCTCGA CCATGTTCTC CATCTGCTCG CTCAACAACG |
|
| 1001 | AGCCGTTTAC CAATGCCCTT TTGGACAACC AGTCCTATAA AGGCTTCAGT |
|
| 1051 | ATGCTGCTCG ACATCCCGCA CTCTCCGGCA GGCGAAAAAA CCTTCGACGA |
|
| 1101 | TTTGTTTATG GATTTGGCGG TACGCCTGTC CGGCCAGTTG AACCTGAATC |
|
| 1151 | TGGTCAACGA CAAAATGGAA GAAGTTTCGA CCCAATGGCT CAAAGACGTG |
|
| 1201 | CGCACTTATG TATTGGCTCG TCAGTCCGAG ATGCTCAAAG TCGGTATCGA |
|
| 1251 | ACCGGGCGGC AAAACCGCAT TGCGCCTGTT CTCCTAA |
[1167]This encodes a protein having amino acid sequence <SEQ ID 528>:
[0000]| 1 | MIYIVLFLAA VLAVVAYNMY QENQYRKKVR DQFGHSDKDA LLNSKTSHVR | |
|
| 51 | DGKPSGGPVM MPKPQPAVKK TAKSQDPAMR NLQEQDAVYI AKQKQAKASP |
|
| 101 | FKTEIETALE ESGIIGNSAH TVPEPQTGHS APKPADAPAK PVPVPQTPAK |
|
| 151 | PLITLKELSK VELPWFDVRF DFISYIALTE AKELHALPRL SNRCRYQIVG |
|
| 201 | CTMDDHFQIA EPIPGIRYQA FIVGIQAVSR NGLASQEELS AFNRQVDAFA |
|
| 251 | HSMGGQTLHT DLAAFIEVAS ALDAFCARVD QTIAIHLVSP TSISGVELRS |
|
| 301 | AVTGVGFVLE DDGAFHYTDT SGSTMFSICS LNNEPFTNAL LDNQSYKGFS |
|
| 351 | MLLDIPHSPA GEKTFDDLFM DLAVRLSGQL NLNLVNDKME EVSTQWLKDV |
|
| 401 | RTYVLARQSE MLKVGIEPGG KTALRLFS* |
[1168]ORF119a and ORF119-1 show 98.6% identity in 428 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1169]ORF119 shows 93.1% identity over a 175aa overlap with a predicted ORF (ORF119ng) from N. gonorrhoeae:
[0000]
[1170]The complete length ORF119ng nucleotide sequence <SEQ ID 529> is:
[0000]| 1 | ATGATTTACA TCGTACTGTT CCTCGCCGCC GTCCTCGCCG TTGTCGCCTA | |
|
| 51 | CAATATGTAT CAGGAAAACC AATACCGCAA AAAAGTGCGC GACCAGTTCG |
|
| 101 | GACACTCCGA CAAAGATGCC CTGCTCAACA GCAAAACCAG CCATGTCCGC |
|
| 151 | GACGGCAAAC CGTCCGGCGG GCCAGTCATG ATGCCGAAAC CCCAACCGGC |
|
| 201 | GGTCAAAAAA CCGGCCAAAC CCCAAGACTC CGCCATGCGC AACCTGCAAG |
|
| 251 | AACAGGATGC CGTCTACATC GCCAAGCAGA AACAGGCAAA AGCCTCCCCG |
|
| 301 | TTCAAAACCG AAATCGAAAC CGCCTTGGAA GAAATCGGCA TTATCGGCAA |
|
| 351 | CTCCGCCCAC ACCGTTTCCG AACCCCAAAC CGGACATTCC GCACCGAAAC |
|
| 401 | CTGCCGACGC GCCGGCAAAA CCCGTTCCCG TTCCGCAAAC GCCGGCAAAA |
|
| 451 | CCGCTGATTA CGCTCAAAGA GCTGTCGAAG GTCGAGCTGC CCTGGTTTGA |
|
| 501 | CGTGCGCTtc gACTTCATCT CCTATATCGC GCTGACCGAA GCCAAAGAAC |
|
| 551 | TGCACGCACT GCCGCGCCTT tccAACCGCT GCCGCTACCA GATTGTCGGC |
|
| 601 | TGCACCATGG ACGACCATTT CCAGATTGCC GAACCCATCC CGGGCATCCG |
|
| 651 | CTATCAGGCA TTTATCGTGG GTATCCAGGC AGTCAGCCGC AACGGACTTG |
|
| 701 | CCTCGCAGGA AGAACTCTCC GCATTCAACC GCCAGGCGGA CGCATTCGCA |
|
| 751 | CAAAGCATGG GCGGTCAGAC GCTGCACACC GACCTTGCCG CCTTTATCGA |
|
| 801 | AGTGGCTTCC GCACTGGACG CATTCTGCGC GCGCGTCGAC CAGACCATCG |
|
| 851 | CCATCCATTT GGTTTCGCCG ACCAGCATCA GCGGCGTAGA ACTGCGTTCC |
|
| 901 | GCCGTAACGG GCGTGGGTTT CGTTTTGGAA GACGACGGCG CGTTCCACTA |
|
| 951 | TACCGACACG TCGGGCTCGA CCATGTTCTC CATCTGCTCG CTCAACAACG |
|
| 1001 | AGCCGTTTAC CAATGCCCTT TTGGACAACC AGTCCTACAA AGGCTTCAGT |
|
| 1051 | ATGCTGCTCG ACATCCCGCA CTCTCCGGCA GGCGAAAAAA CCTTCGACGA |
|
| 1101 | TTTGTTTATG GATTTGGCGG TACGCCTGTC CGGTCAGTTG AACCTGAATC |
|
| 1151 | TGGTCAACGA CAAAATGGAA GAAGTTTCGA CCCAATGGCT CAAAGACGTA |
|
| 1201 | CGCACTTATG TATTGGCGCG TCAGTCCGAG ATGCTCAAAG TCGGTATCGA |
|
| 1251 | ACCGGGCGGC AAAACCGCCC TGCGCCTGTT TTCATAA |
[1171]This encodes a protein having amino acid sequence <SEQ ID 530>:
[0000]| 1 | MIYIVLFLAA VLAVVAYNMY QENQYRKKVR DQFGHSDKDA LLNSKTSHVR | |
|
| 51 | DGKPSGGPVM MPKPQPAVKK PAKPQDSAMR NLQEQDAVYI AKQKQAKASP |
|
| 101 | FKTEIETALE EIGIIGNSAH TVSEPQTGHS APKPADAPAK PVPVPQTPAK |
|
| 151 | PLITLKELSK VELPWFDVRF DFISYIALTE AKELHALPRL SNRCRYQIVG |
|
| 201 | CTMDDHFQIA EPIPGIRYQA FIVGIQAVSR NGLASQEELS AFNRQADAFA |
|
| 251 | QSMGGQTLHT DLAAFIEVAS ALDAFCARVD QTIAIHLVSP TSISGVELRS |
|
| 301 | AVTGVGFVLE DDGAFHYTDT SGSTMFSICS LNNEPFTNAL LDNQSYKGFS |
|
| 351 | MLLDIPHSPA GEKTFDDLFM DLAVRLSGQL NLNLVNDKME EVSTQWLKDV |
|
| 401 | RTYVLARQSE MLKVGIEPGG KTALRLFS* |
[1172]ORF119ng and ORF119-1 show 98.4% identity over 428 aa overlap:
[0000]
[1173]Based on this analysis, including the presence of a putative leader sequence in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 64
[1174]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 531>
[0000]| 1 | ..GCGCGGCACG GCACGGAAGA TTTCTTCATG AACAACAGCG ACAC.ATCAG | |
|
| 51 | GCAGATAGTC GAAAGCACCA CCGGTACGAT GAAGCTGCTG ATTTCCTCCA |
|
| 101 | TCGCCCTGAT TTCATTGGTA GTCGGCGGCA TCGGCGTGAT GAACATCATG |
|
| 151 | CTGGTGTCCG TTACCGAGCG CACCAAAGAA ATCGGCATAC GGATGGCAAT |
|
| 201 | CGGCGCGCGG CGCGGCAATA TTTyGCAGCA GTTTTTGATT GAGGCGGTGT |
|
| 251 | TAATCTGCGT CATCGGCGGT TTGGTCGGCG TGGGTTTGTC CGCCGCCGTC |
|
| 301 | AGCCTCGTGT TCAATCATTT TGTAACCGAC TTCCCGATGG ACATTTCCGC |
|
| 351 | CATGTCCGTC ATCGGCGCGG TCGCCTGTTC GACCGGAATC GGCATCGCGT |
|
| 401 | TCGGCTTTAT GCCTGCCAAT AAAGCAGCCA AACTCAATCC GATAGACGCA |
|
| 451 | TTGGCACAGG ATTGA |
[1175]This corresponds to the amino acid sequence <SEQ ID 532; ORF134>:
[0000]| 1 | ..ARHGTEDFFM NNSDXIRQIV ESTTGTMKLL ISSIALISLV VGGIGVMNIM | |
|
| 51 | LVSVTERTKE IGIRMAIGAR RGNIXQQFLI EAVLICVIGG LVGVGLSAAV |
|
| 101 | SLVFNHFVTD FPMDISAMSV IGAVACSTGI GIAFGFMPAN KAAKLNPIDA |
|
| 151 | LAQD* |
[1176]Further work revealed the complete nucleotide sequence <SEQ ID 533>:
[0000]| 1 | ATGTCGGTGC AAGCAGTATT GGCGCACAAA ATGCGTTCGC TTCTGACGAT | |
|
| 51 | GCTCGGCATC ATCATCGGTA TCGCGTCGGT GGTTTCCGTC GTCGCATTGG |
|
| 101 | GCAATGGTTC GCAGAAAAAA ATCCTTGAAG ACATCAGTTC GATAGGGACG |
|
| 151 | AACACCATCA GCATCTTCCC GGGGCGCGGC TTCGGCGACA GGCGCAGCGG |
|
| 201 | CAGGATTAAA ACCCTGACCA TAGACGACGC AAAAATCATC GCCAAACAAA |
|
| 251 | GCTACGTTGC TTCCGCCACG CCCATGACTT CGAGCGGCGG CACGCTGACT |
|
| 301 | TACCGCAACA CCGACCTGAC CGCCTCGCTT TACGGCGTGG GCGAACAATA |
|
| 351 | TTTCGACGTG CGCGGACTGA AGCTGGAAAC GGGGCGGCTG TTTGACGAAA |
|
| 401 | ACGATGTGAA AGAAGACGCG CAGGTCGTCG TCATCGACCA AAATGTCAAA |
|
| 451 | GACAAACTCT TTGCGGACTC GGATCCGTTG GGTAAAACCA TTTTGTTCAG |
|
| 501 | GAAACGCCCC TTGACCGTCA TCGGCGTGAT GAAAAAAGAC GAAAACGCTT |
|
| 551 | TCGGCAATTC CGACGTGCTG ATGCTTTGGT CGCCCTATAC GACGGTGATG |
|
| 601 | CACCAAATCA CAGGCGAGAG CCACACCAAC TCCATCACCG TCAAAATCAA |
|
| 651 | AGACAATGCC AATACCCAGG TTGCCGAAAA AGGGCTGACC GATCTGCTCA |
|
| 701 | AAGCGCGGCA CGGCACGGAA GATTTCTTCA TGAACAACAG CGACAGCATC |
|
| 751 | AGGCAGATAG TCGAAAGCAC CACCGGTACG ATGAAGCTGC TGATTTCCTC |
|
| 801 | CATCGCCCTG ATTTCATTGG TAGTCGGCGG CATCGGCGTG ATGAACATCA |
|
| 851 | TGCTGGTGTC CGTTACCGAG CGCACCAAAG AAATCGGCAT ACGGATGGCA |
|
| 901 | ATCGGCGCGC GGCGCGGCAA TATTTTGCAG CAGTTTTTGA TTGAGGCGGT |
|
| 951 | GTTAATCTGC GTCATCGGCG GTTTGGTCGG CGTGGGTTTG TCCGCCGCCG |
|
| 1001 | TCAGCCTCGT GTTCAATCAT TTTGTAACCG ACTTCCCGAT GGACATTTCC |
|
| 1051 | GCCATGTCCG TCATCGGCGC GGTCGCCTGT TCGACCGGAA TCGGCATCGC |
|
| 1101 | GTTCGGCTTT ATGCCTGCCA ATAAAGCAGC CAAACTCAAT CCGATAGACG |
|
| 1151 | CATTGGCACA GGATTGA |
[1177]This corresponds to the amino acid sequence <SEQ ID 534; ORF134-1>:
[0000]| 1 | MSVQAVLAHK MRSLLTMLGI IIGIASVVSV VALGNGSQKK ILEDISSIGT | |
|
| 51 | NTISIFPGRG FGDRRSGRIK TLTIDDAKII AKQSYVASAT PMTSSGGTLT |
|
| 101 | YRNTDLTASL YGVGEQYFDV RGLKLETGRL FDENDVKEDA QVVVIDQNVK |
|
| 151 | DKLFADSDPL GKTILFRKRP LTVIGVMKKD ENAFGNSDVL MLWSPYTTVM |
|
| 201 | HQITGESHTN SITVKIKDNA NTQVAEKGLT DLLKARHGTE DFFMNNSDSI |
|
| 251 | RQIVESTTGT MKLLISSIAL ISLVVGGIGV MNIMLVSVTE RTKEIGIRMA |
|
| 301 | IGARRGNILQ QFLIEAVLIC VIGGLVGVGL SAAVSLVFNH FVTDFPMDIS |
|
| 351 | AMSVIGAVAC STGIGIAFGF MPANKAAKLN PIDALAQD* |
[1178]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the Hypothetical Protein o648 of E. coli (Accession Number AE000189)
[1179]ORF134 and o648 protein show 45% aa identity in 153aa overlap:
[0000]| Orf134: | 2 | RHGTEDFFMNNSDXIRQIVESTTGTMKXXXXXXXXXXXVVGGIGVMNIMLVSVTERTKEI | 61 | |
| | RHG +DFF N D + + VE TT T++ VVGGIGVMNIMLVSVTERT+EI |
| o648: | 496 | RHGKKDFFTWNMDGVLKTVEKTTRTLQLFLTLVAVISLVVGGIGVMNIMLVSVTERTREI | 555 |
|
| Orf134: | 62 | GIRMAIGARRGNIXQQFLIEAXXXXXXXXXXXXXXXXXXXXXFNHFVTDFPMDISAMSVI | 121 |
| | GIRMA+GAR ++ QQFLIEA F+ + + S ++++ |
| o648: | 556 | GIRMAVGARASDVLQQFLIEAVLVCLVGGALGITLSLLIAFTLQLFLPGWEIGFSPLALL | 615 |
|
| Orf134: | 122 | GAVACSTGIGIAFGFMPANKAAKLNPIDALAQD | 154 |
| | A CST GI FG++PA AA+L+P+DALA++ |
| o648: | 616 | LAFLCSTVTGILFGWLPARNAARLDPVDALARE | 648 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1180]ORF134 shows 98.7% identity over a 154aa overlap with an ORF (ORF134a) from strain A of N. meningitidis:
[0000]
[1181]The complete length ORF134a nucleotide sequence <SEQ ID 535> is:
[0000]| 1 | ATGTCGGTGC AAGCAGTATT GGCGCACAAA ATGCGTTCGC TTCTGACGAT | |
|
| 51 | GCTCGGCATC ATCATCGGTA TCGCTTCGGT TGTCTCCGTC GTCGCATTGG |
|
| 101 | GCAACGGTTC GCAGAAAAAA ATCCTTGAAG ACATCAGTTC GATAGGGACG |
|
| 151 | AACACCATCA GCATCTTCCC AGGGCGCGGC TTCGGCGACA GGCGCAGCGG |
|
| 201 | CAGGATTAAA ACCCTGACCA TAGACGACGC AAAAATCATC GCCAAACAAA |
|
| 251 | GCTACGTTGC TTCCGCCACG CCCATGACTT CGAGCGGCGG CACGCTGACT |
|
| 301 | TACCGCAATA CCGACCTGAC CGCTTCTTTG TACGGTGTGG GCGAACAATA |
|
| 351 | TTTCGACGTG CGCGGGCTGA AGCTGGAAAC GGGGCGGCTG TTTGACGAAA |
|
| 401 | ACGATGTGAA AGAAGACGCG CAGGTCGTCG TCATCGACCA AAATGTCAAA |
|
| 451 | GACAAACTCT TTGCGGACTC GGATCCGTTG GGTAAAACCA TTTTGTTCAG |
|
| 501 | GAAACGCCCC TTGACCGTCA TCGGCGTGAT GAAAAAAGAC GAAAACGCTT |
|
| 551 | TCGGCAATTC CGACGTGCTG ATGCTTTGGT CGCCCTATAC GACGGTGATG |
|
| 601 | CACCAAATCA CAGGCGAGAG CCACACCAAC TCCATCACCG TCAAAATCAA |
|
| 651 | AGACAATGCC AATACCCAGG TTGCCGAAAA AGGGCTGACC GATCTGCTCA |
|
| 701 | AAGCGCGGCA CGGCACGGAA GATTTCTTCA TGAACAACAG CGACAGCATC |
|
| 751 | AGGCAGATAG TCGAAAGCAC CACCGGTACG ATGAAGCTGC TGATTTCCTC |
|
| 801 | CATCGCCCTG ATTTCATTGG TAGTCGGCGG CATCGGCGTG ATGAACATCA |
|
| 851 | TGCTGGTGTC CGTTACCGAG CGCACCAAAG AAATCGGCAT ACGGATGGCA |
|
| 901 | ATCGGCGCGC GGCGCGGCAA TATTTTGCAG CAGTTTTTGA TTGAGGCGGT |
|
| 951 | GTTAATCTGC GTCATCGGCG GTTTGGTCGG CGTGGGTTTG TCCGCCGCCG |
|
| 1001 | TCAGCCTCGT GTTCAATCAT TTTGTAACCG ACTTCCCGAT GGACATTTCC |
|
| 1051 | GCCATGTCCG TCATCGGCGC GGTCGCCTGT TCGACCGGAA TCGGCATCGC |
|
| 1101 | GTTCGGCTTT ATGCCTGCCA ATAAAGCAGC CAAACTCAAT CCGATAGATG |
|
| 1151 | CATTGGCGCA GGATTGA |
[1182]This encodes a protein having amino acid sequence <SEQ ID 536>:
[0000]| 1 | MSVQAVLAHK MRSLLTMLGI IIGIASVVSV VALGNGSQKK ILEDISSIGT | |
|
| 51 | NTISIFPGRG FGDRRSGRIK TLTIDDAKII AKQSYVASAT PMTSSGGTLT |
|
| 101 | YRNTDLTASL YGVGEQYFDV RGLKLETGRL FDENDVKEDA QVVVIDQNVK |
|
| 151 | DKLFADSDPL GKTILFRKRP LTVIGVMKKD ENAFGNSDVL MLWSPYTTVM |
|
| 201 | HQITGESHTN SITVKIKDNA NTQVAEKGLT DLLKARHGTE DFFMNNSDSI |
|
| 251 | RQIVESTTGT MKLLISSIAL ISLVVGGIGV MNIMLVSVTE RTKEIGIRMA |
|
| 301 | IGARRGNILQ QFLIEAVLIC VIGGLVGVGL SAAVSLVFNH FVTDFPMDIS |
|
| 351 | AMSVIGAVAC STGIGIAFGF MPANKAAKLN PIDALAQD* |
[1183]ORF134a and ORF134-1 show 100.0% identity in 388 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1184]ORF134 shows 96.8% identity over a 154aa overlap with a predicted ORF (ORF134.ng) from N. gonorrhoeae:
[0000]
[1185]The complete length ORF134ng nucleotide sequence <SEQ ID 537> is:
[0000]| 1 | ATGTCGGTGC AAGCAGTATT GGCGCACAAA ATGCGTTCGC TTCTGACCAT | |
|
| 51 | GCTCGGCATC ATCATCGGTA TCGCTTCGGT TGTCTCCGTC GTCGCGCTGG |
|
| 101 | GCAACGGTTC GCAGAAAAAA ATCCTCGAAG ACATCAGTTC GATGGGGACG |
|
| 151 | AACACCATCA GCATCTTCCC CGGGCGCGGC TTCGGCGACA GGCGCAGCGG |
|
| 201 | CAAAATCAAA ACCCTGACCA TAGACGACGC AAAAATCATC GCCAAACAAA |
|
| 251 | GCTACGTTGC CTCCGCCACG CCCATGACTT CGAGCGGCGG CACGCTGACC |
|
| 301 | TACCGCAATA CCGACCTGAC CGCTTCTTTG TACGGTGTGG GCGAACAATA |
|
| 351 | TTTCGACGTG CGCGGGCTGA AGCTGGAAAC GGGGCGGCTG TTTGATGAGA |
|
| 401 | ACGATGTGAA AGAAGACGCG CAAGTCGTCG TCATCGACCA AAATGTCAAA |
|
| 451 | GACAAACTCT TTGCGGACTC GGATCCGTTG GGTAAAACCA TTTTGTTCAG |
|
| 501 | GAAACGCCCC TTGACCGTCA TCGGCGTGAT GAAAAAAGAC GAAAACGCTT |
|
| 551 | TCGGCAATTC CGACGTGCTG ATGCTTTGGT CGCCCTATAC GACGGTGATG |
|
| 601 | CACCAAATCA CAGGCGAGAG CCACACCAAC TCCATCACCG TCAAAATCAA |
|
| 651 | AGACAATGCC AATACCCGGG TTGCCGAAAA AGGGCTGGCC GAGCTGCTCA |
|
| 701 | AAGCACGGCA CGGCACGGAA GACTTCTTTA TGAACAACAG CGACAGCATC |
|
| 751 | AGGCAGATGG TCGAAAGCAC CACCGGTACG ATGAAGCTGC TGATTTCCTC |
|
| 801 | CATCGCCCTG ATTTCATTGG TAGTCGGCGG CATCGGTGTG ATGAACATTA |
|
| 851 | TGCTGGTGTC CGTTACCGAG CGCACCAAAG AAATCGGCAT ACGGATGGCA |
|
| 901 | ATCGGCGCGC GGCGCGGCAA TATTTTGCAG CAGTTTTTGA TTGAGGCGGT |
|
| 951 | GTTAATCTGC ATCATCGGAG GCTTGGTCGG CGTAGGTTTG TCCGCCGCCG |
|
| 1001 | TCAGCCTCGT GTTCAATCAT TTTGTAACCG ATTTCCCGAT GGACATTTCG |
|
| 1051 | GCGGCATCCG TTATCGGGGC GGTCGCCTGT TCGACCGGAA TCGGCATCGC |
|
| 1101 | GTTCGGCTTT ATGCCTGCCA ATAAGGCAGC CAAACTCAAT CCGATAGATG |
|
| 1151 | CATTGGCGCA GGATTGA |
[1186]This encodes a protein having amino acid sequence <SEQ ID 538>:
[0000]| 1 | MSVQAVLAHK MRSLLTMLGI IIGIASVVSV VALGNGSQKK ILEDISSMGT | |
|
| 51 | NTISIFPGRG FGDRRSGKIK TLTIDDAKII AKQSYVASAT PMTSSGGTLT |
|
| 101 | YRNTDLTASL YGVGEQYFDV RGLKLETGRL FDENDVKEDA QVVVIDQNVK |
|
| 151 | DKLFADSDPL GKTILFRKRP LTVIGVMKKD ENAFGNSDVL MLWSPYTTVM |
|
| 201 | HQITGESHTN SITVKIKDNA NTRVAEKGLA ELLKARHGTE DFFMNNSDSI |
|
| 251 | RQMVESTTGT MKLLISSIAL ISLVVGGIGV MNIMLVSVTE RTKEIGIRMA |
|
| 301 | IGARRGNILQ QFLIEAVLIC IIGGLVGVGL SAAVSLVFNH FVTDFPMDIS |
|
| 351 | AASVIGAVAC STGIGIAFGF MPANKAAKLN PIDALAQD* |
[1187]ORF134ng and ORF134-1 show 97.9% identity in 388 aa overlap:
[0000]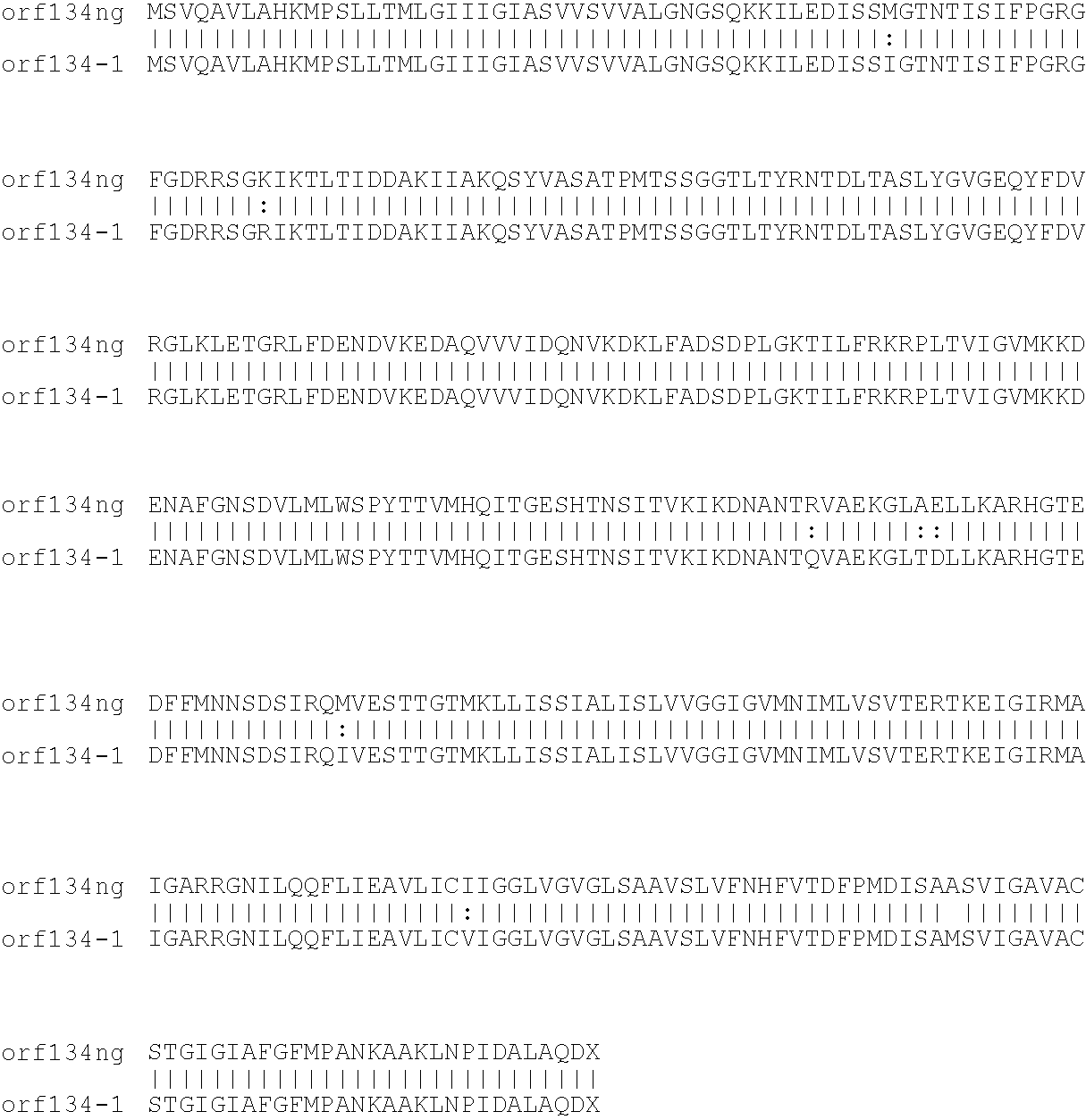
[1188]ORF134ng also shows homology to an E. coli ABC transporter:
[0000]| sp|P75831|YBJZ_ECOLI HYPOTHETICAL ABC TRANSPORTER ATP-BINDING | |
| PROTEIN YBJZ >gi5 (AE000189) 0648; similar to YBBA_HAEIN SW: |
| P45247 [Escherichia coli] Length = 648 |
| Score = 297 bits (753), Expect = 6e−80 |
| Identities = 162/389 (41%), Positives = 230/389 (58%), Gaps = 1/389 (0%) |
|
| Query: | 1 | MSVQAVLAHKMRSLLTMLXXXXXXXXXXXXXXLGNGSQKKILEDISSMGTNTISIFPGRG | 60 | |
| | M+ +A+ A+KMR+LLTML +G+ +++ +L DI S+GTNTI ++PG+ |
| Sbjct: | 260 | MAWRALAANKMRTLLTMLGIIIGIASVVSIVVVGDAAKQMVLADIRSIGTNTIDVYPGKD | 319 |
|
| Query: | 61 | FGDRRSGKIKTLTIDDAKIIAKQSYVASATPMTSSGGTLTYRNTDLTASLYGVGEQYFDV | 120 |
| | FGD + L DD I KQ +VASATP S L Y N D+ AS GV YF+V |
| Sbjct: | 320 | FGDDDPQYQQALKYDDLIAIQKQPWVASATPAVSQNLRLRYNNVDVAASANGVSGDYFNV | 379 |
|
| Query: | 121 | RGLKLETGRLFDENDVKEDAQVVVIDQNVKDKLFAD-SDPLGKTILFRKRPLTVIGVMKK | 179 |
| | G+ G F++ + AQVVV+D N + +LF +D +G+ IL P VIGV ++ |
| Sbjct: | 380 | YGMTFSEGNTFNQEQLNGRAQVVVLDSNTRRQLFPHKADVVGEVILVGNMPARVIGVAEE | 439 |
|
| Query: | 180 | DENAFGNSDVLMLWSPYTTVMHQITGESHTNSITVKIKDNANTRVAEKGLAELLKARHGT | 239 |
| | ++ FG+S VL +W PY+T+ ++ G+S NSITV++K+ ++ AE+ L LL REG |
| Sbjct: | 440 | KQSMFGSSKVLRVWLPYSTMSGRVMGQSWLNSITVRVKEGFDSAEAEQQLTRLLSLRHGK | 499 |
|
| Query: | 240 | EDFFMNNSDSIRQMVESTTGTMKXXXXXXXXXXXVVGGIGVMNIMLVSVTERTKEIGIRM | 299 |
| | +DFF N D + + VE TT T++ VVGGIGVMNIMLVSVTERT+EIGIRM |
| Sbjct: | 500 | KDFFTWNMDGVLKTVEKTTRTLQLFLTLVAVISLVVGGIGVMNIMLVSVTERTREIGIRM | 559 |
|
| Query: | 300 | AIGARRGNILQQFLIEXXXXXXXXXXXXXXXXXXXXXXFNHFVTDFPMDISAASVIGAVA | 359 |
| | A+GAR ++LQQFLIE F+ + + S +++ A |
| Sbjct: | 560 | AVGARASDVLQQFLIEAVLVCLVGGALGITLSLLIAFTLQLFLPGWEIGFSPLALLLAFL | 619 |
|
| Query: | 360 | CSTGIGIAFGFMPANKAAKLNPIDALAQD | 388 |
| | CST GI FG++PA AA+L+P+DALA++ |
| Sbjct: | 620 | CSTVTGILFGWLPARNAARLDPVDALARE | 648 |
[1189]Based on this analysis, including the presence of the leader peptide and transmembrane regions in the gonococcal protein, it is prediceted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 65
[1190]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 539>:
[0000]| 1 | ..GGGACGGGAG CGATGCTGCT GCTGTTTTAC GCGGTAACGA T.CTGCCTTT | |
|
| 51 | GGCCACTGGC GTTACCCTGA GTTACACCTC GTCGATTTTT TTGGCGGTAT |
|
| 101 | TTTCCTTCCT GATTTTGAAA GAACGGATTT CCGTTTACAC GCAGGCGGTG |
|
| 151 | CTGCTCCTTG GTTTTGCCGG CGTGGTATTG CTGCTTAATC CCTCGTTCCG |
|
| 201 | CAGCGGTCAG GAAACGGCGG CACTCGCCGG GCTGGCGGGC GGCGCGATGT |
|
| 251 | CCGGCTGGGC GTATTTGAAA GTGCGCGAAC TGTCTTTGGC GGGCGAACCC |
|
| 301 | GGCTGGCGCG TCGTGTTTTA CCTTTCCGTG ACAGGTGTGG CGATGTCGTC |
|
| 351 | GGTTTGGGCG ACGCTGACCG GCTGGCACAC CCTGTCCTTT CCATCGGCAG |
|
| 401 | TTTATCTGTC GTGCATCGGC GTGTCCGCGC TGATTGCCCA ACTGTCGATG |
|
| 451 | ACGCGCGCCT ACAAAGTCGG CGACAAATTC ACGGTTGCCT CGCTTTCCTA |
|
| 501 | TATGACCGTC GTTTTTTCCG CTCTGTCTGC CGCATTTTTT CTGGGCGAAG |
|
| 551 | AGCTTTTCTG GCAGGAAATA CTCGGTATGT GCATCATCAT CCTCAGCGGT |
|
| 601 | ATTTTGA |
[1191]This corresponds to the amino acid sequence <SEQ ID 540; ORF135>:
[0000]| 1 | ..GTGAMLLLFY AVTILPLATG VTLSYTSSIF LAVFSFLILK ERISVYTQAV | |
|
| 51 | LLLGFAGVVL LLNPSFRSGQ ETAALAGLAG GAMSGWAYLK VRELSLAGEP |
|
| 101 | GWRVVFYLSV TGVAMSSVWA TLTGWHTLSF PSAVYLSCIG VSALIAQLSM |
|
| 151 | TRAYKVGDKF TVASLSYMTV VFSALSAAFF LGEELFWQEI LGMCIIISAV |
|
| 201 | F* |
[1192]Further work revealed the complete nucleotide sequence <SEQ ID 541>:
[0000]| 1 | ATGGATACCG CAAAAAAAGA CATTTTAGGA TCGGGCTGGA TGCTGGTGGC | |
|
| 51 | GGCGGCCTGC TTTACCATTA TGAACGTATT GATTAAAGAG GCATCGGCAA |
|
| 101 | AATTTGCCCT CGGCAGCGGC GAATTGGTCT TTTGGCGCAT GCTGTTTTCA |
|
| 151 | ACCGTTGCGC TCGGGGCTGC CGCCGTATTG CGTCGGGACA mCTTCCGCAC |
|
| 201 | GCCCCATTGG AAAAACCACT TAAACCGCAG TATGGTCGGG ACGGGGGCGA |
|
| 251 | TGCTGCTGCT GTTTTACGCG GTAACGCATC TGCCTTTGGC CACTGGCGTT |
|
| 301 | ACCCTGAGTT ACACCTCGTC GATTTTTTTG GCGGTATTTT CCTTCCTGAT |
|
| 351 | TTTGAAAGAA CGGATTTCCG TTTACACGCA GGCGGTGCTG CTCCTTGGTT |
|
| 401 | TTGCCGGCGT GGTATTGCTG CTTAATCCCT CGTTCCGCAG CGGTCAGGAA |
|
| 451 | ACGGCGGCAC TCGCCGGGCT GGCGGGCGGC GCGATGTCCG GCTGGGCGTA |
|
| 501 | TTTGAAAGTG CGCGAACTGT CTTTGGCGGG CGAACCCGGC TGGCGCGTCG |
|
| 551 | TGTTTTACCT TTCCGTGACA GGTGTGGCGA TGTCGTCGGT TTGGGCGACG |
|
| 601 | CTGACCGGCT GGCACACCCT GTCCTTTCCA TCGGCAGTTT ATCTGTCGTG |
|
| 651 | CATCGGCGTG TCCGCGCTGA TTGCCCAACT GTCGATGACG CGCGCCTACA |
|
| 701 | AAGTCGGCGA CAAATTCACG GTTGCCTCGC TTTCCTATAT GACCGTCGTT |
|
| 751 | TTTTCCGCTC TGTCTGCCGC ATTTTTTCTG GGCGAAGAGC TTTTCTGGCA |
|
| 801 | GGAAATACTC GGTATGTGCA TCATCATCCT CAGCGGTATT TTGAGCAGCA |
|
| 851 | TCCGCCCCAC TGCCTTCAAA CAGCGGCTGC AATCCCTGTT CCGCCAAAGA |
|
| 901 | TAA |
[1193]This corresponds to the amino acid sequence <SEQ ID 542; ORF135-1>:
[0000]| 1 | MDTAKKDILG SGWMLVAAAC FTIMNVLIKE ASAKFALGSG ELVFWRMLFS | |
|
| 51 | TVALGAAAVL RRDXFRTPHW KNHLNRSMVG TGAMLLLFYA VTHLPLATGV |
|
| 101 | TLSYTSSIFL AVFSFLILKE RISVYTQAVL LLGFAGVVLL LNPSFRSGQE |
|
| 151 | TAALAGLAGG AMSGWAYLKV RELSLAGEPG WRVVFYLSVT GVAMSSVWAT |
|
| 201 | LTGWHTLSFP SAVYLSCIGV SALIAQLSMT RAYKVGDKFT VASLSYMTVV |
|
| 251 | FSALSAAFFL GEELFWQEIL GMCIIILSGI LSSIRPTAFK QRLQSLFRQR |
|
| 301 | * |
[1194]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1195]ORF135 shows 99.0% identity over a 197aa overlap with an ORF (ORF135a) from strain A of N. meningitidis:
[0000]
[1196]The complete length ORF135a nucleotide sequence <SEQ ID 543> is:
[0000]| 1 | ATGGATACCG CAAAAAAAGA CATTTTAGGA TCGGGCTGGA TGCTGGTGGC | |
|
| 51 | GGCGGCCTGC TTTACCATTA TGAACGTATT GATTAAAGAG GCATCGGCAA |
|
| 101 | AATTTGCCCT CGGCAGCGGC GAATTGGTCT TTTGGCGCAT GCTGTTTTCA |
|
| 151 | ACCGTTGCGC TCGGGGCTGC CGCCGTATTG CGTCGGGACA CCTTCCGCAC |
|
| 201 | GCCCCATTGG AAAAACCACT TAAACCGCAG TATGGTCGGG ACGGGGGCGA |
|
| 251 | TGCTGCTGCT GTTTTACGCG GTAACGCATC TGCCTTTGGC CACCGGCGTT |
|
| 301 | ACCCTGAGTT ACACCTCGTC GATTTTTTTG GCGGTATTTT CCTTCCTGAT |
|
| 351 | TTTGAAAGAA CGGATTTCCG TTTACACGCA GGCGGTGCTG CTCCTTGGTT |
|
| 401 | TTGCCGGCGT GGTATTGCTG CTTAATCCCT CGTTCCGCAG CGGTCAGGAA |
|
| 451 | ACGGCGGCAC TCGCCGGGCT GGCGGGCGGC GCGATGTCCG GCTGGGCGTA |
|
| 501 | TTTGAAAGTG CGCGAACTGT CTTTGGCGGG CGAACCCGGC TGGCGCGTCG |
|
| 551 | TGTTTTACCT TTCCGTGACA GGTGTGGCGA TGTCATCGGT TTGGGCGACG |
|
| 601 | CTGACCGGCT GGCACACCCT GTCCTTTCCA TCGGCAGTTT ATCTGTCGTG |
|
| 651 | CATCGGCGTG TCCGCGCTGA TTGCCCAACT GTCGATGACG CGCGCCTACA |
|
| 701 | AAGTCGGCGA CAAATTCACG GTTGCCTCGC TTTCCTATAT GACCGTCGTT |
|
| 751 | TTTTCCGCTC TGTCTGCCGC ATTTTTTCTG GCCGAAGAGC TTTTCTGGCA |
|
| 801 | GGAAATACTC GGTATGTGCA TCATCATCCT CAGCGGTATT TTGAGCAGCA |
|
| 851 | TCCGCCCCAC TGCCTTCAAA CAGCGGCTGC AATCCCTGTT CCGCCAAAGA |
|
| 901 | TAA |
[1197]This encodes a protein having amino acid sequence <SEQ ID 544>:
[0000]| 1 | MDTAKKDILG SGWMLVAAAC FTIMNVLIKE ASAKFALGSG ELVFWRMLFS | |
|
| 51 | TVALGAAAVL RRDTFRTPHW KNHLNRSMVG TGAMLLLFYA VTHLPLATGV |
|
| 101 | TLSYTSSIFL AVFSFLILKE RISVYTQAVL LLGFAGVVLL LNPSFRSGQE |
|
| 151 | TAALAGLAGG AMSGWAYLKV RELSLAGEPG WRVVFYLSVT GVAMSSVWAT |
|
| 201 | LTGWHTLSFP SAVYLSCIGV SALIAQLSMT RAYKVGDKFT VASLSYMTVV |
|
| 251 | FSALSAAFFL AEELFWQEIL GMCIIILSGI LSSIRPTAFK QRLQSLFRQR |
|
| 301 | * |
[1198]ORF135a and ORF135-1 show 99.3% identity in 300 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1199]ORF135 shows 97% identity over a 201aa overlap with a predicted ORF (ORF135ng) from N. gonorrhoeae:
[0000]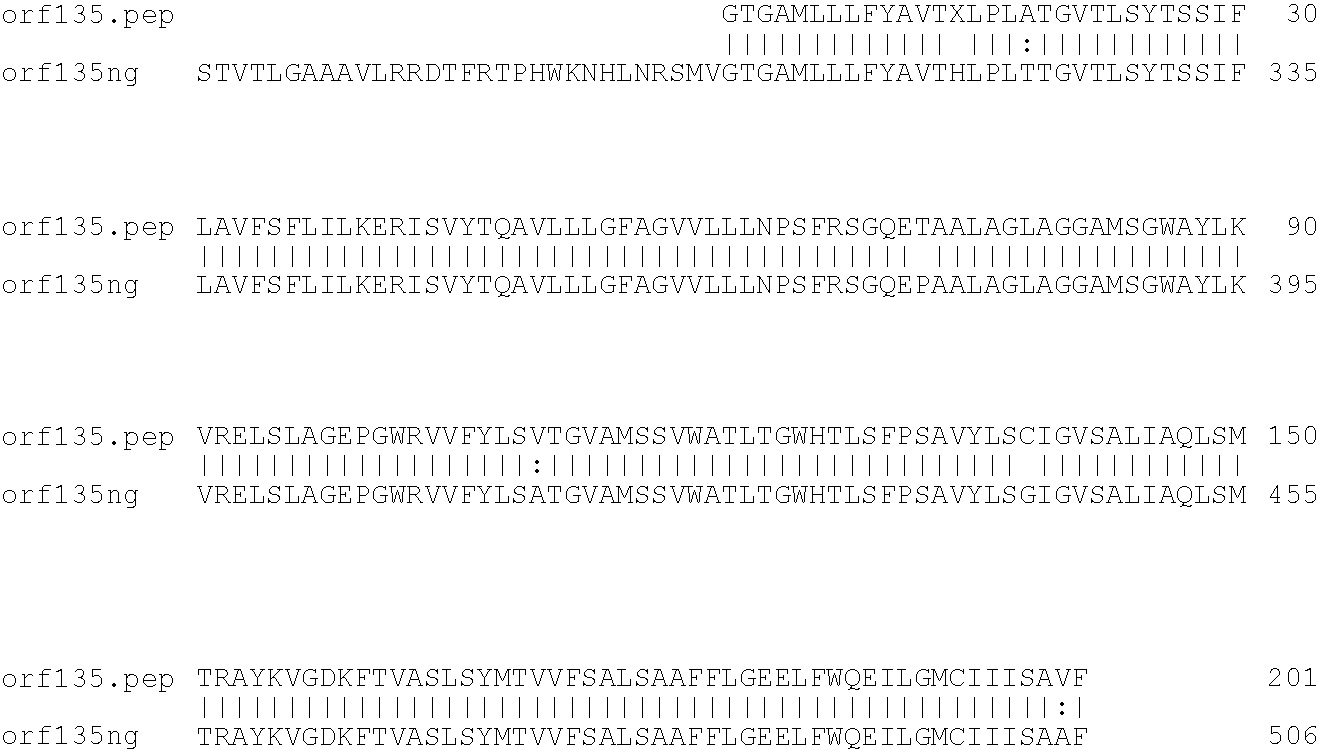
[1200]An ORF135ng nucleotide sequence <SEQ ID 545> was predicted to encode a protein having amino acid sequence <SEQ ID 546>:
[0000]| 1 | MPSEKAFRRH LRTASFQGLH LHHFHQKVGK CGIIGFGIHI FPTLLPAAQG | |
|
| 51 | ILDIQLGLFR IDFAALAVYR RTQVDFIHTV IDGIASDQAF SEVVQILRRL |
|
| 101 | NLGHFTDTHL IAQARRFIAD FGNIRPMRRG EAKTFCRCFR FDGIDGIHGD |
|
| 151 | FRQCGHINRL APGKDCRNGK RDKVFFHTRH YNQVCLEKTN CSARKIKFRH |
|
| 201 | QKQAKTHSTS LAARFTIRPS LSQRPFMDTA KKDILGSGWM LVAAACFTVM |
|
| 251 | NVLIKEASAK FALGSGELVF WRMLFSTVTL GAAAVLRRDT FRTPHWKNHL |
|
| 301 | NRSMVGTGAM LLLFYAVTHL PLTTGVTLSY TSSIFLAVFS FLILKERISV |
|
| 351 | YTQAVLLLGF AGVVLLLNPS FRSGQEPAAL AGLAGGAMSG WAYLKVRELS |
|
| 401 | LAGEPGWRVV FYLSATGVAM SSVWATLTGW HTLSFPSAVY LSGIGVSALI |
|
| 451 | AQLSMTRAYK VGDKFTVASL SYMTVVFSAL SAAFFLGEEL FWQEILGMCI |
|
| 501 | IISAAF* |
[1201]Further work revealed the following gonococcal sequence <SEQ ID 547>:
[0000]| 1 | ATGGATACCG CAAAAAAAGA CATTTTAGGA TCGGGCTGGA TGCTGGTGGC | |
|
| 51 | GGCGGCCTGC TTCACCGTTA TGAACGTATT GATTAAAGAG GCATCGGCAA |
|
| 101 | AATTTGCCCT CGGCAGCGGC GAATTGGTCT TTTGGCGCAT GCTGTTTTCA |
|
| 151 | ACCGTTACGC TCGGTGCTGC CGCCGTATTG CGGCGCGACA CCTTCCGCAC |
|
| 201 | GCCCCATTGG AAAAACCACT TAAACCGCAG TATGGTCGGG ACGGGGGCGA |
|
| 251 | TGCTGCTGCT GTTTTACGCG GTAACGCATC TGCCTTTGAC AACCGGCGTT |
|
| 301 | ACCCTGAGTT ACACCTCGTC GATTTTTttg GCGGTATTTT CCTTCCTCAT |
|
| 351 | TTTGAAAGAA CGGATTTCCG TTTACACGCA GGCGGTGCTG CTCCTTGGTT |
|
| 401 | TTGCCGGCGT GGTATTGCTG CTTAATCCCT CGTTCCGCAG CGGTCAGGAA |
|
| 451 | CCGGCGGCAC TCGCCGGGCT GGCGGGCGGC GCGATGTCCG GCTGGGCGTA |
|
| 501 | TTTGAAAGTG CGCGAACTGT CTTTGGCGGG CGAACCCGGC TGGCGCGTCG |
|
| 551 | TGTTTTACCT TTCCGCAACC GGCGTGGCGA TGTCGTCggt ttgggcgacg |
|
| 601 | Ctgaccggct ggCACAcccT GTCCTTTcca tcggcagttt ATCtgtCGGG |
|
| 651 | CATCGGCGTG tccgcgCtgA TTGCCCAaCT GtcgatgAcg cGCGcctaca |
|
| 701 | aaGTCGGCGA CAAATTCACG GTTGCCTCGC tttcctaTAt gaccgtcGTC |
|
| 751 | TTTTCCGCCC TGTCTGCCGC ATTTTTTCTg ggcgaagagc tttTCtggCA |
|
| 801 | GGAAATACTC GGTATGTGCA TCATTAtccT CAGCGGCATT TTGAGCAGCA |
|
| 851 | TCCGCCCCAT TGCCTTCAAA CAGCGGCTGC AAGCCCTCTT CCGCCAAAGA |
|
| 901 | TAA |
[1202]This corresponds to the amino acid sequence <SEQ ID 548; ORF135ng-1>:
[0000]| 1 | MDTAKKDILG SGWMLVAAAC FTVMNVLIKE ASAKFALGSG ELVFWRMLFS | |
|
| 51 | TVTLGAAAVL RRDTFRTPHW KNHLNRSMVG TGAMLLLFYA VTHLPLTTGV |
|
| 101 | TLSYTSSIFL AVFSFLILKE RISVYTQAVL LLGFAGVVLL LNPSFRSGQE |
|
| 151 | PAALAGLAGG AMSGWAYLKV RELSLAGEPG WRVVFYLSAT GVAMSSVWAT |
|
| 201 | LTGWHTLSFP SAVYLSGIGV SALIAQLSMT RAYKVGDKFT VASLSYMTVV |
|
| 251 | FSALSAAFFL GEELFWQEIL GMCIIILSGI LSSIRPIAFK QRLQALFRQR |
|
| 301 | * |
[1203]ORF135ng-1 and ORF135-1 show 97.0% identity in 300 aa overlap:
[0000]
[1204]Based on this analysis, including the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 66
[1205]The following DNA sequence was identified in N. meningitidis <SEQ ID 549>:
[0000]| 1 | ATGAAGCGGC GTATAGCCGT CTTCGTCCTG TTCCCGCAGA TAATCCGAGT | |
|
| 51 | TTTGGGACAA CTGTTGCCGA AAATCGTCAA TACAGTTCCG GCACATCGGA |
|
| 101 | TGCTCTTCCA GATTTTCGGG ATGTTCTTTT TCTTCATACA CCAGCAATAT |
|
| 151 | CTGCCCGGGA TCGCCGAAAT CGATTCCCCA TGCGGCATCG TGTTCGGTGC |
|
| 201 | GCTCCTCTTC CGTCATCTGC CCGCGCATTG CCTGTATGGT AAAGCCGCCG |
|
| 251 | TAGGGGATGC CgTTGCACAC GAACATCCAG TCGCTGATGT CGTCAACCGG |
|
| 301 | AACGCAAACG cTTTCGCCTT GTTCGACATT GGTCAGTTCG CCsGGTTCAT |
|
| 351 | TGTTCAGCAC ACCGTAAATA TAAAGACCGT CAAAATAAAT ATCGTCGATC |
|
| 401 | CACATATGTT CGCAAATTTC GCCGTCTTCG CCGTCTTGGA AAAAAGGGAC |
|
| 451 | TTTGACCATG GCAAAATCCA AGGCGGAAAT AATGCGGCGG CGTTCCCAAA |
|
| 501 | AAAGcTCGCG CCAAAAATAT TTGAATGTTT TACGGGCGCG TTCGTCGGCA |
|
| 551 | CGGTTTACCG GTTCGTCTGC CTGTTCTACA TAATAAATGA CGGAATCGCC |
|
| 601 | CATCATATCT GCTCCTCAAC GTGTACGGTA TCTGTTTGCA CCTTACTGCG |
|
| 651 | GCTTTCTgcC kTCGGCATCC GATTCGGATT TGAAAAGTTC mmrwyATTCG |
|
| 701 | GAATAG |
[1206]This corresponds to the amino acid sequence <SEQ ID 550; ORF136>:
[0000]| 1 | MKRRIAVFVL FPQIIRVLGQ LLPKIVNTVP AHRMLFQIFG MFFFFIHQQY | |
|
| 51 | LPGIAEIDSP CGIVFGALLF RHLPAHCLYG KAAVGDAVAE EHPVADVVNR |
|
| 101 | NANAFALFDI GQFAXFIVQH TVNIKTVKIN IVDPHMFANF AVFAVLEKRD |
|
| 151 | FDHGKIQGGN NAAAFPKKLA PKIFECFTGA FVGTVYRFVC LFYIINDGIA |
|
| 201 | HHSAPQRVRY LFAPYCGFLP SASDSDLKSS XXSE* |
[1207]Further work revealed the complete nucleotide sequence <SEQ ID 551>:
[0000]| 1 | ATGATGAAGC GGCGTATAGC CGTCTTCGTC CTGTTCCCGC AGATAATCCG | |
|
| 51 | AGTTTTGGGA CAACTGTTGC CGAAAATCGT CAATACAGTT CCGGCACATC |
|
| 101 | GGATGCTCTT CCAGATTTTC GGGATGTTCT TTTTCTTCAT ACACCAGCAA |
|
| 151 | TATCTGCCCG GGATCGCCGA AATCGATTCC CCATGCGGCA TCGTGTTCGG |
|
| 201 | TGCGCTCCTC TTCCGTCATC TGCCCGCGCA TTGCCTGTAT GGTAAAGCCG |
|
| 251 | CCGTAGGGGA TGCCGTTGCA CACGAACATC CAGTCGCTGA TGTCGTCAAC |
|
| 301 | CGGAACGCAA ACGCTTTCGC CTTGTTCGAC ATTGGTCAGT TCGCCGGGTT |
|
| 351 | CATTGTTCAG CACACCGTAA ATATAAAGAC CGTCAAAATA AATATCGTCG |
|
| 401 | ATCCACATAT GTTCGCAAAT TTCGCCGTCT TCGCCGTCTT GGAAAAAAGG |
|
| 451 | GACTTTGACC ATGGCAAAAT CCAAGGCGGA AATAATGCGG CGGCGTTCCC |
|
| 501 | AAAAAAGCTC GCGCCAAAAA TATTTGAATG TTTTACGGGC GCGTTCGTCG |
|
| 551 | GCACGGTTTA CCGGTTCGTC TGCCTGTTCT ACATAATAAA TGACGGAATC |
|
| 601 | GCCCATCATT CTGCTCCTCA ACGTGTACGG TATCTGTTTG CACCTTACTG |
|
| 651 | CGGCTTTCTG CCTTCGGCAT CCGATTCGGA TTTGAAAAGT TCCAAATATT |
|
| 701 | CGGAATAG |
[1208]This corresponds to the amino acid sequence <SEQ ID 552; ORF136-1>:
[0000]| 1 | MMKRRIAVFV LFPQIIRVLG QLLPKIVNTV PAHRMLFQIF GMFFFFIHQQ |
|
| 51 | YLPGIAEIDS PCGIVFGALL FRHLPAHCLY GKAAVGDAVA HEHPVADVVN |
|
| 101 | RNANAFALFD IGQFAGFIVQ HTVNIKTVKI NIVDPHMFAN FAVFAVLEKR |
|
| 151 | DFDHGKIQGG NNAAAFPKKL APKIFECFTG AFVG TVYRFV CLFYIINDGI |
|
| 201 | AHHSAPQRVR YLFAPYCGFL PSASDSDLKS SKYSE* |
[1209]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1210]ORF136 shows 71.7% identity over a 237aa overlap with an ORF (ORF136a) from strain A of N. meningitidis:
[0000]
[1211]The complete length ORF136a nucleotide sequence <SEQ ID 553> is:
[0000]| 1 | ATGATGAAGC GGCGTATAGC CGTCTTCGTC CTGCTCATGC AGAAAATCCG | |
|
| 51 | GATTTTGGGA CAACTGTTGC CGAAAATCGT CAATACAGTT CCGGCACATC |
|
| 101 | GGATGCTCTT CCAGATNTTC GGGATGTTCT TTTTCTTCAT ACACCAGCAA |
|
| 151 | TACCTGCCCG GGATCGCCGA AATCGATTCC CCATGCGGCA TCGTGTTCGG |
|
| 201 | TACGCTCCTC TTCCGTCATC NGTCCACGCA TTGCCTGTAT GGTAAAGCCG |
|
| 251 | CCGTAGGGAA TGCCGTTGCA CACGAACATC CAGTCGCTGA TGTCGTCAAC |
|
| 301 | CGGAACGCAA ACGCTTTCGC CTTGTTCGAC ATTGGTCAGT TCGCCGGGTT |
|
| 351 | CATTGTTCAG CACGCCATAA ATGTAAAGAC CGTCAAAATA AATATCGTCG |
|
| 401 | ATCCACATAT GTTCGCAAAT TTCGCCNTCT TCGCCGTCTT GGAAAAAAGG |
|
| 451 | GCTTTGACCA TGGCAAAATC TAAGGNGNNA NNGATGCGGC GGCGTTCCCA |
|
| 501 | AAAAAGCTCG CGCCAAAAAT ATTTGAATGT TTTGCGGGCG CGTTCGCCGG |
|
| 551 | CACGGTTTAC CGGTTTGTCT GCCTGTTCTA CATAATAAAT GACGGAATCG |
|
| 601 | CCCATCATAT CTGCTCCTCA ACGTGTACGG TATCTGTTTG CACCTTACTG |
|
| 651 | CGGCTTTCTG CCTTCGGCAT CCGATTCGGA TTTGAAAAGT TCCAAATATT |
|
| 701 | CGGAATAG |
[1212]This encodes a protein having amino acid sequence <SEQ ID 554>:
[0000]| 1 | MMKRRIAVFV LLMQKIRILG QLLPKIVNTV PAHRMLFQXF GMFFFFIHQQ | |
|
| 51 | YLPGIAEIDS PCGIVFGTLL FRHXSTHCLY GKAAVGNAVA HEHPVADVVN |
|
| 101 | RNANAFALFD IGQFAGFIVQ HAINVKTVKI NIVDPHMFAN FAXFAVLEKR |
|
| 151 | ALTMAKSKXX XMRRRSQKSS RQKYLNVLRA RSPARFTGLS ACST**MTES |
|
| 201 | PIISAPQRVR YLFAPYCGFL PSASDSDLKS SKYSE* |
[1213]ORF136a and ORF136-1 show 73.1% identity in 238 aa overlap:
[0000]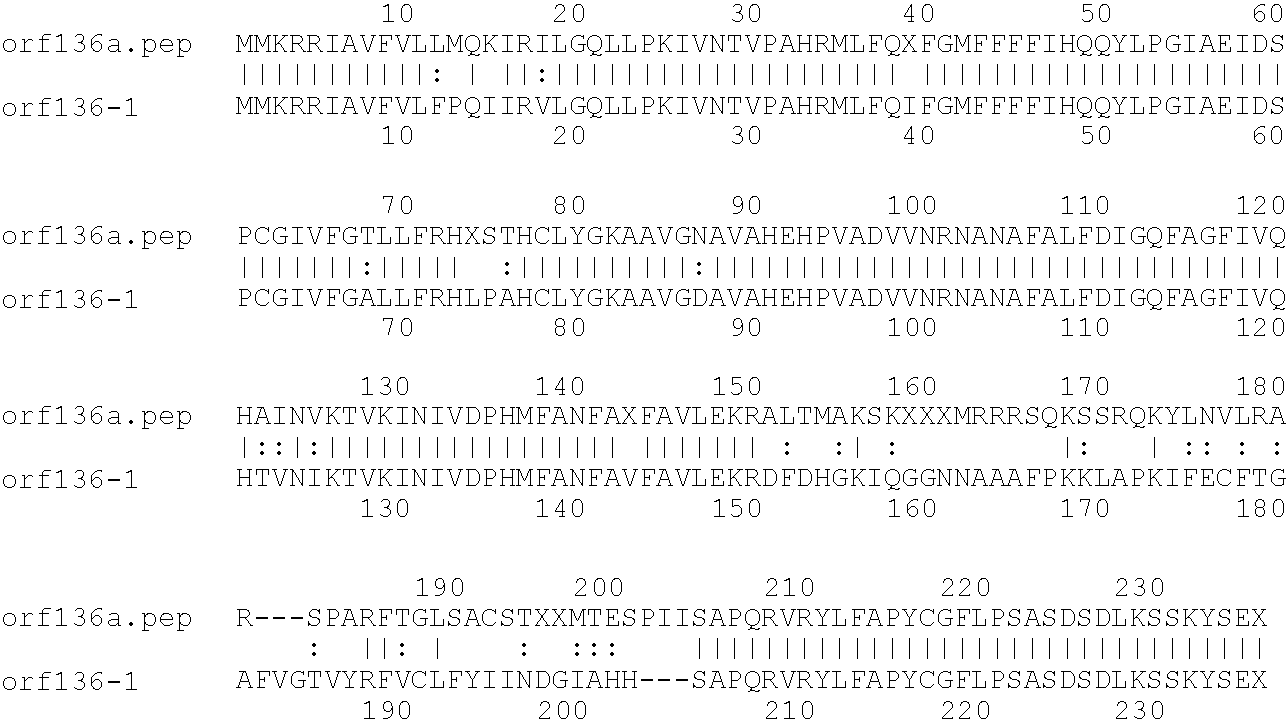
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1214]ORF136 shows 92.3% identity over a 234aa overlap with a predicted ORF (ORF136ng) from N. gonorrhoeae:
[0000]
[1215]The complete length ORF136ng nucleotide sequence <SEQ ID 555> is:
[0000]| 1 | ATGATGAAGC GGCGTATAGC CGTCTTCGTC CTGCTCATGC AGAAAATCCG | |
|
| 51 | GATTTTGGGA CAACTGTTGC CGAAAATCGT CAATACAGTT CCGGCACATC |
|
| 101 | GGATGCTCTT CCAAATTTTC GGGATGTTCT TTTTCTTCAT ACACCGGCAA |
|
| 151 | TACCTGCCCG GGATCGCCGA AATCGATTCC CCAGGCGGTA TCGTGTTCGG |
|
| 201 | TACGCTCCTC TTCCGTCATC TGTCCGCGCA TTGCCTGTAC GGTAAAGCCG |
|
| 251 | CCGTAGGGGA TGCCGTTGCA CACGAACATC CAGTCGCTGA TGTCGCCAAC |
|
| 301 | CGGAACGCAA ACGCTTTCGC CTTGTTCGAC ATTGGTCAGT CCGCCGGGTT |
|
| 351 | CATTGTTCAG CACACCGTAA ATATAAAGAC CGTCAAAATA AATATCGTCG |
|
| 401 | ATCCACATAT GTTCGCAAAT TTCGCCGTCT TCGCCGTCTT GGAAAAAAGG |
|
| 451 | GACTTTGACC ATGGCAAAAT CCAAGGCGGA AATAATGCGG CGGCGTTCCC |
|
| 501 | AAAAAAGCTC GCGCCAAAAG TATTTGAATG TTTTACGGGC GCGTTCGCCG |
|
| 551 | GCACGGTTTA CCGGTTCGTC TGCCTGTTCT ACATAATAAA TGACGGAATC |
|
| 601 | GCCCATCATA CTGCTCCTCA ACGTGTACGG TATCTGTTTG CACCTTACCG |
|
| 651 | CGGTTTTCTA CCTCCGGCAT CCGATTCGGA TTTGAAAAGT TCCAAATATT |
|
| 701 | CGGAATAG |
[1216]This encodes a protein having amino acid sequence <SEQ ID 556>:
[0000]| 1 | MMKRRIAVFV LLMQKIRILG QLLPKIVNTV PAHRMLFQIF GMFFFFIHRQ | |
|
| 51 | YLPGIAEIDS PGGIVFGTLL FRHLSAHCLY GKAAVGDAVA HEHPVADVAN |
|
| 101 | RNANAFALFD IGQSAGFIVQ HTVNIKTVKI NIVDPHMFAN FAVFAVLEKR |
|
| 151 | DFDHGKIQGG NNAAAFPKKL APKVFECFTG AFAGTVYRFVP CLFYIINDGI |
|
| 201 | AHHTAPQRVR YLFAPYRGFL PPASDSDLKS SKYSE* |
[1217]ORF136ng and ORF136-1 show 93.6% identity in 235 aa overlap:
[0000]
[1218]Based on the presence of the putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 67
[1219]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 557>:
[0000]| 1 | ATGGAAAATA TGGTAACGTT TTCAAAAATC AGACCGCTTT TGGCAATCGC | |
|
| 51 | CGCCGCCGCG TTGCTTGCCG CC.TGCGGAC GGCGGGAAAT AATGCTGTCC |
|
| 101 | GCAAGCCGGT GCAAACCGCC AAACCCGCCG CAGTGGTCGG TTTGGCACTC |
|
| 151 | GGTGGCGGCG CATCTAAAGG ATTTGCCCAT GTAGGTATTA TTAAGGTTTT |
|
| 201 | GAAAGAAAAC GGTATTCCTG TGAAGGTGGT TACCGGCACC TCCGCAGGTT |
|
| 251 | CGATTGTCGG CAACCTTTTT GCATCGGGTA TGTCGCCCGA CCGCCTCGAA |
|
| 301 | TTGGAAGCCG AAATTTTAGG CAAAACCGAT TTGGTCGATT TAACCTTGTC |
|
| 351 | CACCAATGGG TTTATCAAAG GCGCAAAGCT GCAAAATTAC ATCAACCGAA |
|
| 401 | AACTCCGCGG CATGCAGATT CAGCAGTTTC CCATCAAATT TGCCGCC.. |
[1220]This corresponds to the amino acid sequence <SEQ ID 558; ORF137>:
[0000]| 1 | MENMVTFSKI RPLLAIAAAA LLAAXRTAGN NAVRKPVQTA KPAAVVGLAL | |
|
| 51 | GGGASKGFAH VGIIKVLKEN GIPVKVVTGT SAGSIVGNLF ASGMSPDRLE |
|
| 101 | LEAEILGKTD LVDLTLSTNG FIKGAKLQNY INRKLRGMQI QQFPIKFAA.. |
[1221]Further work revealed the complete nucleotide sequence <SEQ ID 559>:
[0000]| 1 | ATGGAAAATA TGGTAACGTT TTCAAAAATC AGACCGCTTT TGGCAATCGC | |
|
| 51 | CGCCGCCGCG TTGCTTGCCG CCTGCGGCAC GGCGGGAAAT AATGCTGTCC |
|
| 101 | GCAAGCCGGT GCAAACCGCC AAACCCGCCG CAGTGGTCGG TTTGGCACTC |
|
| 151 | GGTGGCGGCG CATCTAAAGG ATTTGCCCAT GTAGGTATTA TTAAGGTTTT |
|
| 201 | GAAAGAAAAC GGTATTCCTG TGAAGGTGGT TACCGGCACA TCGGCAGGTT |
|
| 251 | CGATTGTCGG CAGCCTTTTT GCATCGGGTA TGTCGCCCGA CCGCCTCGAA |
|
| 301 | TTGGAAGCCG AAATTTTAGG CAAAACCGAT TTGGTCGATT TAACCTTGTC |
|
| 351 | CACCAGTGGT TTTATCAAAG GCGAAAAGCT GCAAAATTAC ATCAACCGAA |
|
| 401 | AAGTCGGCGG CAGGCAGATT CAGCAGTTTC CCATCAAATT TGCCGCCGTT |
|
| 451 | GCTACTGATT TTGAAACCGG CAAGGCCGTC GCTTTCAATC AGGGGAATGC |
|
| 501 | CGGGCAGGCT GTGCGCGCTT CCGCCGCCAT TCCCAATGTG TTCCAACCCG |
|
| 551 | TTATCATCGG CAGGCATACA TATGTTGACG GCGGTCTGTC GCAGCCCGTG |
|
| 601 | CCCGTCAGTG CCGCCCGGCG GCAGGGGGCG AATTTCGTGA TTGCCGTCGA |
|
| 651 | TATTTCCGCC CGTCCGGGCA AAAACATCAG CCAAGGTTTC TTCTCTTATC |
|
| 701 | TCGATCAGAC GCTGAACGTA ATGAGCGTTT CTGCGTTGCA AAATGAGTTG |
|
| 751 | GGGCAGGCGG ATGTGGTTAT CAAACCGCAG GTTTTGGATT TGGGTGCAGT |
|
| 801 | CGGCGGATTC GATCAGAAAA AACGCGCCAT CCGGTTGGGT GAGGAGGCAG |
|
| 851 | CACGTGCCGC ATTGCCTGAA ATCAAACGCA AACTGGCGGC ATACCGTTAT |
|
| 901 | TGA |
[1222]This corresponds to the amino acid sequence <SEQ ID 560; ORF137-1>:
[0000]| 1 | MENMVTFSKI RPLLAIAAAA LLAACGTAGN NAVRKPVQTA KPAAVVGLAL | |
|
| 51 | GGGASKGFAH VGIIKVLKEN GIPVKVVTGT SAGSIVGSLF ASGMSPDRLE |
|
| 101 | LEAEILGKTD LVDLTLSTSG FIKGEKLQNY INRKVGGRQI QQFPIKFAAV |
|
| 151 | ATDFETGKAV AFNQGNAGQA VRASAAIPNV FQPVIIGRHT YVDGGLSQPV |
|
| 201 | PVSAARRQGA NFVIAVDISA RPGKNISQGF FSYLDQTLNV MSVSALQNEL |
|
| 251 | GQADVVIKPQ VLDLGAVGGF DQKKRAIRLG EEAARAALPE IKRKLAAYRY |
|
| 301 | * |
[1223]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1224]ORF137 shows 93.3% identity over a 149aa overlap with an ORF (ORF137a) from strain A of N. meningitidis.
[0000]
[1225]The complete length ORF137a nucleotide sequence <SEQ ID 561> is:
[0000]| 1 | ATGGAAAATA TGGTAACGTT TTCAAAAATC AGACCGCTTT TGGCAATCGC | |
|
| 51 | CGCCGCCGCG TTGCTTGCCG CCTGCGGCAC GGCGGGAAAT AATGCTGCCC |
|
| 101 | GCAAGCCGGT GCAAACCGCC AAACCCGCCG CAGTGGTCGG TTTGGCACTC |
|
| 151 | GGTGGCGGCG CATCTAAAGG ATTTGCCCAT GTAGGTATTA TTAAGGTTTT |
|
| 201 | GAAAGAAAAC GGTATTCCTG TGAAGGTGGT TACCGGCACA TCGGCAGGTT |
|
| 251 | CGATAGTCGG CAGCCTTTTT GCATCGGGTA TGTCGCCCGA CCGCCTCGAA |
|
| 301 | TTGGAAGCCG AAATTTTAGG TAAAACCGAT TTGGTCGATT TAACCTTGTC |
|
| 351 | CACCAGTGGT TTTATCAAAG GCGAAAAGCT GCAAAATTAC ATCAACCGAA |
|
| 401 | AAGTCGGCGG CAGGCGGATT CAGCAGTTTC CCATCAAATT TGCCGCCGTT |
|
| 451 | GCTACTGATT TTGAAACCGG CAAGGCCGTC GCTTTCAATC AAGGGAATGC |
|
| 501 | CGGGCAGGCT GTGCGCGCTT CCGCCGCCAT TCCCAATGTG TTCCAACCCG |
|
| 551 | TTATCATCGG CAGGCATACA TATGTTGACG GCGGTCTGTC GCAGCCCGTG |
|
| 601 | CCCGTCAGTG CCGCCCGGCG GCANGNNNNG NATNTCGTGA TTGCCGTCGA |
|
| 651 | TATTTCCGCC CGTCCGAGCA AAAACATCAG CCAAGGCTTC TTCTCTTATC |
|
| 701 | TCGATCAGAC GCTGAACGTA ATGAGCGTTT CCGCGTTGCA AAATGAGTTG |
|
| 751 | GGGCAGGCGG ATGTGGTTAT CAAACCGCAG GTTTTGGATT TGGGTGCAGT |
|
| 801 | CGGCGGATTC GATCAGAAAA AACGCGCCAT CCGGTTGGGT GAGGAGGCAG |
|
| 851 | CACGTGCCGC ATTGCCTGAA ATCAAACGCA AACTGGCGGC ATACCGTTAT |
|
| 901 | TGA |
[1226]This encodes a protein having amino acid sequence <SEQ ID 562>:
[0000]| 1 | MENMVTFSKI RPLLAIAAAA LLAACGTAGN NAARKPVQTA KPAAVVGLAL | |
|
| 51 | GGGASKGFAH VGIIKVLKEN GIPVKVVTGT SAGSIVGSLF ASGMSPDRLE |
|
| 101 | LEAEILGKTD LVDLTLSTSG FIKGEKLQNY INRKVGGRRI QQFPIKFAAV |
|
| 151 | ATDFETGKAV AFNQGNAGQA VRASAAIPNV FQPVIIGRHT YVDGGLSQPV |
|
| 201 | PVSAARRXXX XXVIAVDISA RPSKNISQGF FSYLDQTLNV MSVSALQNEL |
|
| 251 | GQADVVIKPQ VLDLGAVGGF DQKKRAIRLG EEAARAALPE IKRKLAAYRY |
|
| 301 | * |
[1227]ORF137a and ORF137-1 show 97.3% identity in 300 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1228]ORF137 shows 89.9% identity over a 149aa overlap with a predicted ORF (ORF137ng) from N. gonorrhoeae:
[0000]
[1229]The complete length ORF137ng nucleotide sequence <SEQ ID 563> is:
[0000]| 1 | ATGGAAAATA TGGTAACGTT TTCAAAAATC AGATCATTTT TGGCAATCGC | |
|
| 51 | CGCCGCCGCG TTGCTTGCCG CCTGCGGTAC GGCGGGAAAC AATGCCGCCC |
|
| 101 | GCAAGCCGGT GCAAACCGCC AAACCCGCCG CAGTGGTCGC TTTGGCACTC |
|
| 151 | GGTGGCGGCG CATCTAAAGG ATTTGCCCAT ATAGGAATTG TTAAGGTTTT |
|
| 201 | GAAAGAAAAC GGTATTCCTG TGAAGGTGGT TACCGGCACA TCGGCAGGTT |
|
| 251 | CGATAGTCGG CAGCCTTTTG GCATCGGGTA TGTCGCCCGA CCGCCTCGAA |
|
| 301 | TTGGAAGCCG AGATTTTAGG TAAAACCGAT TTAGTCGATT TAACCTTGTC |
|
| 351 | CACCAGTGGT TTTATCAAAG GCGAAAAGCT GCAAAATTAC ATCAACCGAA |
|
| 401 | AAGTCGGCGG CAGGCAGATT CAGCAGTTTC CCATCAAATT TGCCGCCGTT |
|
| 451 | GCCACTGATT TTGAAACCGG CAAGGCCGTC GCTTTCAATC AAGGGAATGC |
|
| 501 | CGGGCAGGCG GTTCGTGCTT CCGCCGCCAT TCCCAATGTG TTCCAGCCAG |
|
| 551 | TCATCATCGG CAGGCACAAA TATGTTGACG GCGGTCTGTC GCAGCCCGTG |
|
| 601 | CCCGTCAGTG CCGCTCGGCG GCAGGGGGCG AATTTCGTGA TTGCCGTCGA |
|
| 651 | TATTTCCGCA CGTCCGAGCA AAAATGTCGG TCAAGGTTTC TTCTCTTATC |
|
| 701 | TCGATCAGAC GCTGAACGTG ATGAGCGTTT CCGTGTTGCA AAACGAGTTG |
|
| 751 | gggcAGGCGG ATGTGGTTAT CAAACCGCag gtTTTGGATT TGGGTGCAGT |
|
| 801 | CGGCGGATTC GATCAGAAAA AGCGCGCCAT CCGGTTGGGC GAGGAGGCAG |
|
| 851 | CACGTGCCGC ATTGCCTGAA ATCAAACGCA AACTGGCGGC ATACCGTTAT |
|
| 901 | TGA |
[1230]This encodes a protein having amino acid sequence <SEQ ID 564>:
[0000]| 1 | MENMVTFSKI RSFLAIAAAA LLAACGTAGN NAARKPVQTA KPAAVVALAL | |
|
| 51 | GGGASKGFAH IGIVKVLKEN GIPVKVVTGT SAGSIVGSLL ASGMSPDRLE |
|
| 101 | LEAEILGKTD LVDLTLSTSG FIKGEKLQNY INRKVGGRQI QQFPIKFAAV |
|
| 151 | ATDFETGKAV AFNQGNAGQA VRASAAIPNV FQPVIIGRHK YVDGGLSQPV |
|
| 201 | PVSAARRQGA NFVIAVDISA RPSKNVGQGF FSYLDQTLNV MSVSVLQNEL |
|
| 251 | GQADVVIKPQ VLDLGAVGGF DQKKRAIRLG EEAARAALPE IKRKLAAYRY |
|
| 301 | * |
[1231]ORF137ng and ORF137-1 show 96.0% identity in 300 aa overlap:
[0000]
[1232]Based on the presence of a predicted prokaryotic membrane lipoprotein lipid attachment site (underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 68
[1233]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 565>:
[0000]| 1 | ATGTTTCGTT TACAATTCAG GCTGTTTCCC CCTTTGCGAA CCGCCATGCA | |
|
| 51 | CATCCTGTTG ACCGCCCTGC TCAAATGCCT CTCCCTGcTG CCGCTTTCCT |
|
| 101 | GTCTGCACAC GCTGGGAAAC CGGCTCGGAC ATCTGGCGTT TTACCTTTTA |
|
| 151 | AAGGAAGACC GCGCGCGCAT CGTCGCCmAT ATGCGGCAGG CGGGTTTGAA |
|
| 201 | CCCCGACCCC AAAACGGTCA AAGCCGTTTT TGCGGAAACG GCAAAAGGCG |
|
| 251 | GTTTGGAACT TGCCCCCGCG TTTTTCAGAA AACCGGAAGA CATAGAAACA |
|
| 301 | ATGTTCAAAG CGGTACACGG CTGGGAACAT GTGCAGCAGG CTTTGGACAA |
|
| 351 | ACACGAAGGG CTGCTATTC.. |
[1234]This corresponds to the amino acid sequence <SEQ ID 566; ORF138>:
[0000]| 1 | MFRLQFRLFP PLRTAMHILL TALLKCLSLL PLSCLHTLGN RLGHLAFYLL | |
|
| 51 | KEDRARIVAX MRQAGLNPDP KTVKAVFAET AKGGLELAPA FFRKPEDIET |
|
| 101 | MFKAVHGWEH VQQALDKHEG LLF |
[1235]Further work revealed the complete nucleotide sequence <SEQ ID 567>:
[0000]| 1 | ATGTTTCGTT TACAATTCAG GCTGTTTCCC CCTTTGCGAA CCGCCATGCA | |
|
| 51 | CATCCTGTTG ACCGCCCTGC TCAAATGCCT CTCCCTGCTG CCGCTTTCCT |
|
| 101 | GTCTGCACAC GCTGGGAAAC CGGCTCGGAC ATCTGGCGTT TTACCTTTTA |
|
| 151 | AAGGAAGACC GCGCGCGCAT CGTCGCCAAT ATGCGGCAGG CGGGTTTGAA |
|
| 201 | CCCCGACCCC AAAACGGTCA AAGCCGTTTT TGCGGAAACG GCAAAAGGCG |
|
| 251 | GTTTGGAACT TGCCCCCGCG TTTTTCAGAA AACCGGAAGA CATAGAAACA |
|
| 301 | ATGTTCAAAG CGGTACACGG CTGGGAACAT GTGCAGCAGG CTTTGGACAA |
|
| 351 | ACACGAAGGG CTGCTATTCA TCACGCCGCA CATCGGCAGC TACGATTTGG |
|
| 401 | GCGGACGCTA CATCAGCCAG CAGCTTCCGT TCCCGCTGAC CGCCATGTAC |
|
| 451 | AAACCGCCGA AAATCAAAGC GATAGACAAA ATCATGCAGG CGGGCAGGGT |
|
| 501 | TCGCGGCAAA GGAAAAACCG CGCCTACCAG CATACAAGGG GTCAAACAAA |
|
| 551 | TCATCAAAGC CCTGCGTTCG GGCGAAGCAA CCATCGTCCT GCCCGACCAC |
|
| 601 | GTCCCCTCCC CTCAAGAAGG CGGGGAAGGC GTATGGGTGG ATTTCTTCGG |
|
| 651 | CAAACCTGCC TATACCATGA CGCTGGCGGC AAAATTGGCA CACGTCAAAG |
|
| 701 | GCGTGAAAAC CCTGTTTTTC TGCTGCGAAC GCCTGCCTGG CGGACAAGGT |
|
| 751 | TTCGATTTGC ACATCCGCCC CGTCCAAGGG GAATTGAACG GCGACAAAGC |
|
| 801 | CCATGATGCC GCCGTGTTCA ACCGCAATGC CGAATATTGG ATACGCCGTT |
|
| 851 | TTCCGACGCA GTATCTGTTT ATGTACAACC GCTACAAAAT GCCGTAA |
[1236]This corresponds to the amino acid sequence <SEQ ID 568; ORF138-1>:
[0000]| 1 | MFRLQFRLFP PLRTAMHILL TALLKCLSLL PLSCLHTLGN RLGHLAFYLL | |
|
| 51 | KEDRARIVAN MRQAGLNPDP KTVKAVFAET AKGGLELAPA FFRKPEDIET |
|
| 101 | MFKAVHGWEH VQQALDKHEG LLFITPHIGS YDLGGRYISQ QLPFPLTAMY |
|
| 151 | KPPKIKAIDK IMQAGRVRGK GKTAPTSIQG VKQIIKALRS GEATIVLPDH |
|
| 201 | VPSPQEGGEG VWVDFFGKPA YTMTLAAKLA HVKGVKTLFF CCERLPGGQG |
|
| 251 | FDLHIRPVQG ELNGDKAHDA AVFNRNAEYW IRRFPTQYLF MYNRYKMP* |
[1237]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1238]ORF138 shows 99.2% identity over a 123aa overlap with an ORF (ORF138a) from strain A of N. meningitidis.
[0000]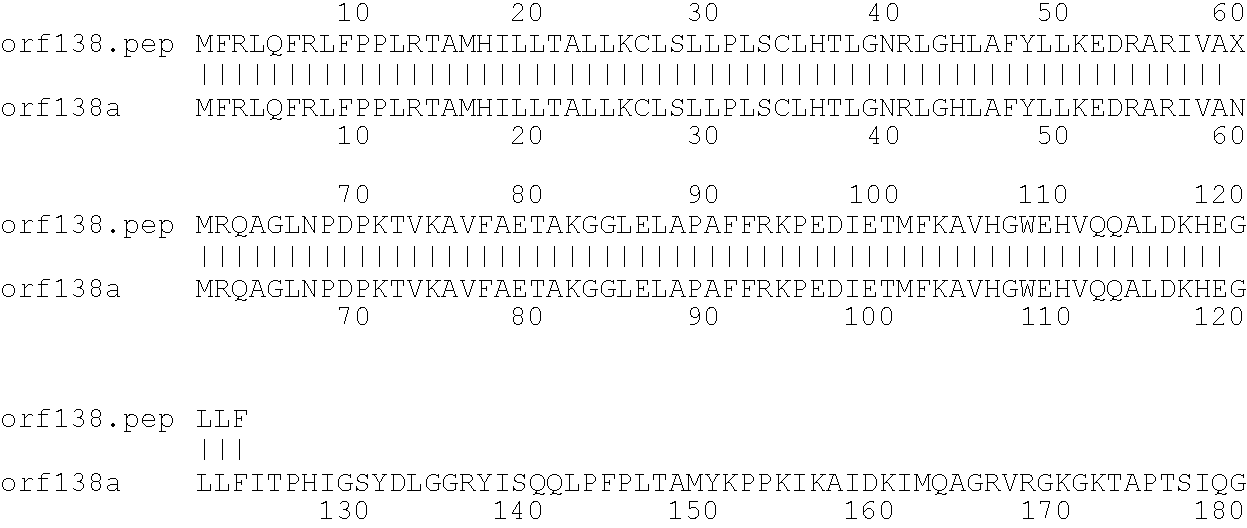
[1239]The complete length ORF138a nucleotide sequence <SEQ ID 569> is:
[0000]| 1 | ATGTTTCGTT TACAATTCAG GCTGTTTCCC CCTTTGCGAA CCGCCATGCA | |
|
| 51 | CATCCTGTTG ACCGCCCTGC TCAAATGCCT CTCCCTGCTG CCGCTTTCCT |
|
| 101 | GTCTGCACAC GCTGGGAAAC CGGCTCGGAC ATCTGGCGTT TTACCTTTTA |
|
| 151 | AAGGAAGACC GCGCGCGCAT CGTCGCCAAT ATGCGTCAGG CAGGCATGAA |
|
| 201 | TCCCGACCCC AAAACGGTCA AAGCCGTTTT TGCGGAAACG GCAAAAGGCG |
|
| 251 | GTTTGGAACT TGCCCCCGCG TTTTTCAGAA AACCGGAAGA CATAGAAACA |
|
| 301 | ATGTTCAAAG CGGTACACGG CTGGGAACAT GTGCAGCAGG CTTTGGACAA |
|
| 351 | ACACGAAGGG CTGCTATTCA TCACGCCGCA CATCGGCAGC TACGATTTGG |
|
| 401 | GCGGACGCTA CATCAGCCAG CAGCTTCCGT TCCCGCTGAC CGCCATGTAC |
|
| 451 | AAACCGCCGA AAATCAAAGC GATAGACAAA ATCATGCAGG CGGGCAGGGT |
|
| 501 | TCGCGGCAAA GGAAAAACCG CGCCTACCAG CATACAAGGG GTCAAACAAA |
|
| 551 | TCATCAAAGC CCTGCGTTCG GGCGAAGCAA CCATCGTCCT GCCCGACCAC |
|
| 601 | GTCCCCTCCC CTCAAGAAGG CGGGGAAGGC GTATGGGTGG ATTTCTTCGG |
|
| 651 | CAAACCTGCC TATACCATGA CGCTGGCGGC AAAATTGGCA CACGTCAAAG |
|
| 701 | GCGTGAAAAC CCTGTTTTTC TGCTGCGAAC GCCTGCCTGG CGGACAAGGT |
|
| 751 | TTCGATTTGC ACATCCGCCC CGTCCAAGGG GAATTGAACG GCGACAAAGC |
|
| 801 | CCATGATGCC GCCGTGTTCA ACCGCAATGC CGAATATTGG ATACGCCGTT |
|
| 851 | TTCCGACGCA GTATCTGTTT ATGTACAACC GCTACAAAAT GCCGTAA |
[1240]This encodes a protein having amino acid sequence <SEQ ID 570>:
[0000]| 1 | MFRLQFRLFP PLRTAMHILL TALLKCLSLL PLSCLHTLGN RLGHLAFYLL | |
|
| 51 | KEDRARIVAN MRQAGLNPDP KTVKAVFAET AKGGLELAPA FFRKPEDIET |
|
| 101 | MFKAVHGWEH VQQALDKHEG LLFITPHIGS YDLGGRYISQ QLPFPLTAMY |
|
| 151 | KPPKIKAIDK IMQAGRVRGK GKTAPTSIQG VKQIIKALRS GEATIVLPDH |
|
| 201 | VPSPQEGGEG VWVDFFGKPA YTMTLAAKLA HVKGVKTLFF CCERLPGGQG |
|
| 251 | FDLHIRPVQG ELNGDKAHDA AVFNRNAEYW IRRFPTQYLF MYNRYKMP* |
[1241]ORF138a and ORF138-1 show 99.7% identity over a 298aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1242]ORF138 shows 94.3% identity over a 123aa overlap with a predicted ORF (ORF138ng) from N. gonorrhoeae:
[0000]
[1243]The complete length ORF138ng nucleotide sequence <SEQ ID 571> is:
[0000]| 1 | ATGTTTCGTT TACAATTCAG GCTGTTTCCC CCTTTGCGAA CCGCCATGCA | |
|
| 51 | CATCCTGTTG ACCGCCCTGC TCAAATGCCT CTCCCTGCTG TCGCTTTCCT |
|
| 101 | GTCTGCACAC GCTGGGAAAC CGGCTCGGAC ATCTGGCGTT TTACCTTTTA |
|
| 151 | AAGGAAGACC GCGCGCGCAT CGTCGCCAAT ATGCGGCAGG CGGGTTTGAA |
|
| 201 | CCCCGACACG CAGACGGTCA AAGCCGTTTT TGCGGAAACG GCAAAATGCG |
|
| 251 | GTTTGGAACT TGCCCCCGCG TTTTTCAAAA AACCGGAAGA CATCGAAACA |
|
| 301 | ATGTTCAAAG CGGTACACGG CTGGGAACAC GTGCAGCAGG CTTTGGACAA |
|
| 351 | GGGCGAAGGG CTGCTGTTCA TCACGCCGCA CATCGGCAGC TACGATTTGG |
|
| 401 | GCGGACGCTA CATCAGCCAG CAGCTTCCGT TCCACCTGAC CGCCATGTAC |
|
| 451 | AAGCCGCCGA AAATCAAAGC GATAGACAAA ATCATGCAGG CGGGCAGGGT |
|
| 501 | GCGCGGCAAA GGCAAAACcg cgcccaccgg catACAAGGG GTCAAACAAA |
|
| 551 | tcatcaAGGC CCTGCGCGCG GGCGAGGCAA CCAtcATCCT GCCCGACCAC |
|
| 601 | GTCCCTTCTC CGCAGGAagg cggCGGCGTG TGGGCGGATT TTTTCGGCAA |
|
| 651 | ACCTGCATAC acCATGACAC TGGCGGCAAA ATTGGCACAC GTCAAAGGCG |
|
| 701 | TGAAAACCCT GTTTTTCTGC TGCGAACGCC TGCCCGACGG ACAAGGCTTC |
|
| 751 | GTGTTGCACA TCCGCCCCGT CCAAGGGGAA TTGAACGGCA ACAAAGCCCA |
|
| 801 | CGATGCCGCC GTGTTCAACC GCAATACCGA ATATTGGATA CGCCGTTTTC |
|
| 851 | CGACGCAGTA TCTGTTTATG TACAACCGCT ATAAAACGCC GTAA |
[1244]This encodes a protein having amino acid sequence <SEQ ID 572>:
[0000]| 1 | MFRLQFRLFP PLRTAMHILL TALLKCLSLL SLSCLHTLGN RLGHLAFYLL | |
|
| 51 | KEDRARIVAN MRQAGLNPDT QTVKAVFAET AKCGLELAPA FFKKPEDIET |
|
| 101 | MFKAVHGWEH VQQALDKGEG LLFITPHIGS YDLGGRYISQ QLPFHLTAMY |
|
| 151 | KPPKIKAIDK IMQAGRVRGK GKTAPTGIQG VKQIIKALRA GEATIILPDH |
|
| 201 | VPSPQEGGGV WADFFGKPAY TMTLAAKLAH VKGVKTLFFC CERLPDGQGF |
|
| 251 | VLHIRPVQGE LNGNKAHDAA VFNRNTEYWI RRFPTQYLFM YNRYKTP* |
[1245]ORF138ng and ORF138-1 show 94.3% identity over 299aa overlap:
[0000]
[1246]In addition, ORF138ng is homologous to htrB protein from Pseudomonas fluorescens:
[0000]| gnl|PID|e334283 (Y14568) htrB [Pseudomonas fluorescens] Length = 253 | |
| Score = 80.8 bits (196), Expect = 9e−15 |
| Identities = 49/151 (32%), Positives = 79/151 (51%), Gaps = 6/151 (3%) |
|
| Query: | 101 | MFKAVHGWEHVQQALDKGEGLLFITPHIGSYD-LGGRYISQQLPFHLTAMYKPPKIKAID | 159 | |
| | + + V G E +++AL G+G++ IT H+G+++ L Y SQ P Y+PPK+KA+D |
| Sbjct: | 94 | LVREVEGLEVLKEALASGKGVVGITSHLGNWEVLNHFYCSQCKPI---IFYRPPKLKAVD | 150 |
|
| Query: | 160 | KIMQAGRVRGKGKTAPTGIQGVKQIIKALRAGEATIILPDHVPSPQEGGGVWADFFGKPA | 219 |
| | ++++ RV+ K A + +G+ +IK +R G I D P P E G++ FF A |
| Sbjct: | 151 | ELLRKQRVQLGNKVAASTKEGILSVIKEVRKGGQVGIPAD--PEPAESAGIFVPFFATQA | 208 |
|
| Query: | 220 | YTMTLAAKLAHVKGVKTLFFCCERLPDGQGF | 250 |
| | T + +F RLPDG G+ |
| Sbjct: | 209 | LTSKFVPNMLAGGKAVGVFLHALRLPDGSGY | 239 |
[1247]Based on this analysis, including the presence of a putative transmembrane domain in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1248]ORF138-1 (57 kDa) was cloned in the pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 14A shows the results of affinity purification of the GST-fusion protein. Purified GST-fusion protein was used to immunise mice, whose sera were used for ELISA (positive result) and FACS analysis (FIG. 14B). These experiments confirm that ORF138-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 69
[1249]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 573>:
[0000]| 1 | ..GCGTGGTCGG CCGGCGAATC GTGGCGTGTG TTAATGGAAA GTGAAACGTG | |
|
| 51 | GCATGCGGTG TGGAATACTT TGCGCTTCTC GGCGGCGGCG GTGTATGCGG |
|
| 101 | CAGCGGTTTT GGGTGTGGTG TATGCGGCGC CGGCGCGGCG GTCGGCGTGG |
|
| 151 | ATGCGCGGGC TGATGTTTTA GCCGTTTATG GTGTCGCCGG TTTGTGTTTC |
|
| 201 | GGCGGGCGTG CTGCTGCTTT ATCCGCAGTG GACGGCTTCG TTGCCGTTGC |
|
| 251 | TGCTGGCGAT GTATGCGCTG CTGGCGTATC CGTTTGTGGC AAAAGATGTT |
|
| 301 | TTATCAGCCT GGGATGCACT GCCGCCGGAT TACGGCAGGG CGGCGGCGGG |
|
| 351 | TTTGGGTGCA AACGGCTTTC AGACGGCATG CCGCATCACG TTCCCCCTCT |
|
| 401 | TGAAACCGGC GTTGCGGCGC GGTCTGACTT TGGCGGCGGC AACCTGCGTG |
|
| 451 | GGCGAATTTG CGGCGACATT GTTTCTGTCG CGTCCGGAAT GGCAGACGCT |
|
| 501 | GACGACTTTG ATTTATGCCT ATTTGGGACG CGCGGGTGAG GATAATTACG |
|
| 551 | CGCGGGCGAT GGTGCTG.. |
[1250]This corresponds to the amino acid sequence <SEQ ID 574; ORF139>:
[0000]| 1 | ..AWSAGESWRV LMESETWHAV WNTLRFSAAA VYAAAVLGVV YAAPARRSAW | |
|
| 51 | MRGLMFXPFM VSPVCVSAGV LLLYPQWTAS LPLLLAMYAL LAYPFVAKDV |
|
| 101 | LSAWDALPPD YGRAAAGLGA NGFQTACRIT FPLLKPALRR GLTLAAATCV |
|
| 151 | GEFAATLFLS RPEWQTLTTL IYAYLGRAGE DNYARAMVL.. |
[1251]Further work revealed the complete nucleotide sequence <SEQ ID 575>:
[0000]| 1 | ATGGATGGAC GGCGTTGGGT GGTATGGGGT GCTTTTGCCC TGCTGCCTTC | |
|
| 51 | GGCTTTTTTG GCGGTAATGG TCGTTGCGCC TTTGTGGGCG GTGGCGGCGT |
|
| 101 | ATGACGGTTT GGCGTGGCGC GCGGTGCTGT CGGATGCCTA TATGCTCAAA |
|
| 151 | CGTTTGGCGT GGACGGTATT TCAGGCAGCG GCAACCTGTG TGCTGGTGCT |
|
| 201 | GCCTTTGGGC GTGCCTGTCG CGTGGGTGCT GGCGCGGCTG GCGTTTCCGG |
|
| 251 | GGCGGGCTTT GGTGCTGCGC CTGCTGATGC TGCCTTTTGT GATGCCCACG |
|
| 301 | TTGGTGGCGG GCGTGGGCGT GCTGGCCCTG TTCGGGGCGG ACGGGCTGTT |
|
| 351 | GTGGCGCGGC AGGCAGGATA CGCCGTATCT GTTGTTGTAC GGCAATGTGT |
|
| 401 | TTTTCAACCT TCCTGTGTTG GTCAGGGCGG CGTATCAGGG GTTTGTGCAA |
|
| 451 | GTGCCTGCGG CACGGCTTCA GACGGCACGG ACGTTGGGCG CGGGGGCGTG |
|
| 501 | GCGGCGGTTT TGGGACATTG AAATGCCCGT TTTGCGCCCG TGGCTTGCCG |
|
| 551 | GCGGCGTGTG CCTTGTCTTT CTGTATTGTT TTTCCGGGTT CGGGCTGGCG |
|
| 601 | CTGCTGCTGG GCGGCAGCCG TTATGCCACG GTCGAAGTGG AAATTTACCA |
|
| 651 | GTTGGTCATG TTCGAACTCG ATATGGCGGT TGCTTCGGTG CTGGTGTGGC |
|
| 701 | TGGTGTTGGG GGTAACGGCG GCGGCAGGGT TGCTGTATGC GTGGTTCGGC |
|
| 751 | AGGCGCGCGG TTTCGGATAA GGCGGTTTCC CCTGTGATGC CGTCGCCGCC |
|
| 801 | GCAGTCGGTC GGGGAATATG TGCTGCTGGC GTTTGCGGCG GCGGTGTTGT |
|
| 851 | CTGTGTGCTG CCTGTTTCCT TTGTTGGCAA TTGTTGTGAA AGCGTGGTCG |
|
| 901 | GCCGGCGAAT CGTGGCGTGT GTTAATGGAA AGTGAAACGT GGCAGGCGGT |
|
| 951 | GTGGAATACT TTGCGCTTCT CGGCGGCGGC GGTGTATGCG GCGGCGGTTT |
|
| 1001 | TGGGTGTGGT GTATGCGGCG GCGGCGCGGC GGTCGGCGTG GATGCGCGGG |
|
| 1051 | CTGATGTTTT TGCCGTTTAT GGTGTCGCCG GTTTGTGTTT CGGCGGGCGT |
|
| 1101 | GCTGCTGCTT TATCCGCAGT GGACGGCTTC GTTGCCGTTG CTGCTGGCGA |
|
| 1151 | TGTATGCGCT GCTGGCGTAT CCGTTTGTGG CAAAAGATGT TTTATCAGCC |
|
| 1201 | TGGGATGCAC TGCCGCCGGA TTACGGCAGG GCGGCGGCGG GTTTGGGTGC |
|
| 1251 | AAACGGCTTT CAGACGGCAT GCCGCATCAC GTTCCCCCTC TTGAAACCGG |
|
| 1301 | CGTTGCGGCG CGGTCTGACT TTGGCGGCGG CAACCTGCGT GGGCGAATTT |
|
| 1351 | GCGGCGACAT TGTTTCTGTC GCGTCCGGAA TGGCAGACGC TGACGACTTT |
|
| 1401 | GATTTATGCC TATTTGGGAC GCGCGGGTGA GGATAATTAC GCGCGGGCGA |
|
| 1451 | TGGTGCTGAC ATTGCTGTTG GCGGCGTTCG CGCTGGGTAT TTTCCTGCTG |
|
| 1501 | TTGGACGGCG GCGAAGGCGG AAAACAGACG GAAACGTTAT AA |
[1252]This corresponds to the amino acid sequence <SEQ ID 576; ORF139-1>:
[0000]| 1 | MDGRRWVVWG AFALLPSAFL AVMVVAPLWA VAAYDGLAWR AVLSDAYMLK | |
|
| 51 | RLAWTVFQAA ATCVLVLPLG VPVAWVLARL AFPGRALVLR LLMLPFVMPT |
|
| 101 | LVAGVGVLAL FGADGLLWRG RQDTPYLLLY GNVFFNLPVL VRAAYQGFVQ |
|
| 151 | VPAARLQTAR TLGAGAWRRF WDIEMPVLRP WLAGGVCLVF LYCFSGFGLA |
|
| 201 | LLLGGSRYAT VEVEIYQLVM FELDMAVASV LVWLVLGVTA AAGLLYAWFG |
|
| 251 | RRAVSDKAVS PVMPSPPQSV GEYVLLAFAA AVLSVCCLFP LLAIVVKAWS |
|
| 301 | AGESWRVLME SETWQAVWNT LRFSAAAVYA AAVLGVVYAA AARRSAWMRG |
|
| 351 | LMFLPFMVSP VCVSAGVLLL YPQWTASLPL LLAMYALLAY PFVAKDVLSA |
|
| 401 | WDALPPDYGR AAAGLGANGF QTACRITFPL LKPALRRGLT LAAATCVGEF |
|
| 451 | AATLFLSRPE WQTLTTLIYA YLGRAGEDNY ARAMVLTLLL AAFALGIFLL |
|
| 501 | LDGGEGGKQT ETL* |
[1253]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1254]ORF139 shows 94.7% identity over a 189aa overlap with an ORF (ORF139a) from strain A of N. meningitidis:
[0000]
[1255]The complete length ORF139a nucleotide sequence <SEQ ID 577> is:
[0000]| 1 | ATGGATGGAC GGCGTTGGGC GGTATGGGGT GCTTTTGCCC TGCTGCCTTC | |
|
| 51 | GGCTTTTTTG GCGGCAATGG TCGTTGCGCC TTTGTGGGCG GTGGCGGCGT |
|
| 101 | ATGACGGTTT GGCGTGGCGC GCGGTGCTGT CGGATGCCTA TATGCTCAAA |
|
| 151 | CGTTTGGCGT GGACGGTATT TCAGGCAGCG GCAACCTGTG TGCTGGTGCT |
|
| 201 | GCCTTTGGGC GTGCCTGTCG CGTGGGTGCT GGCGCGGCTG GCGTTTCCGG |
|
| 251 | GGCGGGCTTT GGTGCTGCGC CTGCTGATGC TGCCTTTTGT GATGCCCACG |
|
| 301 | TTGGTGGCGG GCGTGGGCGT GCTGGCTCTG TTCGGGGCGG ACGGCCTGTN |
|
| 351 | GTGGCGCGGC TGGCAGGATA CGCCGTATCT GTTGTTGTAC GGCAATGTGT |
|
| 401 | TTTTTNACCT TCCTGTGTTG GTCAGGGCGG CATATCAGGG GTTTGTGCAA |
|
| 451 | GTGCCTGCGG CACGGCTTCA GACGGCACNG ACATTGGGCG CGGGGGCGTG |
|
| 501 | GCGGCGGTTT TGGGACATTG AAATGCCCGT TTTGCGCCCG TGGCTTGCCG |
|
| 551 | GCGGCGTGTG CCTTGTCTTC CTGTATTGTT TTTCGGGGTT CGGGCTGGCA |
|
| 601 | TTGCTGCTGG GCGGCAGCCG TTATGCCACG GTCGAAGTGG AAATTTACCA |
|
| 651 | GTTGGTCATG TTCGAACTCG ATATGGCGGT TGCTTCGGTG CTNGTGTGGC |
|
| 701 | TGGTGTNGGG GGTAACNGCG GCGGCAGGGT TGCTGTATGC GTGGTTCGGC |
|
| 751 | AGGCGCGCGG TTTCGGATAA GGCNGTTTCC CCTGTGATGC CGTCGCCGCC |
|
| 801 | GCAGTCGGTC GGGGAATATG TGCTNCTGGC GTTTGCGGCG GCGGTGTNGT |
|
| 851 | CTGTGTGCTG CCTGTTTCNT TTGTTGGCAA TTGTTGTGAA AGCGTGGTCG |
|
| 901 | GCCGGCGAAT CGTGGCGTGT GTTAATGGAA AGTGAAACGT GGCAGGCGGT |
|
| 951 | GTGGAATACT NTGCGCTTCT CGGCGGCGGC GGTGTATGCG GCGGCGGTTT |
|
| 1001 | TGGGTGTGGT GTATGCGGCG GCGGCGCGGC GGTCGGCGTG GATGCGCGGG |
|
| 1051 | CTGATGTTTT TGCCGTTTAT GGTGTCGCCG GTTTGTGTTT CGGCGGGCGT |
|
| 1101 | GCTGCTGCTT NATCCGCAGT GGACGGCTTC GTTGCCGCTG CTGCTGGCGA |
|
| 1151 | TGTATGCGCT GCTGGCGTAT CCGTTTGTGG CAAAAGATGT TTTATCAGCC |
|
| 1201 | TGNGATGCAC TGCCGCCGGA TTACGGCAGG GCGGCGGCGG GTTTGGGTGC |
|
| 1251 | AAACGGCTTT CAGACGGCAT GCCGCATCAC GTTCCCCCTC TTGAAACCGG |
|
| 1301 | CGTTGCGGCG CGGTCTGACT TTGGCGGCGG CAACCTGCGT GGGCGAATTT |
|
| 1351 | GCGGCAACCT TGTTCNTGTC GCGTCNCGAG TGGCAGACGC TGACGACTTT |
|
| 1401 | GATTTATGCC TATNTGGGAC GCGCGGGTGA NGATAATTAC GCGCGGGCGA |
|
| 1451 | TGGTGCTGAC ATTGCTGTTG GCGGCGTTCG CGCTGGGTAT NTTCCTGCTG |
|
| 1501 | TTGGACGGCG GCGAAGGCGG AAAACGGACG GAAACGTTAT AA |
[1256]This encodes a protein having amino acid sequence <SEQ ID 578>:
[0000]| 1 | MDGRRWAVWG AFALLPSAFL AAMVVAPLWA VAAYDGLAWR AVLSDAYMLK | |
|
| 51 | RLAWTVFQAA ATCVLVLPLG VPVAWVLARL AFPGRALVLR LLMLPFVMPT |
|
| 101 | LVAGVGVLAL FGADGLXWRG WQDTPYLLLY GNVFFXLPVL VRAAYQGFVQ |
|
| 151 | VPAARLQTAX TLGAGAWRRF WDIEMPVLRP WLAGGVCLVF LYCFSGFGLA |
|
| 201 | LLLGGSRYAT VEVEIYQLVM FELDMAVSV LVWLVXGVTA AAGLLYAWFG |
|
| 251 | RRAVSDKAVS PVMPSPPQSV GEYVLLAFAA AVXSVCCLFX LLAIVVKAWS |
|
| 301 | AGESWRVLME SETWQAVWNT XRFSAAAVYA AAVLGVVYAA AARRSAWMRG |
|
| 351 | LMFLPFMVSP VCVSAGVLLL XPQWTASLPL LLAMYALLAY PFVAKDVLSA |
|
| 401 | XDALPPDYGR AAAGLGANGF QTACRITFPL LKPALRRGLT LAAATCVGEF |
|
| 451 | AATLFXSRXE WQTLTTLIYA YXGRAGXDNY ARAMVLTLLL AAFALGXFLL |
|
| 501 | LDGGEGGKRT ETL* |
[1257]ORF139a and ORF139-1 show 96.5% homology over a 514aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1258]ORF139 shows 95.2% identity over a 189aa overlap with a predicted ORF (ORF139ng) from N. gonorrhoeae:
[0000]
[1259]The complete length ORF139ng nucleotide sequence <SEQ ID 579> is predicted to encode a protein having amino acid sequence <SEQ ID 580>:
[0000]| 1 | MDGRCWAVRG AFSLLPSAFL AVMVVAPLWA VAAYDGLAWR AVLSDAYMLK | |
|
| 51 | RLAWTVFQAA ATCVLVLPLG VPVAWVLARL AFPGRALVLR LLMLPFVMPT |
|
| 101 | LVAGVGVLAL FGADGLLWRG RQDTPYLLLY GNVFFNLPVL VRAAYQGFAQ |
|
| 151 | VPAARLQTAR TLGAGAWRPF WDIEMPVLRP WLAGGVCLVF LYCFSGFGLA |
|
| 201 | LLLGGSRYAT VEVEIYQLVM FELDMAGASA LVWLVLGVTA AAGLLYAWFG |
|
| 251 | RRAVSDKAVS PVMPSPPQSV GEYVLLAFSV AVLSVCCLFP LSAIVVKAWS |
|
| 301 | AGESRRVLME SETWQAVWNT LRFSAAAVFA AAVLGVVYAA AARRLVWMRG |
|
| 351 | LVFLPFMVSP VCVSAGVLLL YPGWTASLPL LLAMYALLAY PFVAKDVLSA |
|
| 401 | WDALPPDYGR AAAGLGANGF QTACRITFPL LKPALRRGLT LAAATCVGEF |
|
| 451 | AATLFLSRPE WQTLTTLIYA YLGRAGEDNY ARAMVLTLLL SAFAVCIFLL |
|
| 501 | LDNGEGGKRT ETL* |
[1260]Further work revealed a variant gonococcal DNA sequence <SEQ ID 581>:
[0000]| 1 | ATGGATGGAC GGTGTTGGGC GGTACGGGGT GCTTTTTCCC TGCTGCCTTC | |
|
| 51 | GGCTTTTTTG GCGGTAATGG TCGTTGCGCC TTTGTGGGCG GTGGCGGCGT |
|
| 101 | ATGACGGTTT GGCGTGGCGC GCGGTGCTGT CGGATGCCTA TATGCTCAAA |
|
| 151 | CGTTTGGCGT GGACGGTGTT TCAGGCGGCG GCAACCTGTG TGCTGGTGCT |
|
| 201 | GCCTTTGGGC GTGCCTGTCG CGTGGGTGCT GGCGCGGCTG GCGTTCCCGG |
|
| 251 | GGCGGGCTTT GGTGCTGCGC CTGCTGATGC TGCCGTTTGT GATGCCCACG |
|
| 301 | CTGGTGGCGG GCGTGGGCGT GCTGGCTCTG TTCGGGGCGG ACGGGCTGTT |
|
| 351 | GTGGCGCGGC CGGCAGGATA CGCCGTATCT GTTGTTGTAC GGCAATGTGT |
|
| 401 | TTTTCAACCT GCCCGTGTTG GTCAGGGCGG CGTATCAGGG GTTTGCTCAA |
|
| 451 | GTGCCTGCGG CACGGCTTCA GACGGCACGG ACGTTGGGCG CGGGGGCGTG |
|
| 501 | GCGGCGGTTT TGGGACATTG AAATGCCCGT TTTGCGCCCG TGGCTTGCCG |
|
| 551 | GCGGCGTGTG CCTTGTCTTC CTGTATTGTT TTTCGGGGTT CGGGCTGGCA |
|
| 601 | TTGCTGTTGG GCGGCAGCCG TTATGCCACG GTCGAAGTGG AAATTTACCA |
|
| 651 | GTTGGTTATG TTCGAACTCG ATATGGCGGG GGCTTCGGCG CTGGTGTGGC |
|
| 701 | TGGTGTTGGG GGTAACGGCG GCGGCAGGGT TGCTGTATGC GTGGTTCGGC |
|
| 751 | AGGCGCGCGG TTTCGGATAA GGCGGTTTCC CCCGTGATGC CGTCGCCGCC |
|
| 801 | GCAATCGGTG GGGGAATATG TATTGCTGGC ATTTTCGGTG GCGGTGTTGT |
|
| 851 | CCGTGTGCTG CCTGTTTCCT TTGTCGGCAA TTGTTGTGAA AGCGTGGTCG |
|
| 901 | GCCGGCGAAT CGCGGCGTGT GTTAATGGAA AGTGAAACGT GGCAGGCAGT |
|
| 951 | GTGGAATACt ttGCGCTTTT CGGCGGCGGC GGTGTTTGCG GCGGCGGTTT |
|
| 1001 | TGGGTGTGGT GTATGCGGCG GCGGCGCGGC GGCTGGTGTG GATGCGCGGA |
|
| 1051 | CTGGTGTTTT TACCGTTTAT GGTGTCGCCG GTTTGTGTTT CGGCGGGCGT |
|
| 1101 | GCTGCTGCTT TATCCGGGGT GGACGGCTTC GTTACCGCTG CTGCTGGCGA |
|
| 1151 | TGTATGCGCT GCTGGCGTAT CCGTTTGTGG CAAAAGATGT TTTATCGGCC |
|
| 1201 | TGGGATGCAC TGCCGCCGGA TTACGGCAGG GCGGCGGCAG GTTTGGGCGC |
|
| 1251 | AAACGGCTTT CAGACGGCAT GCCGTATCAC GTTCCCCCTC TTGAAACCGG |
|
| 1301 | CGTTGCGGCG CGGTCTGACT TTGGCGGCGG CGACGTGTGT GGGCGAATTT |
|
| 1351 | GCGGCAACCT TGTTCCTGTC GCGTCCGGAA TGGCAGACGT TGACGACTTT |
|
| 1401 | GATTTATGCC TATTTGGGGC GTGCGGGTGA GGACAATTAT GCGCGGGCAA |
|
| 1451 | TGGTGTTGAC ATTGCTGTTG TCGGCATTTG CGGTGTGCAT TTTCCTGCTG |
|
| 1501 | TTGGACAACG GCGAAGGCGg aaaACGGACG GAAACGTTAT AA |
[1261]This corresponds to the amino acid sequence <SEQ ID 582; ORF139ng-1>:
[0000]| 1 | MDGRCWAVRG AFSLLPSAFL AVMVVAPLWA VAAYDGLAWR AVLSDAYMLK | |
|
| 51 | RLAWTVFQAA ATCVLVLPLG VPVAWVLARL AFPGRALVLR LLMLPFVMPT |
|
| 101 | LVAGVGVLAL FGADGLLWRG RQDTPYLLLY GNVFFNLPVL VRAAYQGFAQ |
|
| 151 | VPAARLQTAR TLGAGAWRRF WDIEMPVLRP WLAGGVCLVF LYCFSGFGLA |
|
| 201 | LLLGGSRYAT VEVEIYQLVM FELDMAGASA LVWLVLGVTA AAGLLYAWFG |
|
| 251 | RRAVSDKAVS PVMPSPPQSV GEYVLLAFSV AVLSVCCLFP LSAIVVKAWS |
|
| 301 | AGESRRVLME SETWQAVWNT LRFSAAAVFA AAVLGVVYAA AARRLVWMRG |
|
| 351 | LVFLPFMVSP VCVSAGVLLL YPGWTASLPL LLAMYALLAY PFVAKDVLSA |
|
| 401 | WDALPPDYGR AAAGLGANGF QTACRITFPL LKPALRRGLT LAAATCVGEF |
|
| 451 | AATLFLSRPE WQTLTTLIYA YLGRAGEDNY ARAMVLTLLL SAFAVCIFLL |
|
| 501 | LDNGEGGKRT ETL* |
[1262]ORF139ng-1 and ORF139-1 show 95.9% identity over 513aa overlap:
[0000]
[1263]Based on the presence of a predicted binding-protein-dependent transport systems inner membrane component signature (underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 70
[1264]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 583>:
[0000]| 1 | ATGGACGGCT GGACACAGAC GCTGTCCGCG CAAACCCTGT TGGGCATTTC | |
|
| 51 | GGCGGCGGCA ATCATCCTCA TTCTGATTTT AATCGTCAGA TTCCGCATCC |
|
| 101 | ACGCGCTGCT GACACTGGTC ATCGTCAGCC TGCTGACGGC TTTGGCAACC |
|
| 151 | GGTTTGCCCA CAGGCAGCAT TGTCAAAGAC ATACTGGTCA AAAACTTCGG |
|
| 201 | CGGCACGCTC GGCGGCGTGG CGCTTCTGGT CGGCCTGGGC GCGATGCTCG |
|
| 251 | AACGTTTGGT C... |
[1265]This corresponds to the amino acid sequence <SEQ ID 584; ORF140>:
[0000]| 1 | MDGWTQTLSA QTLLGISAAA IILILILIVR FRIHALLTLV IVSLLTALAT | |
|
| 51 | GLPTGSIVKD ILVKNFGGTL GGVALLVGLG AMLERLV.. |
[1266]Further work revealed the complete nucleotide sequence <SEQ ID 585>:
[0000]| 1 | ATGGACGGCT GGACACAGAC GCTGTCCGCG CAAACCCTGT TGGGCATTTC | |
|
| 51 | GGCGGCGGCA ATCATCCTCA TTCTGATTTT AATCGTCAAA TTCCGCATCC |
|
| 101 | ACGCGCTGCT GACACTGGTC ATCGTCAGCC TGCTGACGGC TTTGGCAACC |
|
| 151 | GGTTTGCCCA CAGGCAGCAT TGTCAACGAC ATACTGGTCA AAAACTTCGG |
|
| 201 | CGGCACGCTC GGCGGCGTGG CGCTTCTGGT CGGCCTGGGC GCGATGCTCG |
|
| 251 | GACGTTTGGT CGAAACATCC GGCGGCGCAC AGTCGCTGGC GGACGCGCTG |
|
| 301 | ATCCGGATGT TCGGCGAAAA ACGCGCACCG TTCGCGCTGG GCGTTGCCTC |
|
| 351 | GCTGATTTTC GGCTTCCCGA TTTTCTTCGA TGCCGGACTA ATCGTCATGC |
|
| 401 | TGCCCATCGT GTTCGCCACC GCACGGCGCA TGAAACAGGA CGTACTGCCC |
|
| 451 | TTCGCGCTTG CCTCCATCGG CGCATTTTCC GTCATGCACG TCTTCCTGCC |
|
| 501 | GCCCCATCCG GGCCCGATTG CCGCTTCCGA ATTTTACGGC GCGAACATCG |
|
| 551 | GCCAAGTTTT GATTTTGGGT CTGCCGACCG CCTTCATCAC ATGGTATTTC |
|
| 601 | AGCGGCTATA TGCTCGGCAA AGTGTTGGGG CGCACCATCC ATGTTCCCGT |
|
| 651 | TCCCGAACTG CTCAGCGGCG GCACGCAAGA CAACGACCTG CCGAAAGAAC |
|
| 701 | CTGCCAAAGC AGGAACGGTC GTCGCCATCA TGCTGATTCC CATGCTGCTG |
|
| 751 | ATTTTCCTGA ATACCGGCGT ATCGGCCCTC ATCAGCGAAA AACTCGTAAG |
|
| 801 | TGCGGACGAA ACCTGGGTTC AGACGGCAAA AATAATCGGT TCGACACCGA |
|
| 851 | TCGCCCTTCT GATTTCCGTA TTGGTCGCAC TGTTTGTCTT GGGACGCAAA |
|
| 901 | CGCGGCGAAA GCGGCAGCGC GTTGGAAAAA ACCGTGGACG GCGCACTCGC |
|
| 951 | CCCCGTCTGT TCCGTGATTC TGATTACCGG CGCGGGCGGT ATGTTCGGCG |
|
| 1001 | GCGTTTTGCG CGCTTCCGGC ATCGGCAAGG CACTCGCCGA CAGCATGGCG |
|
| 1051 | GATTTGGGCA TTCCCGTCCT TTTGGGCTGT TTCCTTGTCG CCTTGGCACT |
|
| 1101 | GCGTATCGCG CAAGGTTCGG CAACCGTCGC CCTGACCACC GCCGCCGCGC |
|
| 1151 | TGATGGCTCC TGCCGTTGCC GCCGCCGGCT TTACCGACTG GCAGCTCGCC |
|
| 1201 | TGTATCGTAT TGGCAACGGC GGCAGGTTCG GTCGGTTGCA GCCACTTCAA |
|
| 1251 | CGACTCCGGC TTCTGGCTGG TCGGCCGTCT CTTGGACATG GACGTACCGA |
|
| 1301 | CCACGCTGAA AACCTGGACG GTCAACCAAA CCCTCATCGC ACTCATCGGC |
|
| 1351 | TTTGCCTTGT CCGCACTGCT GTTCGCCATC GTCTGA |
[1267]This corresponds to the amino acid sequence <SEQ ID 586; ORF140-1>:
[0000]| 1 | MDGWTQTLSA QTLLGISAAA IILILILIVK FRIHALLTLV IVSLLTALAT | |
|
| 51 | GLPTGSIVND ILVKNFGGTL GGVALLVGLG AMLGRLVETS GGAQSLADAL |
|
| 101 | IRMFGEKRAP FALGVASLIF GFPIFFDAGL IVMLPIVFAT ARRMKQDVLP |
|
| 151 | FALASIGAFS VMHVFLPPHP GPIAASEFYG ANIGQVLILG LPTAFITWYF |
|
| 201 | SGYMLGKVLG RTIHVPVPEL LSGGTQDNDL PKEPAKAGTV VAIMLIPMLL |
|
| 251 | IFLNTGVSAL ISEKLVSADE TWVQTAKIIG STPIALLISV LVALFVLGRK |
|
| 301 | RGESGSALEK TVDGALAPVC SVILITGAGG MFGGVLRASG IGKALADSMA |
|
| 351 | DLGIPVLLGC FLVALALRIA QGSATVALTT AAALMAPAVA AAGFTDWQLA |
|
| 401 | CIVLATAAGS VGCSHFNDSG FWLVGRLLDM DVPTTLKTWT VNQTLIALIG |
|
| 451 | FALSALLFAI V* |
[1268]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1269]ORF140 shows 95.4% identity over a 87aa overlap with an ORF (ORF140a) from strain A of N. meningitidis.
[0000]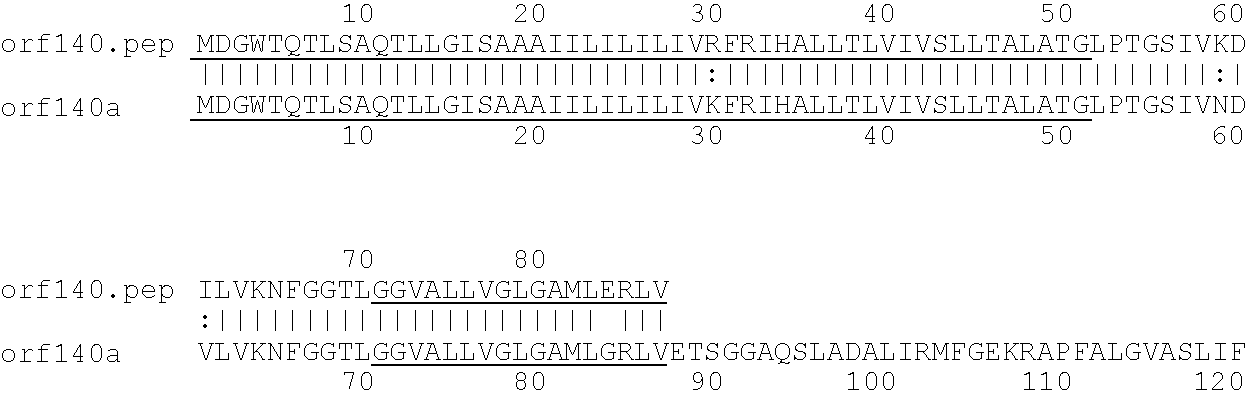
[1270]The complete length ORF140a nucleotide sequence <SEQ ID 587> is:
[0000]| 1 | ATGGACGGCT GGACACAGAC GCTGTCCGCG CAAACCCTGT TGGGCATTTC | |
|
| 51 | GGCGGCGGCA ATCATCCTCA TTCTGATTTT AATCGTCAAA TTCCGCATCC |
|
| 101 | ACGCGCTGCT GACACTGGTC ATCGTCAGCC TGCTGACGGC TTTGGCAACC |
|
| 151 | GGTTTGCCCA CAGGCAGCAT TGTCAACGAC GTACTGGTCA AAAACTTCGG |
|
| 201 | CGGCACGCTC GGCGGCGTGG CGCTTCTGGT CGGCCTGGGC GCGATGCTCG |
|
| 251 | GACGTTTGGT CGAAACATCC GGCGGCGCAC AGTCGCTGGC GGACGCGCTG |
|
| 301 | ATCCGGATGT TCGGCGAAAA ACGCGCACCG TTCGCGCTGG GCGTTGCCTC |
|
| 351 | GCTGATTTTC GGCTTCCCGA TTTTCTTCGA TGCCGGACTA ATCGTCATGC |
|
| 401 | TGCCCATCGT GTTCGCCACC GCACGGCGCA TGAAACAGGA CGTACTGCCC |
|
| 451 | TTCGCGCTTG CCTCCATCGG CGCATTTTCC GTCATGCACG TCTTCCTGCC |
|
| 501 | GCCCCATCCG GGCCCGATTG CCGCTTCCGA ATTTTACGGC GCGAACATCG |
|
| 551 | GCCAAGTTTT GATTTTGGGT CTGCCGACCG CCTTCATCAC ATGGTATTTC |
|
| 601 | AGCGGCTATA TGCTCGGCAA AGTGTTGGGG CGCACCATCC ATGTTCCCGT |
|
| 651 | TCCCGAACTG CTCAGCGGCG GCACGCAAGA CAACGACCTG CCGAAAGAAC |
|
| 701 | CTGCCAAAGC AGGAACGGTC GTCGCCATCA TGCTGATTCC CATGCTGCTG |
|
| 751 | ATTTTCCTGA ATACCGGCGT ATCGGCCCTC ATCAGCGAAA AACTCGTAAG |
|
| 801 | TGCGGACGAA ACCTGGGTTC AGACGGCAAA AATAATCGGT TCGACACCGA |
|
| 851 | TCGCCCTTCT GATTTCCGTA TTGGTCGCAC TGTTTGTCTT GGGACGCAAA |
|
| 901 | CGCGGCGAAA GCGGCAGCGC GTTGGAAAAA ACCGTGGACG GCGCACTCGC |
|
| 951 | CCCCGTCTGT TCCGTGATTC TGATTACCGG CGCGGGCGGT ATGTTCGGCG |
|
| 1001 | GCGTTTTGCG CGCTTCCGGC ATCGGCAAGG CACTCGCCGA CAGCATGGCG |
|
| 1051 | GATTTGGGCA TTCCCGTCCT TTTGGGCTGT TTCCTTGTCG CCTTGGCACT |
|
| 1101 | GCGTATCGCG CAAGGTTCGG CAACCGTCGC CCTGACCACC GCCGCCGCGC |
|
| 1151 | TGATGGCTCC TGCCGTTGCC GCCGCCGGCT TTACCGACTG GCAGCTCGCC |
|
| 1201 | TGTATCGTAT TGGCAACGGC GGCAGGTTCG GTCGGTTGCA GCCACTTCAA |
|
| 1251 | CGACTCCGGC TTCTGGCTGG TCGGCCGCCT CTTGGACATG GACGTACCGA |
|
| 1301 | CCACGCTGAA AACCTGGACG GTCAACCAAA CCCTCATCGC ACTCATCGGC |
|
| 1351 | TTTGCCTTGT CCGCACTGCT GTTCGCCATC GTCTGA |
[1271]This encodes a protein having amino acid sequence <SEQ ID 588>:
[0000]| 1 | MDGWTQTLSA QTLLGISAAA IILILILIVK FRIHALLTLV IVSLLTALAT | |
|
| 51 | GLPTGSIVND VLVKNFGGTL GGVALLVGLG AMLGRLVETS GGAQSLADAL |
|
| 101 | IRMFGEKRAP FALGVASLIF GFPIFFDAGL IVMLPIVFAT ARRMKQDVLP |
|
| 151 | FALASIGAFS VMHVFLPPHP GPIAASEFYG ANIGQVLILG LPTAFITWYF |
|
| 201 | SGYMLGKVLG RTIHVPVPEL LSGGTQDNDL PKEPAKAGTV VAIMLIPMLL |
|
| 251 | IFLNTGVSAL ISEKLVSADE TWVQTAKIIG STPIALLISV LVALFVLGRK |
|
| 301 | RGESGSALEK TVDGALAPVC SVILITGAGG MFGGVLRASG IGKALADSMA |
|
| 351 | DLGIPVLLGC FLVALALRIA QGSATVALTT AAALMAPAVA AAGFTDWQLA |
|
| 401 | CIVLATAAGS VGCSHFNDSG FWLVGRLLDM DVPTTLKTWT VNQTLIALIG |
|
| 451 | FALSALLFAI V* |
[1272]ORF140a and ORF140-1 show 99.8% identity over a 461aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1273]ORF140 shows 92% identity over a 87aa overlap with a predicted ORF (ORF140ng) from N. gonorrhoeae:
[0000]
[1274]The complete length ORF140ng nucleotide sequence <SEQ ID 589> was predicted to encode a protein having amino acid sequence <SEQ ID 590>:
[0000]| 1 | MDGRTQTLSA QTLLGISAAA IILILILIVK FRIRALLTLV IASLLTALAT | |
|
| 51 | GLPTGSIVND VLVKNFGGTL GGVALLVGLG AMLGRLVETS GGAQSLADAL |
|
| 101 | IRMFGEKRAP FAPGVASLIF GFPIFFDAGL IVMLPIVFAT ARRMKQDVLP |
|
| 151 | FALASVGAFS VMHVFLPPHP GPIAASEFYG ANIGQVLILG LPTAFITWYF |
|
| 201 | SGYMLGKVLG RAIHVPVPEL LSGGTQDSDP PKEPAKAGTV VAVMLIPMLL |
|
| 251 | IFLNTGVSAL ISEKLVSADE TWVQTAKMIG STPVALLISV LAALLVLGRK |
|
| 301 | RGESGSTLEK TVDGALAPAC SVILITGAGG MFGGVLRASG IGKALADSMA |
|
| 351 | DLGIPVLLGC FLVALALRIA QGSATVALTT AAALMAPAVA AAGFTDWQLA |
|
| 401 | CIVLATAAGS VGCSHFNDSG FWLVGRLSDM DVPTTLKTWT VNQTLIAFIG |
|
| 451 | FALSALLFAI V* |
[1275]Further work revealed a variant gonococcal DNA sequence <SEQ ID 591>:
[0000]| 1 | ATGGACGGCC GGACACAGAC GCTGTCCGCG CAAACCTTGT TGGGCATTTC | |
|
| 51 | GGCGGCGGCA ATCATCCTCA TTCTGATTTT AATCGTCAAA TTCCGCATCC |
|
| 101 | GCGCGCTGCT GACACTGGTC ATCGCCAGCC TGCTGACGGC TTTGGCAACC |
|
| 151 | GGTTTGCCCA CAGGCAGCAT CGTCAACGAC GTACTGGTCA AAAACTTCGG |
|
| 201 | CGGCACGCTC GGCGGCGTGG CGCTTCTGGT CGGTCTGGGC GCAATGCTCG |
|
| 251 | GACGTTTGGT AGAAACATCC GGCGGCGCAC AGTCGCTGGC GGACGCGCTG |
|
| 301 | ATCCGGATGT TCGGCGAAAA ACGCGCACCG TTCGCTCCGG GCGTTGCCTC |
|
| 351 | GCTGATTTTC GGCTTCCCGA TTTTCTTCGA TGCCGGACTA ATCGTCATGC |
|
| 401 | TGCCCATCGT ATTCGCCACC GCACGGCGCA TGAAACAGGA CGTACTGCCC |
|
| 451 | TTCGCGCTTG CCTCCGTCGG CGCATTTTCC GTCATGCACG TCTTCCTGCC |
|
| 501 | GCCCCATCCG GGCCCGATTG CCGCTTCCGA ATTTTACGGC GCGAACATCG |
|
| 551 | GCCAGGTTTT GATTTTGGGT CTGCCGACCG CCTTCATCAC ATGGTATTTC |
|
| 601 | AGCGGCTATA TGCTCGGCAA AGTGTTGGGG CGCGCCATCC ATGTTCCCGT |
|
| 651 | TCCCGAACTG CTCAGCGGCG GCACGCAAGA CAGCGACCCG CCGAAAGAAC |
|
| 701 | CTGCCAAAGC AGGAACGGTC GTCGCCGTCA TGCTGATTCC CATGCTGCTG |
|
| 751 | ATTTTCCTGA ATACCGGCGT ATCAGCCCTC ATCAGCGAAA AACTCGTAAG |
|
| 801 | TGCGGACGAA ACTTGGGTTC AGACGGCAAA AATGATCGGT TCGACACCTG |
|
| 851 | TCGCCCTTCT GATTTCCGTA TTGGCCGCAC TGTTGGTCTT GGGACGCAAA |
|
| 901 | CGCGGCGAAA GCGGCAGCAC GTTGGAAAAA ACCGTGGACG GCGCACTCGC |
|
| 951 | CCCCGCCTGT TCCGTGATTC TGATTACCGG CGCGGGCGGT ATGTTCGGCG |
|
| 1001 | GCGTTTTGCG CGCTTCCGGC ATCGGCAAGG CACTCGCCGA CAGCATGGCG |
|
| 1051 | GATTTGGGCA TTCCCGTCCT TTTGGGCTGC TTCCTTGTCG CCTTGGCACT |
|
| 1101 | GCGTATCGCG CAAGGTTCGG CAACCGTCGC CCTGACCACA GCCGCCGCGC |
|
| 1151 | TGATGGCTCC TGCCGTTGCC GCCGCCGGCT TTACCGACTG GCAGCTCGCC |
|
| 1201 | TGTATCGTAT TGGCAACGGC GGCAGGTTCG GTCGGTTGCA GCCACTTCAA |
|
| 1251 | CGACTCCGGC TTCTGGCTGG TCGGCCGCCT CTTGGATATG GACGTACCGA |
|
| 1301 | CCACGCTGAA AACCTGGACG GTCAACCAAA CCCTCATCGC ATTCATCGGC |
|
| 1351 | TTTGCCTTGT CCGCACTGCT GTTTGCCATC GTCTGA |
[1276]This corresponds to the amino acid sequence <SEQ ID 592; ORF140ng-1>:
[0000]| 1 | MDGRTQTLSA QTLLGISAAA IILILILIVK FRIRALLTLV IASLLTALAT | |
|
| 51 | GLPTGSIVND VLVKNFGGTL GGVALLVGLG AMLGRLVETS GGAQSLADAL |
|
| 101 | IRMFGEKRAP FAPGVASLIF GFPIFFDAGL IVMLPIVFAT ARRMKQDVLP |
|
| 151 | FALASVGAFS VMHVFLPPHP GPIAASEFYG ANIGQVLILG LPTAFITWYF |
|
| 201 | SGYMLGKVLG RAIHVPVPEL LSGGTQDSDP PKEPAKAGTV VAVMLIPMLL |
|
| 251 | IFLNTGVSAL ISEKLVSADE TWVQTAKMIG STPVALLISV LAALLVLGRK |
|
| 301 | RGESGSTLEK TVDGALAPAC SVILITGAGG MFGGVLRASG IGKALADSMA |
|
| 351 | DLGIPVLLGC FLVALALRIA QGSATVALTT AAALMAPAVA AAGFTDWQLA |
|
| 401 | CIVLATAAGS VGCSHFNDSG FWLVGRLLDM DVPTTLKTWT VNQTLIAFIG |
|
| 451 | FALSALLFAI V* |
[1277]ORF140ng-1 and ORF140-1 show 96.3% identity over 461aa overlap:
[0000]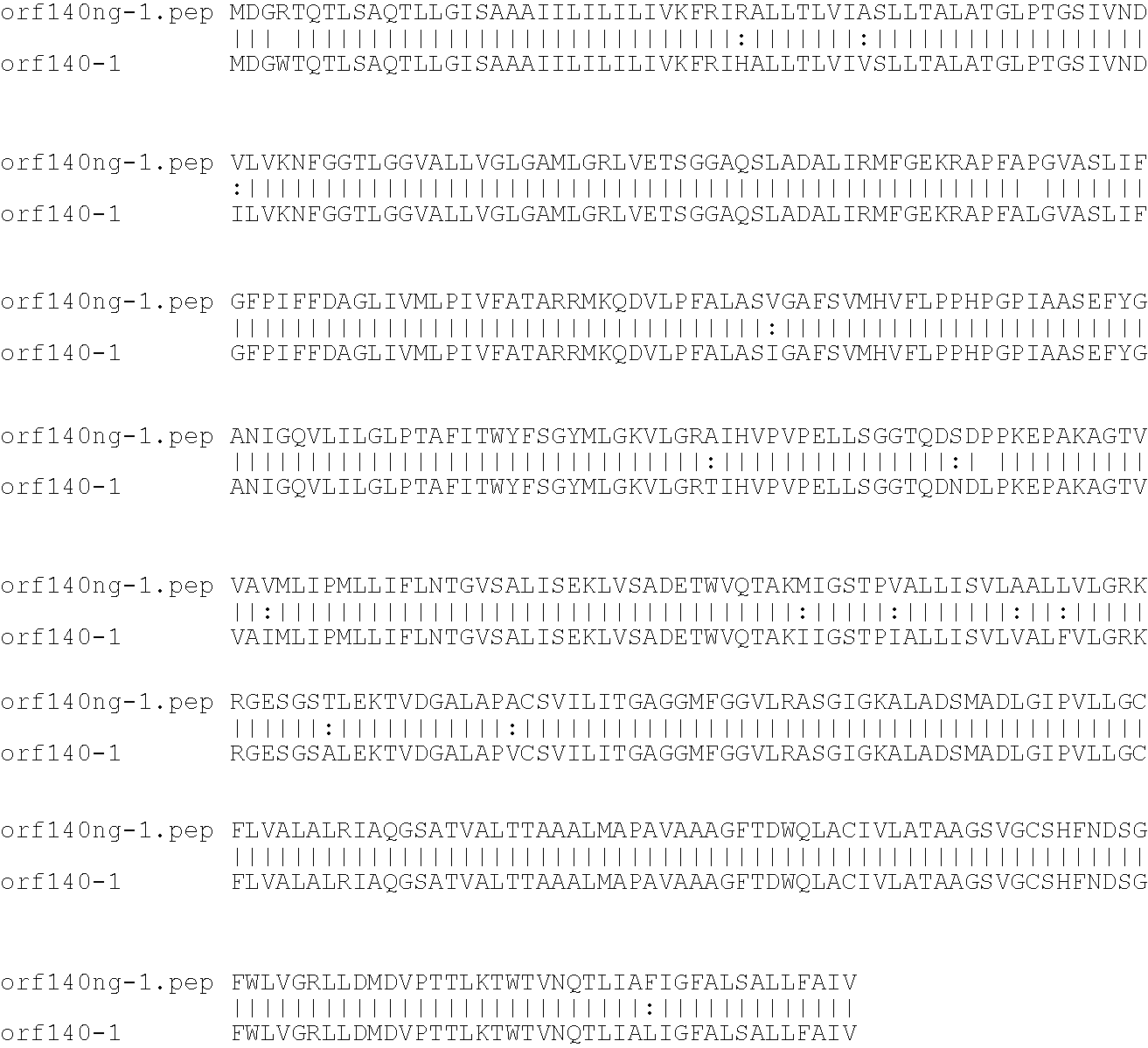
[1278]Furthermore, ORF140ng-1 is homologous to an E. coli protein:
[0000]| gi|882633 (U29579) ORF_o454 [Escherichia coli] >gi|1789097 (AE000358) o454; | |
| This 454 aa ORF is 34% identical (9 gaps) to 444 residues of an approx. 456 aa |
| protein GNTP_BACLI SW: P46832 [Escherichia coli] Length = 454 |
| Score = 210 bits (529), Expect = 1e−53 |
| Identities = 130/384 (33%), Positives = 194/384 (49%), Gaps = 19/384 (4%) |
|
| Query: | 88 | ETSGGAQSLADALIRMFGEKRAPFAPGVASLIFGFPIFFDAGLIVMLPIVFATARRMKQD | 147 | |
| | E SGGA+SLA+ R G+KR A +A+ G P+FFD G I++ PI++ A+ K |
| Sbjct: | 80 | EHSGGAESLANYFSRKLGDKRTIAALTLAAFFLGIPVFFDVGFIILAPIIYGFAKVAKIS | 139 |
|
| Query: | 148 | VLPFALASVGAFSVMHVFLPPHPGPIAASEFYGANIGQVLILGLPTAFITWYFSGYMLGK | 207 |
| | L F L G +HV +PPHPGP+AA+ A+IG + I+G+ + I GY K |
| Sbjct: | 140 | PLKFGLPVAGIMLTVHVAVPPHPGPVAAAGLLHADIGWLTIIGIAIS-IPVGVVGYFAAK | 198 |
|
| Query: | 208 | VLGRAIHVPVPELL----------SGGTQDSDPPKEPAKAGTVVAVMLIPMLLIFLNTGV | 257 |
| | ++ + + E+L G T+ SD P A V ++++IP+ +I T |
| Sbjct: | 199 | IINKRQYAMSVEVLEQMQLAPASEEGATKLSDKINPPGVA-LVTSLIVIPIAIIMAGT-- | 255 |
|
| Query: | 258 | SALISEKLVSADETWVQTAKMIGSTPXXXXXXXXXXXXXXGRKRGESGSTLEKTVDGALA | 317 |
| | +S L+ + T ++IGS +RG S + AL |
| Sbjct: | 256 | ---VSATLMPPSHPLLGTLQLIGSPMVALMIALVLAFWLLALRRGWSLQHTSDIMGSALP | 312 |
|
| Query: | 318 | PACSVILITGAGGMFGGVLRASGIGKALADSMADLGIPVLLGCFLVALALRIAQGSXXXX | 377 |
| | A VIL+TGAGG+FG VL SG+GKALA+ + + +P+L F+++LALR +QGS |
| Sbjct: | 313 | TAAVVILVTGAGGVFGKVLVESGVGKALANMLQMIDLPLLPAAFIISLALRASQGS--AT | 370 |
|
| Query: | 378 | XXXXXXXXXXXXXXXGFTDWQLACIVLATAAGSVGCSHFNDSGFWLVGRLLDMDVPTTLK | 437 |
| | G Q + LA G +G SH NDSGFW+V + L +V LK |
| Sbjct: | 371 | VAILTTGGLLSEAVMGLNPIQCVLVTLAACFGGLGASHINDSGFWIVTKYLGLSVADGLK | 430 |
|
| Query: | 438 | TWTVNQTLIAFIGFALSALLFAIV | 461 |
| | TWTV T++ F GF ++ ++A++ |
| Sbjct: | 431 | TWTVLTTILGFTGFLITWCVWAVI | 454 |
[1279]Based on this analysis, including the identification of the presence of a putative leader sequence (double-underlined) and several putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 71
[1280]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 593>:
[0000]| 1 | ..GATTTCGGCA TATCGCCCGT GTATCTTTGG GTTGCCGCCG CGTTCAAACA | |
|
| 51 | TTTGCTGTCG CCGTGGGCTG CCGACTCATA CGATGTCGCA CGCTTTGCAG |
|
| 101 | GCGTATTTTT TGCCGTTATC GGACTGACTT CCTGCGGCTT TGCCGGTTTC |
|
| 151 | AACTTTTTGG GCAGACACCA CGGGCGCAC. GTCGTCCTGA TTCTCATCGG |
|
| 201 | CTGTATCGGG CTGATTCCAG TTGCCCATTT CCTCAACCCC GCTGCCGCCG |
|
| 251 | CCTTTGCCGC CGCCGGACTG GTGCTGCACG GTTATTCTTT GGCTCGCCGG |
|
| 301 | CGCGTGATTG CCGCCTCTTT TCTGCTCGGT ACGGGCTGGA CGCTGATGTC |
|
| 351 | GTTGGCAGCA GCTTATCCGG CAGCATTTGC CCTGATGCTG CCCTTGCCCG |
|
| 401 | TACTGATGTT TTTCCGTCCG .. |
[1281]This corresponds to the amino acid sequence <SEQ ID 594; ORF141>:
[0000]| 1 | ..DFGISPVYLW VAAAFKHLLS PWAADSYDVA RFAGVFFAVI GLTSCGFAGF | |
|
| 51 | NFLGRHHGRX VVLILIGCIG LIPVAHFLNP AAAAFAAAGL VLHGYSLARR |
|
| 101 | RVIAASFLLG TGWTLMSLAA AYPAAFALML PLPVLMFFRP .. |
[1282]Further work revealed the complete nucleotide sequence <SEQ ID 595>:
[0000]| 1 | ATGCTGACCT ATACCCCGCC CGATGCCCGC CCGCCCGCCA AAACCCACGA | |
|
| 51 | AAAGCCGTGG CTGCTGCTGT TGATGGCGTT TGCCTGGTTG TGGCCCGGCG |
|
| 101 | TGTTTTCCCA CGATTTGTGG AATCCTGACG AACCTGCCGT CTATACCGCC |
|
| 151 | GTCGAAGCAC TGGCAGGCAG CCCCACCCCC TTGGTTGCCC ATCTGTTCGG |
|
| 201 | TCAAACCGAT TTCGGCATAC CGCCCGTGTA TCTTTGGGTT GCCGCCGCGT |
|
| 251 | TCAAACATTT GCTGTCGCCG TGGGCTGCCG ACTCATACGA TGCCGCACGC |
|
| 301 | TTTGCAGGCG TATTTTTTGC CGTTATCGGA CTGACTTCCT GCGGCTTTGC |
|
| 351 | CGGTTTCAAC TTTTTGGGCA GACACCACGG GCGCAgCGTC GTCCTGATTC |
|
| 401 | TCATCGGCTG TATCGGGCTG ATTCCAGTTG CCCATTTCCT CAACCCCGCT |
|
| 451 | GCCGCCGCCT TTGCCGCCGC CGGACTGGTG CTGCACGGTT ATTCTTTGGC |
|
| 501 | TCGCCGGCGC GTGATTGCCG CCTCTTTTCT GCTCGGTACG GGCTGGACGC |
|
| 551 | TGATGTCGTT GGCAGCAGCT TATCCGGCAG CATTTGCCCT GATGCTGCCC |
|
| 601 | TTGCCCGTAC TGATGTTTTT CCGTCCGTGG CAAAGCAGGC GTTTGATGTT |
|
| 651 | GACGGCAGTC GCCTCACTTG CCTTTGCCCT GCCGCTTATG ACCGTTTACC |
|
| 701 | CGCTGCTCTT GGCAAAAACG CAGCCCGCGC TGTTCGCGCA ATGGCTCGAC |
|
| 751 | TATCACGTTT TCGGTACGTT CGGCGGCGTG CGGCACGTTC AGACGGCATT |
|
| 801 | CAGTTTGTTT TACTATCTGA AAAACCTGCT TTGGTTTGCA TTGCCCGCGC |
|
| 851 | TGCCGCTGGC GGTTTGGACG GTTTGCCGCA CGCGCCTGTT TTCGACCGAC |
|
| 901 | TGGGGGATTT TGGGCGTCGT CTGGATGCTT GCCGTTTTGG TGCTGCTTGC |
|
| 951 | CGTCAATCCG CAGCGTTTTC AGGATAACCT CGTCTGGCTG CTTCCGCCGC |
|
| 1001 | TTGCCCTGTT CGGCGCGGCG CAACTGGACA GCCTGAGGCG CGGCGCGGCG |
|
| 1051 | GCGTTTGTCA ACTGGTTCGG CATTATGGCG TTCGGACTGT TTGCCGTGTT |
|
| 1101 | CCTGTGGACG GGCTTTTTCG CCATGAATTA CGGCTGGCCC GCCAAGCTTG |
|
| 1151 | CCGAACGCGC CGCCTATTTC AGCCCGTATT ATGTTCCTGA TATCGATCCC |
|
| 1201 | ATTCCGATGG CGGTTGCCGT ACTGTTCACA CCCTTGTGGC TGTGGGCGAT |
|
| 1251 | TACCCGGAAA AACATACGCG GCAGGCAGGC GGTTACCAAC TGGGCGGCAG |
|
| 1301 | GCGTTACCCT GACCTGGGCT TTGCTGATGA CGCTGTTCCT GCCGTGGCTG |
|
| 1351 | GACGCGGCGA AAAGCCACGC GCCGGTCGTC CGGAGTATGG AGGCATCGCT |
|
| 1401 | TTCCCCGGAA TTGAAACGGG AGCTTTCAGA CGGCATCGAG TGTATCGGCA |
|
| 1451 | TAGGCGGCGG CGACCTGCAC ACGCGGATTG TTTGGACGCA GTACGGCACA |
|
| 1501 | TTGCCGCACC GCGTCGGCGA TGTACAATGC CGCTACCGCA TCGTCCTCCT |
|
| 1551 | GCCCCAAAAT GCGGATGCGC CGCAAGGCTG GCAGACGGTT TGGCAGGGTG |
|
| 1601 | CGCGTCCGCG CAACAAAGAC AGTAAGTTCG CACTGATACG GAAAATCGGG |
|
| 1651 | GAAAATATAT AA |
[1283]This corresponds to the amino acid sequence <SEQ ID 596; ORF141-1>:
[0000]| 1 | MLTYTPPDAR PPAKTHEKPW LLLLMAFAWL WPGVFSHDLW NPDEPAVYTA | |
|
| 51 | VEALAGSPTP LVAHLFGQTD FGIPPVYLWV AAAFKHLLSP WAADSYDAAR |
|
| 101 | FAGVFFAVIGLTSCGFAGFN FLGRHHGRSV VLILIGCIGL IPVAHFLNPA |
|
| 151 | AAAFAAAGLV LHGYSLARRR VIAASFLLGTGWTLMSLAAA YPAAFALMLP |
|
| 201 | LPVLMFFRPW QSRRLMLTAV ASLAFALPLM TVYPLLLAKT QPALFAQWLD |
|
| 251 | YHVFGTFGGV RHVQTAFSLF YYLKNLLWFA LPALPLAVWT VCRTRLFSTD |
|
| 301 | WGILGVVWML AVLVLLAVNP QRFQDNLVWL LPPLALFGAA QLDSLRRGAA |
|
| 351 | AFVNWFGIMA FGLFAVFLWT GFFAMNYGWP AKLAERAAYF SPYYVPDIDP |
|
| 401 | IPMAVAVLFT PLWLWAITRK NIRGRQAVTN WAAGVTLTWA LLMTLFLPWL |
|
| 451 | DAAKSRAPVV RSMEASLSPE LKRELSDGIE CIGIGGGDLH TRIVWTQYGT |
|
| 501 | LPHRVGDVQC RYRIVLLPQN ADAPQGWQTV WQGARPRNKD SKFALIRKIG |
|
| 551 | ENI* |
[1284]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1285]ORF141 shows 95.0% identity over a 140aa overlap with an ORF (ORF141a) from strain A of N. meningitidis.
[0000]
[1286]The complete length ORF141a nucleotide sequence <SEQ ID 597> is:
[0000]| 1 | ATGCTGACCT ATACCCCGCC CGATGCCCGC CCGCCCGCCA AAACCCACGA | |
|
| 51 | AAAGCCGTGG CTGTTGCTGT TGATGGCGTT TGCCTGGTTG TGGCCCGGCG |
|
| 101 | TGTTTTCCCA CGATTTGTGG AATCCTGACG AACCTGCCGT CTATACCGCC |
|
| 151 | GTCGAAGCAC TGGCAGGCAG CCCCACCCCT TTGGTTGCCC ATCTGTTCGG |
|
| 201 | TCAAATCGAT TTCGGCATAC CGCCCGTGTA TCTTTGGGTT GCCGCCGCGT |
|
| 251 | TCAAACATTT GCTGTCGCCG TGGGCTGCCG ACCCGTATGA TGCCGCACGC |
|
| 301 | TTTGCCGGCG TGTTTTTCGC CGTTGTCGGA CTGACTTCCT GCGGCTTTGC |
|
| 351 | CGGTTTCAAC TTTTTGGGCA GACACCACGG GCGCAGCGTC GTCCTGATTC |
|
| 401 | TCATCGGCTG TATCGGGCTG ATTCCGACCG TACACTTTCT CAACCCCGCT |
|
| 451 | GCCGCCGCCT TTGCCGCCGC CGGACTGGTG CTGCACGGTT ATTCTTTGGC |
|
| 501 | TCGCCGGCGC GTGATTGCCG CCTCTTTTCT GCTCGGTACG GGTTGGACGC |
|
| 551 | TGATGTCGTT GGCAGCAGCT TATCCGGCGG CATTTGCCCT GATGCTGCCC |
|
| 601 | CTGCCCGTGC TGATGTTTTT CCGTCCGTGG CAAAGCAGGC GTTTGATGTT |
|
| 651 | GACGGCAGTC GCCTCGCTTG CCTTTGCCCT GCCGCTTATG ACCGTTTACC |
|
| 701 | CGCTGCTCTT GGCAAAAACG CAGCCCGCGC TGTTCGCGCA ATGGCTCGAC |
|
| 751 | GATCACGTTT TCGGTACGTT CGGCGGCGTG CGGCACATTC AGACGGCATT |
|
| 801 | CAGTTTGTTT TACTATCTGA AAAACCTGCT TTGGTTTGCA TTGCCTGCGC |
|
| 851 | TGCCGCTGGC GGTTTGGACG GTTTGCCGCA CGCGCCTGTT TTCGACCGAC |
|
| 901 | TGGGGGATTT TGGGCGTCGT CTGGATGCTT GCCGTTTTGG TGCTGCTTGC |
|
| 951 | CGTCAATCCG CAGCGTTTTC AGGATAACCT CGTCTGGCTG CTTCCGCCGC |
|
| 1001 | TTGCCCTGTT CGGCGCGGCG CAACTGGACA GCCTGAGACG CGGCGCGGCG |
|
| 1051 | GCGTTTGTCA ACTGGTTCGG CATTATGGCG TTCGGACTGT TTGCCGTGTT |
|
| 1101 | CCTGTGGACG GGCTTTTTCG CCATGAATTA CGGCTGGCCC GCCAAGCTTG |
|
| 1151 | CCGAACGCGC CGCCTATTTC AGCCCGTATT ATGTTCCTGA TATCGATCCC |
|
| 1201 | ATTCCGATGG CGGTTGCCGT ACTGTTCACA CCCTTGTGGC TGTGGGCGAT |
|
| 1251 | TACCCGCAAA AACATACGCG GCAGGCAGGC GGTTACCAAC TGGGCGGCAG |
|
| 1301 | GCGTTACCCT GACCTGGGCT TTGCTGATGA CGCTGTTCCT GCCGTGGCTG |
|
| 1351 | GACGCGGCGA AAAGCCACGC GCCCGTCGTC CGGAGTATGG AGGCATCGCT |
|
| 1401 | TTCCCCGGAA TTAAAACGGG AGCTTTCAGA CGGCATCGAG TGTATCGACA |
|
| 1451 | TAGGCGGCGG CGACCTACAC ACGCGGATTG TTTGGACGCA GTACGGCACA |
|
| 1501 | TTGCCGCACC GCGTCGGCGA TGTACAATGC CGCTACCGCA TCGTCCGCTT |
|
| 1551 | GCCCCAAAAC GCGGATGCGC CGCAAGGCTG GCAGACGGTC TGGCAGGGTG |
|
| 1601 | CGCGCCCGCG CAACAAAGAC AGTAAGTTCG CACTGATACG GAAAACCGGG |
|
| 1651 | GAAAATATAT TAAAAACAAC AGATTGA |
[1287]This encodes a protein having amino acid sequence <SEQ ID 598>:
[0000]| 1 | MLTYTPPDAR PPAKTHEKPW LLLLMAFAWL WPGVFSHDLW NPDEPAVYTA | |
|
| 51 | VEALAGSPTP LVAHLFGQID FGIPPVYLWV AAAFKHLLSP WAADPYDAAR |
|
| 101 | FAGVFFAWG LTSCGFAGFN FLGRHHGRSV VLILIGCIGL IPTVHFLNPA |
|
| 151 | AAAFAAAGLV LHGYSLARRR VIAASFLLGT GWTLMSLAAA YPAAFALMLP |
|
| 201 | LPVLMFFRPW QSRRLMLTAV ASLAFALPLM TVYPLLLAKT QPALFAQWLD |
|
| 251 | DHVFGTFGGV RHIQTAFSLF YYLKNLLWFA LPALPLAVWT VCRTRLFSTD |
|
| 301 | WGILGVVWML AVLVLLAVNP QRFQDNLVWL LPPLALFGAA QLDSLRRGAA |
|
| 351 | AFVNWFGIMA FGLFAVFLWT GFFAMNYGWP AKLAERAAYF SPYYVPDIDP |
|
| 401 | IPMAVAVLFT PLWLWAITRK NIRGRQAVTN WAAGVTLTWA LLMTLFLPWL |
|
| 451 | DAAKSHAPVV RSMEASLSPE LKRELSDGIE CIDIGGGDLH TRIVWTQYGT |
|
| 501 | LPHRVGDVQC RYRIVRLPQN ADAPQGWQTV WQGARPRNKD SKFALIRKTG |
|
| 551 | ENILKTTD* |
[1288]ORF141a and ORF141-1 show 98.2% identity in 553 aa overlap:
[0000]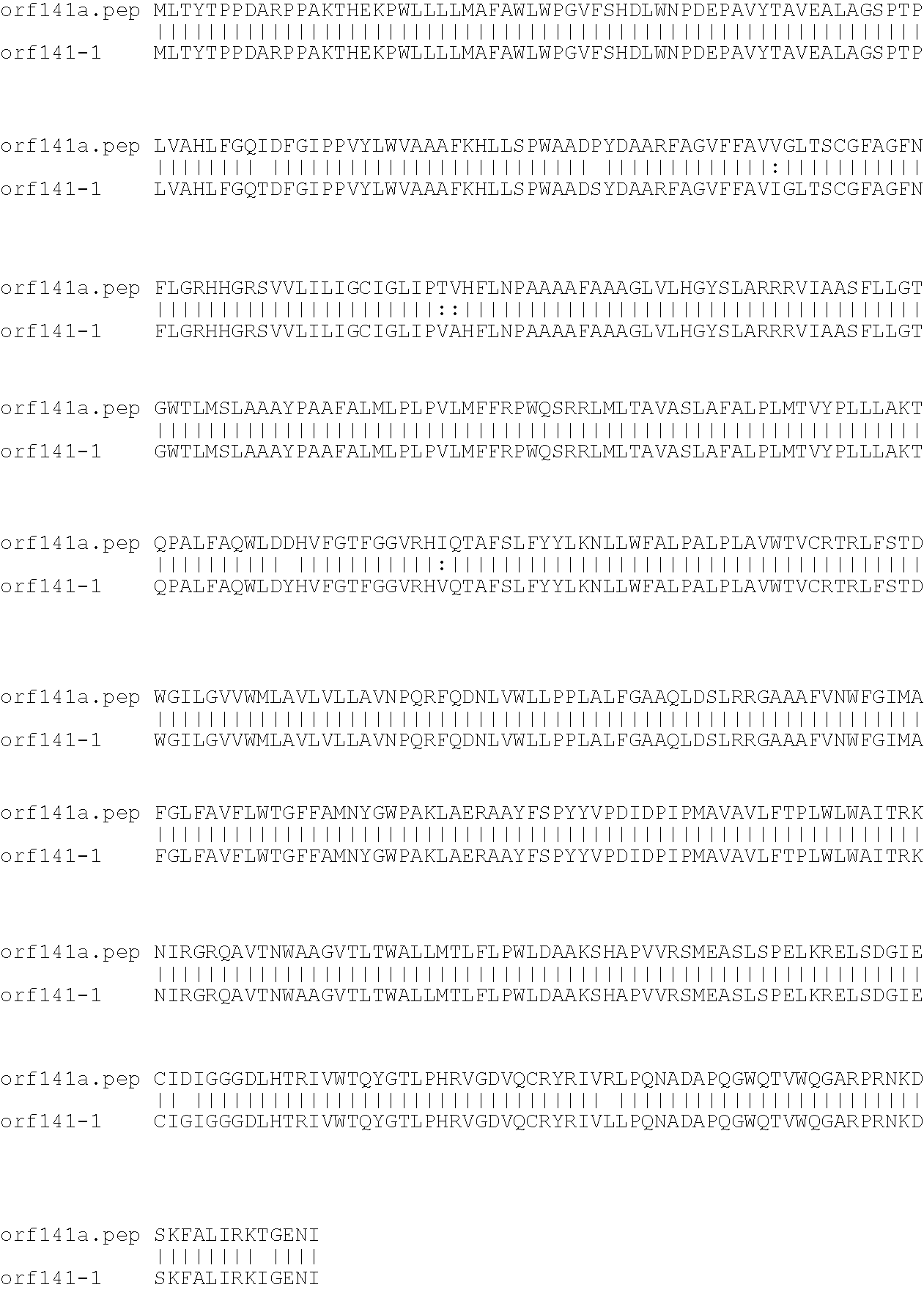
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1289]ORF141 shows 95% identity over a 140aa overlap with a predicted ORF (ORF141ng) from N. gonorrhoeae:
[0000]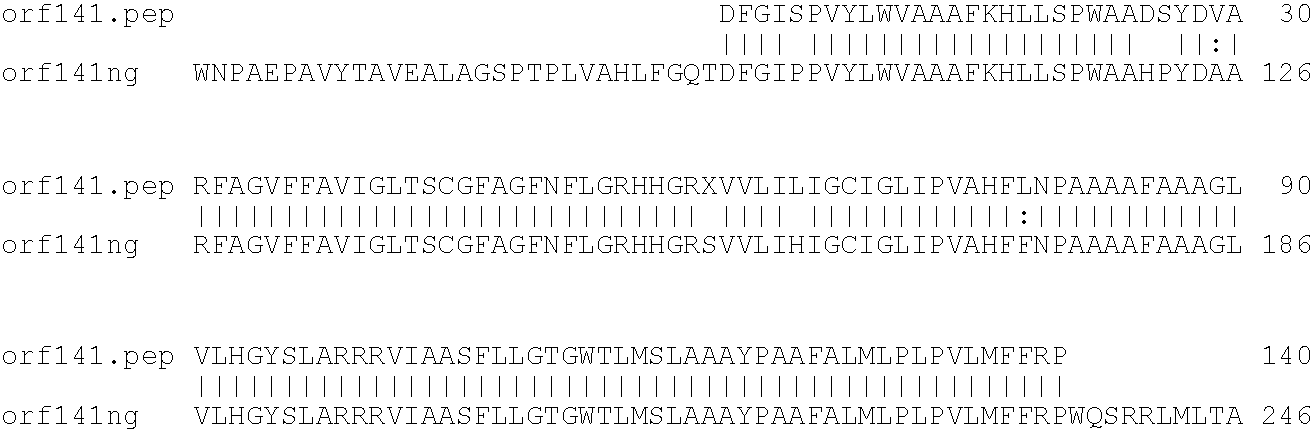
[1290]An ORF141ng nucleotide sequence <SEQ ID 599> was predicted to encode a protein having amino acid sequence <SEQ ID 600>:
[0000]| 1 | MPSEAVSARP LCEYLLHLAI RPFLLTLMLT YTPPDARPPA KTHEKPWLLL | |
|
| 51 | LMAFAWLWFG VFSHDLWNPA EPAVYTAVEA LAGSPTPLVA HLFGQTDFGI |
|
| 101 | PPVYLWVAAA FKHLLSPWAA HPYDAARFAG VFFAVIGLTS CGFAGFNFLG |
|
| 151 | RHHGRSVVLI HIGCIGLIPV AHFFNPAAAA FAAAGLVLHG YSLARRRVIA |
|
| 201 | ASFLLGTGWT LMSLAAAYPA AFALMLPLPV LMFFRPWQSR RLMLTAVASL |
|
| 251 | AFALPLMTVY PLLLAKTQPA LFAQWLNYHV FGTFGGVRHI QRAFSLFHYL |
|
| 301 | KNLLWFAPPG LPLAVWTVCR TRLFSTDWGI LGIVWMLAVL VLLAFNPQRF |
|
| 351 | QDNLVWLLPP LALFGAAQLD SLRRGAAAFV NWFGIMAFGL FAVFLWTGFF |
|
| 401 | AMNYGWPAKL AERAAYFSPY YVPDIDPIPM AVAVLFTPLW LWAITRKNIR |
|
| 451 | GRQAVTNWAA GVTLTWALLM TLFLPWLDAA KSHAPVVRSM EASFSPELKR |
|
| 501 | ELSDGIECIG IGGGDLHTRI VWTQYGTLPH RVGDVRCRYR IVRLPQNADA |
|
| 551 | PQGWQTVWQG ARPRNKDSKF ALIRKIGENI LKTTD* |
[1291]Further work revealed the following gonococcal DNA sequence <SEQ ID 601>:
[0000]| 1 | ATGCTGACCT ATACCCCGCC CGATGCCCGC CCGCCCGCCA AAACCCACGA | |
|
| 51 | AAAACCGTGG CTGCTGCTGT TGATGGCGTT TGCCTGGCTG TGGCCCGGCG |
|
| 101 | TGTTTTCCCA CGATTTGTGG AATCCTGCCG AACCTGCCGT CTATACCGCC |
|
| 151 | GTCGAAGCAC TGGCAGGCAG CCCCACCCCC TTGGTTGCCC ATCTGTTCGG |
|
| 201 | TCAAACCGAT TTCGGCATAC CGCCCGTGTA TCTTTGGGTT GCCGCCGCAT |
|
| 251 | TCAAACATTT GCTGTCGCCG TGGGCAGCCG ACCCGTATGA TGCCGCACGC |
|
| 301 | TTTGCAGGCG TATTTTTTGC CGTTATCGGA CTGACTTCTT GCGGCTTTGC |
|
| 351 | CGGTTTCAAC TTTTTGGGCA GACACCACGG GCGCAGCGTT GTTTTAATCC |
|
| 401 | ATATCGGCTG TATCGGGCTG ATTCCGGTTG CCCATTTCCT CAATCCcgcc |
|
| 451 | gccgccgcct tTGCCGCCGC CGGACTGGTG CTGCacggct actcgctgGC |
|
| 501 | ACGCCGGCGC GTGATtgccg cctctTtccT GCTCGGTACG GGTTGGACGT |
|
| 551 | TGATGTCGCT GGCGGCAGCT TATCCGGCGG CGTTTGCGCT GATGCTGCCC |
|
| 601 | CTGCCCGTGC TGATGTTTTT CCGTCCGTGG CAAAGCAGGC GTTTGATGTT |
|
| 651 | GACGGCAGTC GCCTCGCTTG CCTTTGCCCT GCCGCTTATG ACCGTTTACC |
|
| 701 | CGCTGCTCtt gGCAAAAACG CAGCCCGCGC TGTTTGCGCA ATGGCTCAAC |
|
| 751 | TATCACGTTT TCGGTACGTt cggcgGCGTG CGGCAcaTTC AGAggGCatT |
|
| 801 | Cagtttgttt cactatctgA AAaatctgct ttggttcgca ccgcccgggC |
|
| 851 | TGCCGCTGGC GGTTTGGACG GTTTGCCGCA CACGCCTGTT TTCGACCGAC |
|
| 901 | TGGGGGATTT TGGGCATTGT CTGGATGCTT GCCGTTTTGG TGCTGCTCGC |
|
| 951 | CTTTAATCCG CAGCGTTTTC AAGACAACCT CGTCTGGCTG CTGCCGCCGC |
|
| 1001 | TTGCCCTGTT CGGCGCGGCG CAACTGGACA GCCTGAGGCG CGGCGCGGCG |
|
| 1051 | GCTTTTGTCA ACTGGTTCGG CATTATGGCG TTCGGGCTGT TTGCCGTGTT |
|
| 1101 | CCTGTGGACG GGCTTTTTCG CCATGAATTA CGGCTGGCCC GCCAAGCTTG |
|
| 1151 | CCGAACGCGC CGCCTACTTC AGCCCGTATT ACGTTCCCGA CATCGATCCC |
|
| 1201 | ATTCCGATGG CGGTTGCCGT ACTGTTCACA CCCTTGTGGC TGTGGGCGAT |
|
| 1251 | TACCCGGAAA AACATACGCG GCAGGCAGGC GGTTACCAAC TGGGCGGCAG |
|
| 1301 | GCGTTACCCT GACCTGGGCT TTGCTGATGA CGCTGTTCCT GCCGTGGCTG |
|
| 1351 | GACGCGGCGA AAAGCCACGC GCCCGTCGTC CGGAGTATGG AGGCATCGTT |
|
| 1401 | TTCCCCGGAA TTAAAACGGG AGCTTTCAGA CGGCATCGAG TGTATCGGCA |
|
| 1451 | TAGGCGGCGG CGACCTGCAC ACGCGGATTG TTTGGACGCA GTACGGCACA |
|
| 1501 | TTGCCGCACC GCGTCGGCGA TGTCCGTTGC CGCTACCGTA TCGTCCGCCT |
|
| 1551 | GCCCCAAAAC GCGGATGCGC CGCAAGGCTG GCAGACGGTC TGGCAGGGTG |
|
| 1601 | CGCGCCCGCG CAACAAAGAC AGTAAGTTTG CACTGATACG GAAAATCGGG |
|
| 1651 | GAAAATATAT TAAAAACAAC AGATTGA |
[1292]This corresponds to the amino acid sequence <SEQ ID 602; ORF141ng-1>:
[0000]| 1 | MLTYTPPDAR PPAKTHEKPW LLLLMAFAWL WPGVFSHDLW NPAEPAVYTA | |
|
| 51 | VEALAGSPTP LVAHLFGQTD FGIPPVYLWV AAAFKHLLSP WAADPYDAAR |
|
| 101 | FAGVFFAVIG LTSCGFAGFN FLGRHHGRSV VLIHIGCIGL IPVAHFLNPA |
|
| 151 | AAAFAAAGLV LHGYSLARRR VIAASFLLGT GWTLMSLAAA YPAAFALMLP |
|
| 201 | LPVLMFFRPW QSRRLMLTAV ASLAFALPLM TVYPLLLAKT QPALFAQWLN |
|
| 251 | YHVFGTFGGV RHIQRAFSLF HYLKNLLWFA PPGLPLAVWT VCRTRLFSTD |
|
| 301 | WGILGIVWML AVLVLLAFNP QRFQDNLVWL LPPLALFGAA QLDSLRRGAA |
|
| 351 | AFVNWFGIMA FGLFAVFLWT GFFAMNYGWP AKLAERAAYF SPYYVPDIDP |
|
| 401 | IPMAVAVLFT PLWLWAITRK NIRGRQAVTN WAAGVTLTWA LLMTLFLPWL |
|
| 451 | DAAKSHAPVV RSMEASFSPE LKRELSDGIE CIGIGGGDLH TRIVWTQYGT |
|
| 501 | LPHRVGDVRC RYRIVRLPQN ADAPQGWQTV WQGARPRNKD SKFALIRKIG |
|
| 551 | ENILKTTD* |
[1293]ORF141ng-1 and ORF141-1 show 97.5% identity in 553 aa overlap:
[0000]
[1294]Based on the presence of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 72
[1295]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 603>:
[0000]| 1 | ..CAATCCGCCA AATGGTTATC GGGCCAAACT CTAGTCGGCA CAGCAATTGG | |
|
| 51 | GATACGCGGG CAGATAAAGC TTGGCGGCAA CCTGCATTAC GATATATTTA |
|
| 101 | CCGGCCGCGC ATTGAAAAAG CCCGAATTTT TCCAATCAAG GAAATGGGCA |
|
| 151 | AGCGGTTTTC AGGTAGGCTA TACGTTTTAA |
[1296]This corresponds to the amino acid sequence <SEQ ID 604; ORF142>:
[0000]| 1 | ..QSAKWLSGOT LVGTAIGIRG QIKLGGNLHY DIFTGRALKK PEFFQSRKWA | |
|
| 51 | SGFQVGYTF* |
[1297]Further work revealed the complete nucleotide sequence <SEQ ID 605>:
[0000]| 1 | ATGGATAATT CGGGTAGTGA GGCGACAGGA AAATACCAAG GAAATATCAC | |
|
| 51 | TTTCTCTGCC GACAATCCTT TGGGACTGAG TGATATGTTC TATGTAAATT |
|
| 101 | ATGGACGTTC GATTGGCGGT ACGCCCGATG AGGAAAGTTT TGACGGCCAT |
|
| 151 | CGCAAAGAAG GCGGATCAAA CAATTACGCC GTACATTATT CAGCCCCTTT |
|
| 201 | CGGTAAATGG ACATGGGCAT TCAATCACAA TGGCTACCGT TACCATCAGG |
|
| 251 | CAGTTTCCGG ATTATCGGAA GTCTATGACT ATAATGGAAA AAGTTACAAT |
|
| 301 | ACTGATTTCG GCTTCAACCG CCTGTTGTAT CGTGATGCCA AACGCAAAAC |
|
| 351 | CTATCTCGGT GTAAAACTGT GGATGAGGGA AACAAAAAGT TACATTGATG |
|
| 401 | ATGCCGAACT GACTGTACAA CGGCGTAAAA CTGCGGGTTG GTTGGCAGAA |
|
| 451 | CTTTCCCACA AAGAATATAT CGGTCGCAGT ACGGCAGATT TTAAGTTGAA |
|
| 501 | ATATAAACGC GGCACCGGCA TGAAAGATGC TCTGCGCGCG CCTGAAGAAG |
|
| 551 | CCTTTGGCGA AGGCACGTCA CGTATGAAAA TTTGGACGGC ATCGGCTGAT |
|
| 601 | GTAAATACTC CTTTTCAAAT CGGTAAACAG CTATTTGCCT ATGACACATC |
|
| 651 | CGTTCATGCA CAATGGAACA AAACCCCGCT AACATCGCAA GACAAACTGG |
|
| 701 | CTATCGGCGG ACACCACACC GTACGTGGCT TCGACGGTGA AATGAGTTTG |
|
| 751 | TCTGCCGAGC GGGGATGGTA TTGGCGCAAC GATTTGAGCT GGCAATTTAA |
|
| 801 | ACCAGGCCAT CAGCTTTATC TTGGGGCTGA TGTAGGACAT GTTTCAGGAC |
|
| 851 | AATCCGCCAA ATGGTTATCG GGCCAAACTC TAGTCGGCAC AGCAATTGGG |
|
| 901 | ATACGCGGGC AGATAAAGCT TGGCGGCAAC CTGCATTACG ATATATTTAC |
|
| 951 | CGGCCGCGCA TTGAAAAAGC CCGAATTTTT CCAATCAAGG AAATGGGCAA |
|
| 1001 | GCGGTTTTCA GGTAGGCTAT ACGTTTTAA |
[1298]This corresponds to the amino acid sequence <SEQ ID 606; ORF142-1>:
[0000]| 1 | MDNSGSEATG KYQGNITFSA DNPLGLSDMF YVNYGRSIGG TPDEESFDGH | |
|
| 51 | RKEGGSNNYA VHYSAPFGKW TWAFNHNGYR YHQAVSGLSE VYDYNGKSYN |
|
| 101 | TDFGFNRLLY RDAKRKTYLG VKLWMRETKS YIDDAELTVQ RRKTAGWLAE |
|
| 151 | LSHKEYIGRS TADFKLKYKR GTGMKDALRA PEEAFGEGTS RMKIWTASAD |
|
| 201 | VNTPFQIGKQ LFAYDTSVHA QWNKTPLTSQ DKLAIGGHHT VRGFDGEMSL |
|
| 251 | SAERGWYWRN DLSWQFKPGH QLYLGADVGH VSGQSAKWLS GQTLVGTAIG |
|
| 301 | IRGQIKLGGN LHYDIFTGRA LKKPEFFQSR KWASGFQVGY TF* |
[1299]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1300]ORF142 shows 88.1% identity over a 59aa overlap with a predicted ORF (ORF142ng) from N. gonorrhoeae:
[0000]
[1301]The complete length ORF142ng nucleotide sequence <SEQ ID 607> is:
[0000]| 1 | ATGGATAATT CGGGTAGTGA GGCGACAGGA AAATACCAAG GAAATATCAC | |
|
| 51 | TTTCTCTGCC GACAATCCTT TTGGACTGAG TGATATGTTC TATGTAAATT |
|
| 101 | ATGGACGTTC AATTGGCGGT ACGCCCGATG AGGAAAATTT TGACGGCCAT |
|
| 151 | CGCAAAGAAG GCGGATCAAA CAATTACGCC GTACATTATT CAGCCCCTTT |
|
| 201 | CGGTAAATGG ACATGGGCAT TCAATCACAA TGGCTACCGT TACCATCAGG |
|
| 251 | CGGTTTCCGG ATTATCGGAA GTCTATGACT ATAATGGAAA AAGTTACAAC |
|
| 301 | ACTGATTTCG GCTTCAACCG CCTGTTGTAT CGTGATGCCA AACGCAAAAC |
|
| 351 | CTATCTCAGT GTAAAACTGT GGACGAGGGA AACAAAAAGT TACATTGATG |
|
| 401 | ATGCCGAACT GACTGTACAA CGGCGTAAAA CCACAGGTTG GTTGGCAGAA |
|
| 451 | CTTTCCCACA AAGGATATAT CGGTCGCAGT ACGGCAGATT TTAAGTTGAA |
|
| 501 | ATATAAACAC GGCACCGGCA TGAAAGATGC TCTGCGCGCG CCTGAAGAAG |
|
| 551 | CCTTTGGCGA AGGCACGTCA CGTATGAAAA TTTGGACGGC ATCGGCTGAT |
|
| 601 | GTAAATACTC CTTTTCAAAT CGGTAAACAG CTATTTGCCT ATGACACATC |
|
| 651 | CGTTCATGCA CAATGGAACA AAACCCCGCT AACATCGCAA GACAAACTGG |
|
| 701 | CTATCGGCGG ACACCACACC GTACGTGGCT TCGACGGTGA AATGAGTTTG |
|
| 751 | CCTGCCGAGC GGGGATGGTA TTGGCGCAAC GATTTGAGCT GGCAATTTAA |
|
| 801 | ACCAGGCCAT CAGCTTTATC TTGGGGCTGA TGTAGGACAT GTTTCAGGAC |
|
| 851 | AATCCGCCAA ATGGTTATCG GGCCAAACTC TAGCCGGCAC AGCAATTGGG |
|
| 901 | ATACGCGGGC AGATAAAGCT TGGCGGCAAC CTGCATTACG ATATATTTAC |
|
| 951 | CGGCCGTGCA TTGAAAAAGC CCGAATATTT TCAGACGAAG AAATGGGTAA |
|
| 1001 | CGGGGTTTCA GGTGGGTTAT TCGTTTTGA |
[1302]This encodes a protein having amino acid sequence <SEQ ID 608>:
[0000]| 1 | MDNSGSEATG KYQGNITFSA DNPFGLSDMF YVNYGRSIGG TPDEENFDGH | |
|
| 51 | RKEGGSNNYA VHYSAPFGKW TWAFNHNGYR YHQAVSGLSE VYDYNGKSYN |
|
| 101 | TDFGFNRLLY RDAKRKTYLS VKLWTRETKS YIDDAELTVQ RRKTTGWLAE |
|
| 151 | LSHKGYIGRS TADFKLKYKH GTGMKDALRA PEEAFGEGTS RMKIWTASAD |
|
| 201 | VNTPFQIGKQ LFAYDTSVHA QWNKTPLTSQ DKLAIGGHHT VRGFDGEMSL |
|
| 251 | PAERGWYWRN DLSWQFKPGH QLYLGADVGH VSGQSAKWLS GQTLAGTAIG |
|
| 301 | IRGQIKLGGN LHYDIFTGRA LKKPEYFQTK KWVTGFQVGY SF* |
[1303]The underlined sequence (aromatic-Xaa-aromatic amino acid motif) is usually found at the C-terminal end of outer membrane proteins.
[1304]ORF142ng and ORF142-1 show 95.6% identity over 342aa overlap:
[0000]
[1305]In addition, ORF142ng is homologous to the HecB protein of E. chrysanthemi:
[0000]| gi|1772622 (L39897) HecB [Erwinia chrysanthemi] Length = 558 | |
| Score = 119 bits (295), Expect = 3e−26 |
| Identities = 88/346 (25%), Positives = 151/346 (43%), Gaps = 22/346 (6%) |
|
| Query: | 2 | DNSGSEATGKYQGNITFSADNPFGLSDMFYVNYGRSIGGTPDEENFDGHRKEGGSNNYAV | 61 | |
| | DNSG ++TG+ Q N + + DN FGL+D ++++ G S + + D + G | |
| Sbjct: | 230 | DNSGQKSTGEEQLNGSLALDNVFGLADQWFISAGHS---SRFATSHDAESLQAG------ | 280 |
|
| Query: | 62 | HYSAPFGKWTWAFNHNGYRYHQAVSGLSEVYDYNGKSYNTDFGFNRLLYRDAKRKTYLSV | 121 |
| | +S P+G W +N++ RY + G S F +R+++RD KT ++ | |
| Sbjct: | 281 | -FSMPYGYWNLGYNYSQSRYRNTFINRDFPWHSTGDSDTHRFSLSRVVFRDGTMKTAIAG | 339 |
|
| Query: | 122 | KLWTRETKSYIDDAELTVQRRKTTGWLAELSHKGYIGRSTADFKLKYKHGTGMKDALRAP | 181 |
| | R +Y++ + L RK + ++H + A F Y G + | |
| Sbjct: | 340 | TFSQRTGNNYLNGSLLPSSSRKLSSVSLGVNHSQKLWGGLATFNPTYNRGVRWLGSETDT | 399 |
|
| Query: | 182 | EEAFGEGTSRMKIWTASADVNTPFQIGKQLFAYDTSVHAQWNKTPLTSQDKLAIGGHHTV | 241 |
| | +++ E + WT SA P Y S++ Q++ L ++L +GG ++ | |
| Sbjct: | 400 | DKSADEPRAEFNKWTLSASYYHPV---TDSITYLGSLYGQYSARALYGSEQLTLGGESSI | 456 |
|
| Query: | 242 | RGFDGEMSLPAERGWYWRNDLSWQFKP----GHQLYLGA-DVGHVSGQSAKWLSGQTLAG | 296 |
| | RGF E RG YWRN+L+WQ G+ ++ A D GH+ + +L G | |
| Sbjct: | 457 | RGF-REQYTSGNRGAYWRNELNWQAWQLPVLGNVTFMAAVDGGHLYNHKQDNSTAASLWG | 515 |
|
| Query: | 297 | TAIGIRGQIKLGGNLHYDIFTGRALKKPEYFQTKKWVTGFQVGYSF | 342 |
| | A+G+ + L + G + P + Q V G++VG SF | |
| Sbjct: | 516 | GAVGMTVASRW---LSQQVTVGWPISYPAWLQPDTMVVGYRVGLSF | 558 |
[1306]On the basis of this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 73
[1307]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 609>:
[0000]| 1 | ATGCGGACGA AATGGTCAGC AGTGAGAAGC TGCTTACTTG GgCGGACACC | |
|
| 51 | GCCGACATCG ATACCGCTTT GAACCTGTTG TACCGTTTGC AAAAACTCGA |
|
| 101 | ATTCCTCTAT GGCGATGAAA ACGGTCATTC AGACGGCATC AATTTGwCGG |
|
| 151 | ACGAGCAATT GCCGTTGCTG ATGGAACAAT TGTCCGGCAG CGGTAAGGCG |
|
| 201 | TTATTGGTCG ATCGGAACGG TCTGTATCTT GCCAACGCCA ATTTCCATCA |
|
| 251 | TGAGGCGGCG GAAGAGTTGG GGTTGTTGGC GGCAGAAGTC GCACAGATGG |
|
| 301 | AAAAGAAATA CCGGCTGCTG ATTAAGAACA AC.. |
[1308]This corresponds to the amino acid sequence <SEQ ID 610; ORF143>:
[0000]| 1 | MRTKWSAVRS CTWADTADID TALNLLYRLQ KLEFLYGDEN GHSDGINLXD | |
|
| 51 | EQLPLLMEQL SGSGKALLVD RNGLYLANAN FHHEAAEELG LLAAEVAQME |
|
| 101 | KKYRLLIKNN .. |
[1309]Further work revealed the complete nucleotide sequence <SEQ ID 611>:
[0000]| 1 | ATGGAATCAA CACTTTCACT ACAAGCAAAT TTATATCCCC GCCTGACTCC | |
|
| 51 | TGCCGGTGCA TTTTATGCCG TATCCAGCGA TGCCCCCAGT GCCGGTAAAA |
|
| 101 | CTTTGTTGCA CAGCCTGTTG AAAGCAGATG CGGACGAAAT GGTCAGCAGT |
|
| 151 | GAGAAGCTGC TTACTTGGGC GGACACCGCC GACATCGATA CCGCTTTGAA |
|
| 201 | CCTGTTGTAC CGTTTGCAAA AACTCGAATT CCTCTATGGC GATGAAAACG |
|
| 251 | GTCATTCAGA CGGCATCAAT TTGTCGGACG AGCAATTGCC GTTGCTGATG |
|
| 301 | GAACAATTGT CCGGCAGCGG TAAGGCGTTA TTGGTCGATC GGAACGGTCT |
|
| 351 | GTATCTTGCC AACGCCAATT TCCATCATGA GGCGGCGGAA GAGTTGGGGT |
|
| 401 | TGTTGGCGGC AGAAGTCGCA CAGATGGAAA AGAAATACCG GCTGCTGATT |
|
| 451 | AAGAACAACC TGTATATCAA CAATAACGCT TGGGGCGTTT GCGATCCTTC |
|
| 501 | CGGTCAGAGC GAATTGACAT TTTTCCCATT GTATATCGGT TCAACCAAAT |
|
| 551 | TTATTTTGGT TATCGGCGGC ATTCCCGATT TGGGCAAAGA GGCATTTGTT |
|
| 601 | ACTTTGGTAA GGATTTTATA CCGCCGTTAC AGCAACCGCG TGTAA |
[1310]This corresponds to the amino acid sequence <SEQ ID 612; ORF143-1>:
[0000]| 1 | MESTLSLQAN LYPRLTPAGA FYAVSSDAPS AGKTLLHSLL KADADEMVSS | |
|
| 51 | EKLLTWADTA DIDTALNLLY RLQKLEFLYG DENGHSDGIN LSDEQLPLLM |
|
| 101 | EQLSGSGKAL LVDRNGLYLA NANFHHEAAE ELGLLAAEVA QMEKKYRLLI |
|
| 151 | KNNLYINNNA WGVCDPSGQS ELTFFPLYIG STKFILVIGG IPDLGKEAFV |
|
| 201 | TLVRILYRRY SNRV* |
[1311]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1312]ORF143 shows 92.4% identity over a 105aa overlap with an ORF (ORF143a) from strain A of N. meningitidis:
[0000]
[1313]The complete length ORF143a nucleotide sequence <SEQ ID 613> is:
[0000]| 1 | ATGGAATCAA CANTTTCACT ACAAGCAAAT TTATATCNCC GCCTGACTCC | |
|
| 51 | TGCCGGTGCA TTTTATGCCG TATCCAGCGA TGNCCCCAGT GCCGGTAAAA |
|
| 101 | CTTTGTTGCA CAGCCTGTTG AAAGCGGATG CGGACGAAAT GGTNAGCAGT |
|
| 151 | GAGAAGCTGC TTACCTGGGC GGANACCGCC GACATCGATA CCGCTTTGAA |
|
| 201 | CCTGTTGTAC CGTTTGCAAA AACTCGAATT CCTCTATGGC GATGAAAACG |
|
| 251 | GTCATTCAGA CGGCATCAAT TTGTCGGACG AGCAATTGCC GTTGCTGATG |
|
| 301 | GAACAATTGT CCGGCAGCGG TAAGGCGTTA TTGGTCGATC GGAACGGTCT |
|
| 351 | GTATCTTGCC AACGCCAATT TCCATCATGA GGCGGCGGAA GAGTTGGGGT |
|
| 401 | TGTTGGCGGC AGAAGTCGCA CAGATGGAAA AGAAATACCG GCTGCNNATT |
|
| 451 | AAGAACAACC TGTATATCAA CAATAACGCT TGGGGCGTTT GCGATCCTTC |
|
| 501 | CGGTCAGAGC GAATTGACAT TTTTCCCATT GTATATCGGT TCAACCAAAT |
|
| 551 | TTATTTTGGT TATCGGCGGC ATTCCCGATT TGGGCAAAGA GGCATTTGTT |
|
| 601 | ACTTTGGTAA GGATNTTATA CCNCCNGTTA CAGCAACCGC GTGTAAAACT |
|
| 651 | TGGGAGAGAG GANGGGTTAT GCAGCAATTA TTGA |
[1314]This encodes a protein having amino acid sequence <SEQ ID 614>:
[0000]| 1 | MESTXSLQAN LYXRLTPAGA FYAVSSDXPS AGKTLLHSLL KADADEMVSS | |
|
| 51 | EKLLTWAXTA DIDTALNLLY RLQKLEFLYG DENGHSDGIN LSDEQLPLLM |
|
| 101 | EQLSGSGKAL LVDRNGLYLA NANFHHEAAE ELGLLAAEVA QMEKKYRLXI |
|
| 151 | KNNLYINNNA WGVCDPSGQS ELTFFPLYIG STKFILVIGG IPDLGKEAFV |
|
| 201 | TLVRXLYXXL QQPRVKLGRE XGLCSNY* |
[1315]ORF143a and ORF143-1 show 97.1% identity in 207 aa overlap:
[0000]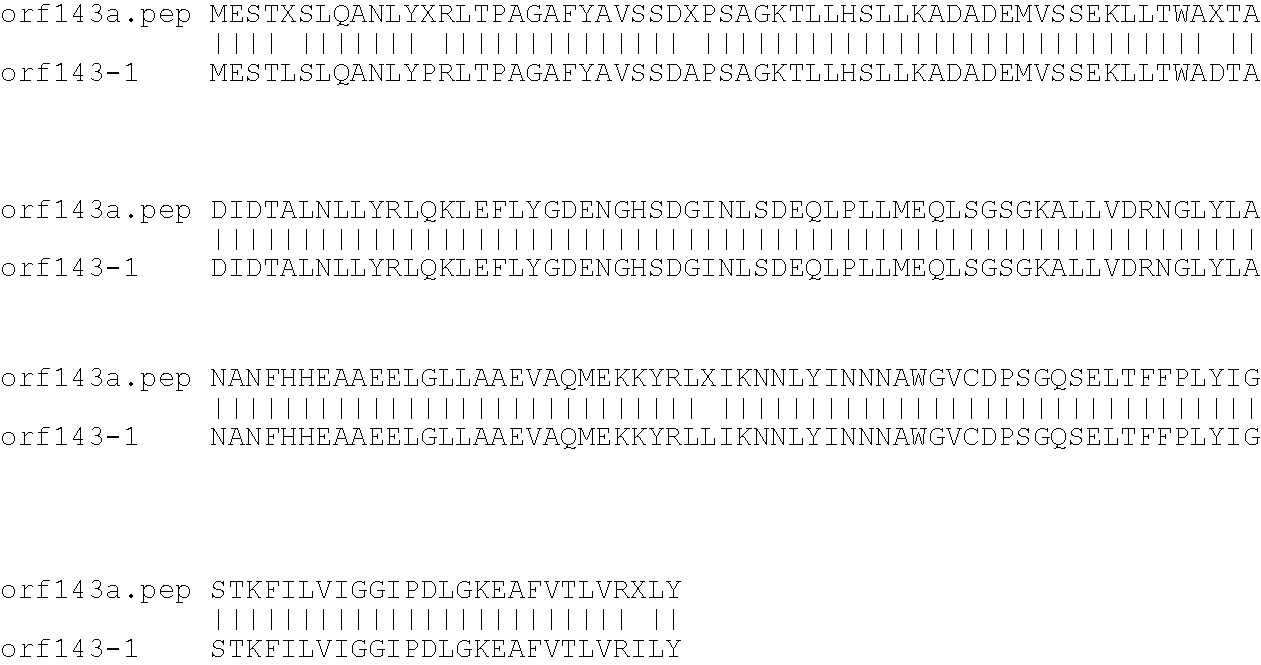
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1316]ORF143 shows 95.5% identity over a 110aa overlap with a predicted ORF (ORF143ng) from N. gonorrhoeae:
[0000]
[1317]An ORF143ng nucleotide sequence <SEQ ID 615> was predicted to encode a protein having amino acid sequence <SEQ ID 616>:
[0000]| 1 | MRTKWSAVRS CSRADTADID TALNLLYRLQ KLEFLYGDEN GHSDGINLSD | |
|
| 51 | EQLPLLMEQL SGSGKALLVD RNGLYLANAN FHHESAEELG LLAAEVAQME |
|
| 101 | KKYRLLIRNN LYINNNAWGV CDPSGQSELT FFPLYIGSTK FILVIAGIPD |
|
| 151 | LSKGGICYFG KDFIPPLQQP RVKLGTGGIM RQLLISILED LNNTSTDIIA |
|
| 201 | SAVISTDGLP MATMLPSHLN SDRVGAISAT LLALGSRSVQ ELACGELEQV |
|
| 251 | MIKGKSGYIL LSQAGKDAVL VLVAKETGRL GLILLDAKRA ARHIAEAI* |
[1318]Further work revealed the following gonococcal DNA sequence <SEQ ID 617>:
[0000]| 1 | ATGGAATCAA CACTTTCACT ACAAGCGAAT TTATATCCCT GCCTGACTCC | |
|
| 51 | TGCCGGTGCA TTTTATGCCG TATCCAGCGA TGCCCCCAGT GCCGGTAAAA |
|
| 101 | CTTTGTTGCG CAGCCTGTTG AAAGCGGATG CGGACGAAGT GGTCAGCAGT |
|
| 151 | GAGAAGCTGC TCGCGGCGGA CACCGCCGAC ATCGATACCG CTTTGAACCT |
|
| 201 | GTTGTACCGT TTGCAAAAAC TCGAATTCCT CTATGGCGAT GAAAACGGTC |
|
| 251 | ATTCAGACGG CATCAATTTG TCGGACGAGC AATTGCCGTT GCTGATGGAA |
|
| 301 | CAATTGTCCG GCAGCGGTAA GGCATTATTG GTCGATCGGA ACGGTCTGTA |
|
| 351 | TCTTGCCAAC GCCAATTTCC ATCATGAGTC GGCGGAAGAG TTGGGGTTGT |
|
| 401 | TGGCGGCAGA AGTCGCACAG ATGGAAAAGA AATACCGGCT GCTGATTAGG |
|
| 451 | AACAACCTGT ATATCAACAA TAACGCTTGG GGCGTTTGCG ATCCTTCCGG |
|
| 501 | TCAGAGCGAA TTGACATTTT TCCCATTGTA TATCGGTTCA ACCAAATTTA |
|
| 551 | TTTTGGTTAT CGCCGGCATT CCCGATTTGA GCAAAGAGGC ATTTGTTACT |
|
| 601 | TTGGTAAGGA TTTTATACCG CCGTTACAGC AACCGCGTGT AA |
[1319]This corresponds to the amino acid sequence <SEQ ID 618; ORF143ng-1>:
[0000]| 1 | MESTLSLQAN LYPCLTPAGA FYAVSSDAPS AGKTLLRSLL KADADEVVSS | |
|
| 51 | EKLLAADTAD IDTALNLLYR LQKLEFLYGD ENGHSDGINL SDEQLPLLME |
|
| 101 | QLSGSGKALL VDRNGLYLAN ANFHHESAEE LGLLAAEVAQ MEKKYRLLIR |
|
| 151 | NNLYINNNAW GVCDPSGQSE LTFFPLYIGS TKFILVIAGI PDLSKEAFVT |
|
| 201 | LVRILYRRYS NRV* |
[1320]ORF143ng-1 and ORF143-1 show 95.8% identity in 214 aa overlap:
[0000]
[1321]Based on the presence of the putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 74
[1322]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 619>:
[0000]| 1 | ATGACCTTTT TACAACGTTT GCAAGGTTTG GCAGACAATA AAATCTGTGC | |
|
| 51 | GTTTGCATGG TTCGTCGTCC GCCGCTTTGA TGAAGAACGC GTACCGCAGr |
|
| 101 | CGGCGGCAAG CATGACGTTT ACGACGCTGC TGGCACTCGT CCCCGTGCTG |
|
| 151 | ACCGTGATGG TGGCGGTCGC TTCGATTTTC CCCGTGTTCG ACCGCTGGTC |
|
| 201 | GGATTCGTTC GTCTCCTTCG TCAACCAAAC CATTGTGCCG CA.GGCGCGG |
|
| 251 | ACATGGTGTT CGACTATATC AATGCGTTCC GCGAGCAGGC GAACCGGCTG |
|
| 301 | ACGGCAATCG GCAGCGTGAT GCTGGTCGTT ACCTCGCTGA TGCTGATTCG |
|
| 351 | GACGATAGAC AATACGTTCA ACCGCATCTG GaCGGGTCAA wTyCCAGCGT |
|
| 401 | CCGTGGATG.. |
[1323]This corresponds to the amino acid sequence <SEQ ID 620; ORF144>:
[0000]| 1 | MTFLQRLQGL ADNKICAFAW FVVRRFDEER VPQXAASMTF TTLLALVPVL | |
|
| 51 | TVMVAVASIF PVFDRWSDSF VSFVNQTIVP XGADMVFDYI NAFREQANRL |
|
| 101 | TAIGSVMLVV TSLMLIRTID NTFNRIWRVX XQRPWM... |
[1324]Further work revealed the complete nucleotide sequence <SEQ ID 621>:
[0000]| 1 | ATGACCTTTT TACAACGTTT GCAAGGTTTG GCAGACAATA AAATCTGTGC | |
|
| 51 | GTTTGCATGG TTCGTCGTCC GCCGCTTTGA TGAAGAACGC GTACCGCAGG |
|
| 101 | CGGCGGCAAG CATGACGTTT ACGACGCTGC TGGCACTCGT CCCCGTGCTG |
|
| 151 | ACCGTGATGG TGGCGGTCGC TTCGATTTTC CCCGTGTTCG ACCGCTGGTC |
|
| 201 | GGATTCGTTC GTCTCCTTCG TCAACCAAAC CATTGTGCCG CAGGGCGCGG |
|
| 251 | ACATGGTGTT CGACTATATC AATGCGTTCC GCGAGCAGGC GAACCGGCTG |
|
| 301 | ACGGCAATCG GCAGCGTGAT GCTGGTCGTT ACCTCGCTGA TGCTGATTCG |
|
| 351 | GACGATAGAC AATACGTTCA ACCGCATCTG GCGGGTCAAT TCCCAGCGTC |
|
| 401 | CGTGGATGAT GCAGTTTCTC GTCTATTGGG CTTTACTGAC GTTCGGGCCG |
|
| 451 | CTGTCTTTGG GCGTGGGCAT TTCCTTTATG GTCGGCTCGG TACAGGATGC |
|
| 501 | CGCGCTTGCC TCAGGTGCGC CGCAGTGGTC GGGCGCGTTG CGAACGGCGG |
|
| 551 | CGACGCTGAC CTTCATGACG CTTTTGCTGT GGGGGCTGTA CCGCTTCGTG |
|
| 601 | CCAAACCGCT TCGTTCCCGC GCGGCAGGCG TTTGTCGGGG CTTTGGCAAC |
|
| 651 | AGCGTTTTGT CTGGAAACCG CGCGCTCCCT CTTCACTTGG TATATGGGCA |
|
| 701 | ATTTCGACGG CTACCGCTCG ATTTACGGCG CGTTTGCCGC CGTGCCGTTT |
|
| 751 | TTTCTGTTGT GGCTGAACCT GTTGTGGACG CTGGTCTTGG GCGGCGCGGT |
|
| 801 | GCTGACTTCT TCACTCTCCT ACTGGCAGGG AGAAGCGTTC CGCAGGGGCT |
|
| 851 | TCGACTCGCG CGGACGGTTT GACGACGTGT TGAAAATCCT GCTGCTTCTG |
|
| 901 | GATGCGGCGC AAAAAGAAGG CAAAGCCTTG CCTGTTCAGG AGTTCAGACG |
|
| 951 | GCATATCAAT ATGGGCTACG ACGAGTTGGG CGAGCTTTTG GAAAAGCTGG |
|
| 1001 | CGCGGCACGG CTACATCTAT TCCGGCAGAC AGGGTTGGGT GTTGAAAACG |
|
| 1051 | GGGGCGGATT CGATTGAGTT GAACGAACTC TTCAAGCTCT TCGTTTACCG |
|
| 1101 | TCCGTTGCCT GTGGAAAGGG ATCATGTGAA CCAAGCTGTC GATGCGGTAA |
|
| 1151 | TGACACCGTG TTTGCAGACT TTGAACATGA CGCTGGCAGA GTTTGACGCT |
|
| 1201 | CAGGCGAAAA AACGGCAGTA G |
[1325]This corresponds to the amino acid sequence <SEQ ID 622; ORF144-1>:
[0000]| 1 | MTFLQRLQGL ADNKICAFAW FVVRRFDEER VPQAAASMTF TTLLALVPVL | |
|
| 51 | TVMVAVASIF PVFDRWSDSF VSFVNQTIVP QGADMVFDYI NAFREQANRL |
|
| 101 | TAIGSVMLVV TSLMLIRTID NTFNRIWRVN SQRPWMMQFL VYWALLTFGP |
|
| 151 | LSLGVGISFM VGSVQDAALA SGAPQWSGAL RTAATLTFMT LLLWGLYRFV |
|
| 201 | PNRFVPARQA FVGALATAFC LETARSLFTW YMGNFDGYRS IYGAFAAVPF |
|
| 251 | FLLWLNLLWT LVLGGAVLTS SLSYWQGEAF RRGFDSRGRF DDVLKILLLL |
|
| 301 | DAAQKEGKAL PVQEFRRHIN MGYDELGELL EKLARHGYIY SGRQGWVLKT |
|
| 351 | GADSIELNEL FKLFVYRPLP VERDHVNQAV DAVMTPCLQT LNMTLAEFDA |
|
| 401 | QAKKRQ* |
[1326]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1327]ORF144 shows 96.3% identity over a 136aa overlap with an ORF (ORF144a) from strain A of N. meningitidis:
[0000]
[1328]The complete length ORF144a nucleotide sequence <SEQ ID 623> is:
[0000]| 1 | ATGACCTTTT TACAACGTTT GCAAGGTTTG GCAGACAATA AAATCTGTGC | |
|
| 51 | GTTTGCATGG TTCGTCGTCC GCCGCTTTGA TGAAGAACGC GTACCGCAGG |
|
| 101 | CGGCGGCAAG CATGACGTTT ACGACACTGC TGGCACTCGT CCCCGTGCTG |
|
| 151 | ACCGTGATGG TGGCGGTCGC TTCGATTTTC CCCGTGTTCG ACCGNTGGTC |
|
| 201 | GGATTCGTTC GTCTCCTTCG TCAACCAAAC CATTGTGCCG CAGGGCGCGG |
|
| 251 | ACATGGTNTT CGACTATATC AATGCGTTCC GCGAGCAGGC GAACCGGCTG |
|
| 301 | ACGGCAATCG GCAGCGTGAT GCTGGTCGTT ACCTCGCNGA TGCTGATTCG |
|
| 351 | GACGATAGAC AATACGTTCA ACCGCATCTG GCGGGTCAAT TCCCAGCGTC |
|
| 401 | CGTGGATGAT GCAGTTTCTC GTCTATTGGG CTTTACTGAC GTTCGGGCCG |
|
| 451 | CTGTCTTTGG GCGTGGGCAT TTCCTTTATN GTCGGCTCGG TACAGGATGC |
|
| 501 | CGCGCTTGCC TCAGGTGCGC CGCAGTGGTC GGGCGCGTTG CGAACGGCGG |
|
| 551 | CGACGCTGAN CTTCATGACG CTTTTGCTGT GGGGGCTGTA CCGCTNCGTG |
|
| 601 | CCAAACCGCT TCGTTCCCGC GCGGCANGCG TTTGTCGGGG CTTTGGCAAC |
|
| 651 | AGCGTTCTGT CTGGAAACCG CGCGTTCCCT CTTTACTTGG TATATGGGCA |
|
| 701 | ATTTCGACGG CTACCGCTCG ATTTACGGNG CGTTTGCCGC CGTGCCGTTT |
|
| 751 | TTTCTGTTGT GGCTGAACCT GTTGTGGACG CTGGTCTTGG GCGGCGCGGT |
|
| 801 | GCTGACTTCT TCACTCTCCT ACTGGCAGGG AGAAGCGTTC CGCAGGGNCT |
|
| 851 | TCGACTCGCG CGGACGGTTT GACGACGTGT TGAAAATCCT GCTGCTTCTG |
|
| 901 | GATGCGGCGC AAAAAGAAGG CNAAGCCTTG CCTGTTCAGG AGTTCAGACG |
|
| 951 | GCATATCAAT ATGGGCTACG ACGAGTTGGG CGAGCTTTTG GAAAAGCTGG |
|
| 1001 | CGCGGCACGG CTACATCTAT TCCGGCAGAC AGGGTTGGGT GTTGAAAACG |
|
| 1051 | GGGGCGGATT CGATTGAGTT GAACGAACTC TTCAAGCTCT TCGTTTACCG |
|
| 1101 | TCCGTTGCCT GTGGAAAGGG ATCATGTGAA CCAAGCTGTC GATGCGGTAA |
|
| 1151 | TGATGCCGTG TTTGCAGACT TTGAACATGA CGCTGGCAGA GTTTGACGCT |
|
| 1201 | CAGGCGAAAA AACAGCAGCA ATCTTGA |
[1329]This encodes a protein having amino acid sequence <SEQ ID 624>:
[0000]| 1 | MTFLQRLQGL ADNKICAFAW FVVRRFDEER VPQAAASMTF TTLLALVPVL | |
|
| 51 | TVMVAVASIF PVFDRWSDSF VSFVNQTIVP QGADMVFDYI NAFREQANRL |
|
| 101 | TAIGSVMLVV TSXMLIRTID NTFNRIWRVN SQRPWMMQFL VYWALLTFGP |
|
| 151 | LSLGVGISFX VGSVQDAALA SGAPQWSGAL RTAATLXFMT LLLWGLYRXV |
|
| 201 | PNRFVPARXA FVGALATAFC LETARSLFTW YMGNFDGYRS IYGAFAAVPF |
|
| 251 | FLLWLNLLWT LVLGGAVLTS SLSYWQGEAF RRXFDSRGRF DDVLKILLLL |
|
| 301 | DAAQKEGXAL PVQEFRRHIN MGYDELGELL EKLARHGYIY SGRQGWVLKT |
|
| 351 | GADSIELNEL FKLFVYRPLP VERDHVNQAV DAVMMPCLQT LNMTLAEFDA |
|
| 401 | QAKKQQQS* |
[1330]ORF144a and ORF144-1 show 97.8% identity in 406 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1331]ORF144 shows 91.2% identity over a 136aa overlap with a predicted ORF (ORF144ng) from N. gonorrhoeae:
[0000]
[1332]The complete length ORF144ng nucleotide sequence <SEQ ID 625> is predicted to encode a protein having amino acid sequence <SEQ ID 626>:
[0000]| 1 | MTFLQCWQGS ADNKICAFAW FVIRRFSEER VPQAAASMTF TTLLALVPVL | |
|
| 51 | TVMVAVASIF PVFDRWSDSF VSFVNQTIVP QGADMVFDYI DAFRDQANRL |
|
| 101 | TAIGSVMLVV TSLMLIRTID NAFNRIWRVN TQRPWMMQFL VYWALLTFGP |
|
| 151 | LSLGVGISFM VGSVQDSVLS SGAQQWADAL KTAARLAFMT LLLWGLYRFV |
|
| 201 | PNRFVPARQA FVGALITAFC LETARFLFTW YMGNFDGYRS IYGAFAAVPF |
|
| 251 | FLLWLNLLWT LVLGGAVLTS SLSYWQGEAF RRGFDSRGRF DDVLKILLLL |
|
| 301 | DAAQKEGRTL SVQEFRRHIN MGYDELGELL EKLARYGYIY SGRQGWVLKT |
|
| 351 | GADSIELSEL FKLFVYRPLP VERDHVNQAV DAVMTPCLQT LNMTLAEFDA |
|
| 401 | QAKKQQQS* |
[1333]Further work revealed the following gonococcal DNA sequence <SEQ ID 627>:
[0000]| 1 | ATGACCTTTT TACAACGTTG GCAAGGTTTG GCGGACAATA AAATCTGTGC | |
|
| 51 | ATTTGCATGG TTCGTCATCC GCCGTTTCAG TGAAGAGCGC GTACCGCAGG |
|
| 101 | CAGCGGCGAG CATGACGTTT ACGACACTGC TGGCACTCGT CCCCGTACTG |
|
| 151 | ACCGTAATGG TCGCGGTCGC TTCGATTTTC CCCGTGTTCG ACCGCTGGTC |
|
| 201 | GGATTCGTTC GTCTCCTTCG TCAACCAAAC CATTGTGCCG CAGGGCGCGG |
|
| 251 | ATATGGTGTT CGACTATATC GACGCATTCC GCGATCAGGC AAACCGGCTG |
|
| 301 | ACCGCCATCG GCAGCGTGAT GCTGGTCGTA ACCTCGCTGA TGCTGATTCG |
|
| 351 | GACGATAGAC AATGCGTTCA ACCGCATCTG GCGGGTTAAC ACGCAACGCC |
|
| 401 | CCTGGATGAT GCAGTTCCTC GTTTATTGGG CGTTGCTGAC TTTCGGGCCT |
|
| 451 | TTGTCTTTGG GTGTGGGCAT TTCCTTTATG GTCGGGTCGG TTCAAGACTC |
|
| 501 | CGTACTCTCC TCCGGAGCGC AACAATGGGC GGACGCGTTG AAGACGGCGG |
|
| 551 | CAAGGCTGGC TTTCATGACG CTTTTGCTGT GGGGGCTGTA CCGCTTCGTG |
|
| 601 | CCCAACCGCT TCGTGCCCGC CCGGCAGGCG TTTGTCGGAG CTTTGATTAC |
|
| 651 | GGCATTCTGC CTGGAGACGG CACGTTTCCT GTTCACCTGG TATATGGGCA |
|
| 701 | ATTTCGACGG CTACCGCTCG ATTTACGGCG CATTTGCCGC CGTGCCGTTT |
|
| 751 | TTCCTGCTGT GGTTAAACCT GCTGTGGACG CTGGTCTTGG GCGGGGCGGT |
|
| 801 | GCTGACTTCG TCGCTGTCTT ATTGGCAGGG CGAGGCCTTC CGCAGGGGAT |
|
| 851 | TCGACTCGCG CGGACGGTTT GACGACGTGT TGAAAATCCT GCTGCTTCTG |
|
| 901 | GATGCGGCGC AAAAAGAAGG CCGAACCCTG TCCGTTCAGG AGTTCAGACG |
|
| 951 | GCATATCAAT ATGGGTTACG ATGAATTGGG CGAGCTTTTG GAAAAGCTGG |
|
| 1001 | CGCGGTACGG CTATATCTAT TCCGGCAGAC AGGGCTGGGT TTTGAAAACG |
|
| 1051 | GGGGCGGATT CGATTGAGTT GAGCGAACTC TTCAAGCTCT TCGTGTACCG |
|
| 1101 | CCCGTTGCct gtggaAAGGG ATCATGTGAA CCAAGCTGtc gaTGCGGTAA |
|
| 1151 | TGAcgccgtG TTTGCAGACT TTGAACATGA CGCTGGCGGA GTTTGACGCT |
|
| 1201 | CAGgcgAAAA AACAGCAGCA GTCTTGA |
[1334]This encodes a variant of ORF144ng, having the amino acid sequence <SEQ ID 628; ORF144ng-1>:
[0000]| 1 | MTFLQRWQGL ADNKICAFAW FVIRRFSEER VPQAAASMTF TTLLALVPVL | |
|
| 51 | TVMVAVASIF PVFDRWSDSF VSFVNQTIVP QGADMVFDYI DAFRDQANRL |
|
| 101 | TAIGSVMLVV TSLMLIRTID NAFNRIWRVN TQRPWMMQFL VYWALLTFGP |
|
| 151 | LSLGVGISFM VGSVQDSVLS SGAQQWADAL KTAARLAFMT LLLWGLYRFV |
|
| 201 | PNRFVPARQA FVGALITAFC LETARFLFTW YMGNFDGYRS IYGAFAAVPF |
|
| 251 | FLLWLNLLWT LVLGGAVLTS SLSYWQGEAF RRGFDSRGRF DDVLKILLLL |
|
| 301 | DAAQKEGRTL SVQEFRRHIN MGYDELGELL EKLARYGYIY SGRQGWVLKT |
|
| 351 | GADSIELSEL FKLFVYRPLP VERDHVNQAV DAVMTPCLQT LNMTLAEFDA |
|
| 401 | QAKKQQQS* |
[1335]ORF144ng-1 and ORF144-1 show 94.1% identity in 406 aa overlap:
[0000]
[1336]On this basis of this analysis, including the identification of several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 75
[1337]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 629>:
[0000]| 1 | ..AGACACGCCC GCCGCATCCG CATCGACACC GCCATCAACC CCGAACTGGA | |
|
| 51 | AGCCCTCGCC GAACACCTCC ACTACCAATG GCAGGGCTTC CTCTGGCTCA |
|
| 101 | GCACCGATAT GCGTCAGGAA ATTTCCGCCC TCGTCATCCT GCTGCAACGC |
|
| 151 | ACCCGCCGCA AATGGCTGGA TGCCCACGAA CGCCAACACC TGCGCCAAAG |
|
| 201 | CCTGCTTGAA ACACGGGAAC ACGGCTGA |
[1338]This corresponds to the amino acid sequence <SEQ ID 630; ORF146>:
[0000]| 1 | ..RHARRIRIDT AINPELEALA EHLHYQWQGF LWLSTDMRQE ISALVILLQR | |
|
| 51 | TRRKWLDAHE RQHLRQSLLE TREHG* |
[1339]Further work revealed the complete nucleotide sequence <SEQ ID 631>:
[0000]| 1 | ATGAACACCT CGCAACGCAA CCGCCTCGTC AGCCGCTGGC TCAACTCCTA | |
|
| 51 | CGAACGCTAC CGCTACCGCC GCCTCATCCA CGCCGTCCGG CTCGGCGGGG |
|
| 101 | CCGTCCTGTT CGCCACCGCC TCCGCCCGGC TGCTCCACCT CCAACACGGC |
|
| 151 | GAGTGGATAG GGATGACCGT CTTCGTCGTC CTCGGCATGC TCCAGTTTCA |
|
| 201 | AGGGGCGATT TACTCCAAGG CGGTGGAACG TATGCTCGGC ACGGTCATCG |
|
| 251 | GGCTGGGCGC GGGTTTGGGC GTTTTATGGC TGAACCAGCA TTATTTCCAC |
|
| 301 | GGCAACCTCC TCTTCTACCT CACCGTCGGC ACGGCAAGCG CACTGGCCGG |
|
| 351 | CTGGGCGGCG GTCGGCAAAA ACGGCTACGT CCCTATGCTG GCAGGGCTGA |
|
| 401 | CGATGTGTAT GCTCATCGGC GACAACGGCA GCGAATGGCT CGACAGCGGA |
|
| 451 | CTCATGCGCG CCATGAACGT CCTCATCGGC GCGGCCATCG CCATCGCCGC |
|
| 501 | CGCCAAACTG CTGCCGCTGA AATCCACACT GATGTGGCGT TTCATGCTTG |
|
| 551 | CCGACAACCT GGCCGACTGC AGCAAAATGA TTGCCGAAAT CAGCAACGGC |
|
| 601 | AGGCGCATGA CCCGCGAACG CCTCGAGGAG AACATGGCGA AAATGCGCCA |
|
| 651 | AATCAACGCA CGCATGGTCA AAAGCCGCAG CCATCTCGCC GCCACATCGG |
|
| 701 | GCGAAAGCCG CATCAGCCCC GCCATGATGG AAGCCATGCA GCACGCCCAC |
|
| 751 | CGTAAAATCG TCAACACCAC CGAGCTGCTC CTGACCACCG CCGCCAAGCT |
|
| 801 | GCAATCTCCC AAACTCAACG GCAGCGAAAT CCGGCTGCTT GACCGCCACT |
|
| 851 | TCACACTGCT CCAAACCGAC CTGCAACAAA CCGTCGCCCT TATCAACGGC |
|
| 901 | AGACACGCCC GCCGCATCCG CATCGACACC GCCATCAACC CCGAACTGGA |
|
| 951 | AGCCCTCGCC GAACACCTCC ACTACCAATG GCAGGGCTTC CTCTGGCTCA |
|
| 1001 | GCACCAATAT GCGTCAGGAA ATTTCCGCCC TCGTCATCCT GCTGCAACGC |
|
| 1051 | ACCCGCCGCA AATGGCTGGA TGCCCACGAA CGCCAACACC TGCGCCAAAG |
|
| 1101 | CCTGCTTGAA ACACGGGAAC ACGGCTGA |
[1340]This corresponds to the amino acid sequence <SEQ ID 632; ORF146-1>:
[0000]| 1 | MNTSQRNRLV SRWLNSYERY RYRRLIHAVR LGGAVLFATA SARLLHLQHG | |
|
| 51 | EWIGMTVFVV LGMLQFQGAI YSKAVERMLG TVIGLGAGLG VLWLNQHYFH |
|
| 101 | GNLLFYLTVG TASALAGWAA VGKNGYVPML AGLTMCMLIG DNGSEWLDSG |
|
| 151 | LMRAMNVLIG AAIAIAAAKL LPLKSTLMWR FMLADNLADC SKMIAEISNG |
|
| 201 | RRMTRERLEE NMAKMRQINA RMVKSRSHLA ATSGESRISP AMMEAMQHAH |
|
| 251 | RKIVNTTELL LTTAAKLQSP KLNGSEIRLL DRHFTLLQTD LQQTVALING |
|
| 301 | RHARRIRIDT AINPELEALA EHLHYQWQGF LWLSTNMRQE ISALVILLQR |
|
| 351 | TRRKWLDAHE RQHLRQSLLE TREHG* |
[1341]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1342]ORF146 shows 98.6% identity over a 74aa overlap with an ORF (ORF146a) from strain A of N. meningitidis:
[0000]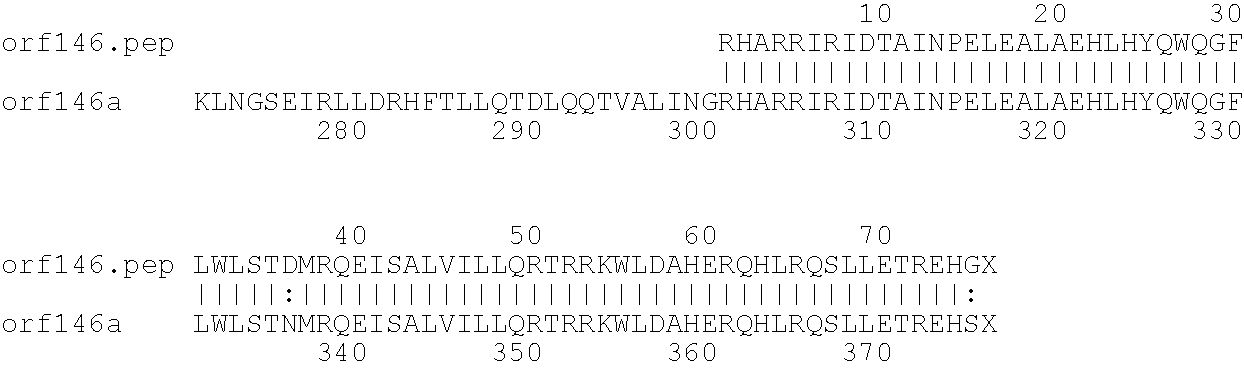
[1343]The complete length ORF146a nucleotide sequence <SEQ ID 633> is:
[0000]| 1 | ATGAACACCT CGCAACGCAA CCGCCTCGTC AGCCGCTGGC TCAACTCCTA | |
|
| 51 | CGAACGCTAC CGCTACCGCC GCCTCATCCA CGCCGTCCGG CTCGGCGGGG |
|
| 101 | CCGTCCTGTT CGCCACCGCC TCCGCCCGGC TGCTCCACCT CCAACACGGC |
|
| 151 | GAGTGGATAG GGATGACCGT CTTCGTCGTC CTCGGCATGC TCCAGTTTCA |
|
| 201 | AGGGGCGATT TACTCCAAGG CGGTGGAACG TATGCTCGGC ACGGTCATCG |
|
| 251 | GGCTGGGCGC GGGTTTGGGC GTTTTATGGC TGAACCAGCA TTATTTCCAC |
|
| 301 | GGCAACCTCC TCTTCTACCT CACCGTCGGC ACGGCAAGCG CACTGGCCGG |
|
| 351 | CTGGGCGGCG GTCGGCAAAA ACGGCTACGT CCCTATGCTG GCGGGGCTGA |
|
| 401 | CGATGTGCAT GCTCATCGGC GACAACGGCA GCGAATGGTT CGACAGCGGC |
|
| 451 | CTGATGCGCG CGATGAACGT CCTCATCGGC GCGGCCATCG CCATCGCCGC |
|
| 501 | CGCCAAACTG CTGCCGCTGA AATCCACACT GATGTGGCGT TTCATGCTTG |
|
| 551 | CCGACAACCT GACCGACTGC AGCAAAATGA TTGCCGAAAT CAGCAACGGC |
|
| 601 | AGGCGCATGA CCCGCGAACG CCTCGAAGAG AACATGGCGA AAATGCGCCA |
|
| 651 | AATCAACGCA CGCATGGTCA AAAGCCGCAG CCACCTCGCC GCCACATCGG |
|
| 701 | GCGAAAGCCG CATCAGCCCC GCCATGATGG AAGCCATGCA GCACGCCCAC |
|
| 751 | CGTAAAATTG TCAACACCAC CGAGCTGCTC CTGACCACCG CCGCCAAGCT |
|
| 801 | GCAATCTCCC AAACTCAACG GCAGCGAAAT CCGGCTGCTT GACCGCCACT |
|
| 851 | TCACACTGCT CCAAACCGAC CTGCAACAAA CCGTCGCCCT TATCAACGGC |
|
| 901 | AGACACGCCC GCCGCATCCG CATCGACACC GCCATCAACC CCGAACTGGA |
|
| 951 | AGCCCTCGCC GAACACCTCC ACTACCAATG GCAGGGCTTC CTCTGGCTCA |
|
| 1001 | GCACCAATAT GCGTCAGGAA ATTTCCGCCC TCGTCATCCT GCTGCAACGC |
|
| 1051 | ACCCGCCGCA AATGGCTGGA TGCCCACGAA CGCCAACACC TGCGCCAAAG |
|
| 1101 | CCTGCTTGAA ACACGGGAAC ACAGTTGA |
[1344]This encodes a protein having amino acid sequence <SEQ ID 634>:
[0000]| 1 | MNTSQRNRLV SRWLNSYERY RYRRLIHAVR LGGAVLFATA SARLLHLQHG | |
|
| 51 | EWIGMTVFVV LGMLQFQGAI YSKAVERMLG TVIGLGAGLG VLWLNQHYFH |
|
| 101 | GNLLFYLTVG TASALAGWAA VGKNGYVPML AGLTMCMLIG DNGSEWFDSG |
|
| 151 | LMRAMNVLIG AAIAIAAAKL LPLKSTLMWR FMLADNLTDC SKMIAEISNG |
|
| 201 | RRMTRERLEE NMAKMRQINA RMVKSRSHLA ATSGESRISP AMMEAMQHAH |
|
| 251 | RKIVNTTELL LTTAAKLQSP KLNGSEIRLL DRHFTLLQTD LQQTVALING |
|
| 301 | RHARRIRIDT AINPELEALA EHLHYQWQGF LWLSTNMRQE ISALVILLQR |
|
| 351 | TRRKWLDAHE RQHLRQSLLE TREHS* |
[1345]ORF146a and ORF146-1 show 99.5% identity in 374 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1346]ORF146 shows 97.3% identity over a 75aa overlap with a predicted ORF (ORF146ng) from N. gonorrhoeae:
[0000]
[1347]An ORF146ng nucleotide sequence <SEQ ID 635> was predicted to encode a protein having amino acid sequence <SEQ ID 636>:
[0000]| 1 | MSGVRFPSPA PIPSTDPPSG SLCFFTFPLQ TASDMNSSQR KRLSGRWLNS | |
|
| 51 | YERYRHRRLI HAVRLGGTVL FATALARLLH LQHGEWIGMT VFVVLGMLQF |
|
| 101 | QGAIYSNAVE RMLGTVIGLG AGLGVLWLNQ HYFHGNLLFY LTIGTASALA |
|
| 151 | GWAAVGKNGY VPMLAGLTMC MLIGDNGSEW LDSGLMRAMN VLIGAAIAIA |
|
| 201 | AAKLLPLKST LMWRFMLADN LADCSKMIAE ISNGRRMTRE RLEQNMVKMR |
|
| 251 | QINARMVKSR SHLAATSGES RISPSMMEAM QHAHRKIVNT TELLLTTAAK |
|
| 301 | LQSPKLNGSE IRLLDRHFTL LQTDLQQTAA LINGRHARRI RIDTAINPEL |
|
| 351 | EALAEHLHYQ WQGFLWLSTN MRQEISALVI PLQRTRRKWL DAHERQHLRQ |
|
| 401 | SLLETREHG* |
[1348]Further work revealed the following gonococcal DNA sequence <SEQ ID 637>:
[0000]| 1 | ATGAACTCCT CGCAACGCAA ACGCCTTTCC GgccGCTGGC TCAACTCCTA | |
|
| 51 | CGAACGCTac cGCCaccGCC GCCTCATACA TGCCGTGCGG CTCGGCggaa |
|
| 101 | ccgtCCTGTT CGCCACCGCA CTCGCCCGgc tACTCCACCT CCAacacggc |
|
| 151 | gAATGGATAG GGAtgaCCGT CTTCGTCGTC CTCGGCATGC TCCAGTTCCA |
|
| 201 | AGGCgcgatt tActccaacg cggtgGAacg taTGctcggt acggtcatcg |
|
| 251 | ggctgGGCGC GGGTTTGGgc gTTTTATGGC TGAACCAGCA TTAtttccac |
|
| 301 | ggcaacCTcc tcttctacct gaccatcggc acggcaagcg cactggccgg |
|
| 351 | ctGGGCGGCG GTCGGCAAAA acggctacgt ccctatgctg GCGGGGctgA |
|
| 401 | CGATGTGCAT gctcatcggc gACAACGGCA GCGAATGGCT CGACAGCGGC |
|
| 451 | CTGATGCGCG CGATGAACGT CCTCATCGGC GCCGCCATCG CCATTGCCGC |
|
| 501 | CGCCAAACTG CTGCCGCTGA AATCCACACT GATGTGGCGT TTCATGCTTG |
|
| 551 | CCGACAACCT GGCCGACTGC AGCAAAATGA TTGCCGAAAT CAGCAACGGC |
|
| 601 | AGGCGTATGA CGCGCGAACG TTTGGAGCAG AATATGGTCA AAATGCGCCA |
|
| 651 | AATCAACGCA CGCATGGTCA AAAGCCGCAG CCACCTCGCC GCCACATCGG |
|
| 701 | GCGAAAGCCG CATCAGCCCC TCCATGATGG AAGCCATGCA GCACGCCCAC |
|
| 751 | CGCAAAATCG TCAACACCAC CGAGCTGCTC CTGACCACCG CCGCCAAGCT |
|
| 801 | GCAATCTCCC AAACTCAACG GCAGCGAAAT CCGGCTGCTC GACCGCCACT |
|
| 851 | TCACACTGCT CCAAACCGAC CTGCAACAAA CCGCCGCCCT CATCAACGGC |
|
| 901 | AGACACGCCC GCCGCATCCG CATCGACACC GCCATCAACC CCGAACTGGA |
|
| 951 | AGCCCTCGCC GAACACCTCC ACTACCAATG GCAGGGCTTC CTCTGGCTCA |
|
| 1001 | GCACCAATAT GCGTCAGGAA ATTTCCGCCC TCGTCATCCT GCTGCAACGC |
|
| 1051 | ACCCGCCGCA AATGGCTGGA TGCCCACGAA CGCCAACACC TGCGCCAAAG |
|
| 1101 | CCTGCTTGAA ACACGGGAAC ACGGCTGA |
[1349]This corresponds to the amino acid sequence <SEQ ID 638; ORF146ng-1>:
[0000]| 1 | MNSSQRKRLS GRWLNSYERY RHRRLIHAVR LGGTVLFATA LARLLHLQHG | |
|
| 51 | EWIGMTVFVV LGMLQFQGAI YSNAVERMLG TVIGLGAGLG VLWLNQHYFH |
|
| 101 | GNLLFYLTIG TASALAGWAA VGKNGYVPML AGLTMCMLIG DNGSEWLDSG |
|
| 151 | LMRAMNVLIG AAIAIAAAKL LPLKSTLMWR FMLADNLADC SKMIAEISNG |
|
| 201 | RRMTRERLEQ NMVKMRQINA RMVKSRSHLA ATSGESRISP SMMEAMQHAH |
|
| 251 | RKIVNTTELL LTTAAKLQSP KLNGSEIRLL DRHFTLLQTD LQQTAALING |
|
| 301 | RHARRIRIDT AINPELEALA EHLHYQWQGF LWLSTNMRQE ISALVILLQR |
|
| 351 | TRRKWLDAHE RQHLRQSLLE TREHG* |
[1350]ORF146ng-1 and ORF146-1 show 96.5% identity in 375 aa overlap
[0000]
[1351]Furthermore, ORF146ng-1 shows homology with a hypothetical E. coli protein:
[0000]| sp|P33011|YEEA_ECOLI HYPOTHETICAL 40.0 KD PROTEIN IN COBU-SBMC INTERGENIC | |
| REGION >gi|1736674|gnl|PID|d1016553 (D90838) ORF_ID: o348#20; similar to |
| [SwissProt Accession Number P33011] [Escherichia coli] |
| >gi|1736682|gnl|PID|d1016560 (D90839) ORF_ID: o348#20; similar to |
| [SwissProt Accession Number P33011] [Escherichia coli] >gi|1788318 |
| (AE000292) f352; 100% identical to fragment YEEA_ECOLI SW: P33011 but has |
| 203 additional C-terminal residues [Escherichia coli] Length = 352 |
| Score = 109 bits (271), Expect = 2e−23 |
| Identities = 89/347 (25%), | Positives = 150/347 (42%), Gaps = 21/347 (6%) | |
|
| Query: | 20 | YRHRRLIHAVRLGGTVLFATALARLLHLQHGEWIGMTVFVVLGMLQFQGAIYSNAVERML | 79 | |
| | YRH R++H R+ L + RL + W +T+ V++G + F G + A ER+ | |
| Sbjct: | 15 | YRHYRIVHGTRVALAFLLTFLIIRLFTIPESTWPLVTMVVIMGPISFWGNVVPRAFERIG | 74 |
|
| Query: | 80 | GTVIGLGAGLGVLWLNQHYFHGNLLFYLTIGTASALAGWAAVGKNGYVPMLAGLTMCMLI | 139 |
| | GTV+G GL L L L + A L GW A+GK Y +L G+T+ +++ | |
| Sbjct: | 75 | GTVLGSILGLIALQLE---LISLPLMLVWCAAAMFLCGWLALGKKPYQGLLIGVTLAIVV | 131 |
|
| Query: | 140 | GDNGSEWLDSGLMRAMNVLIGXXXXXXXXKLLPLKSTLMWRFMLADNLADCSKMIAEISN | 199 |
| | G E +D+ L R+ +V++G + P ++ + WR LA +L + +++ + | |
| Sbjct: | 132 | GSPTGE-IDTALWRSGDVILGSLLAMLFTGIWPQRAFIHWRIQLAKSLTEYNRVYQSAFS | 190 |
|
| Query: | 200 | GRRMTRERLEQNMVKMRQINARMVKSRSHLAATSGESRISPSMMEAMQHAHRKIVNXXXX | 259 |
| | + R RLE ++ K+ VK R +A S E+RI S+ E +Q +R +V | |
| Sbjct: | 191 | PNLLERPRLESHLQKLL---TDAVKMRGLIAPASKETRIPKSIYEGIQTINRNLVCMLEL | 247 |
|
| Query: | 260 | XXXXXXXXQSPK---LNGSEIRLLDRHFXXXXXXXXXXAALINGRHARRIRIDTAINPEL | 316 |
| | + LN ++R D AL G +N + | |
| Sbjct: | 248 | QINAYWATRPSHFVLLNAQKLR--DTQHMMQQILLSLVHALYEGNPQPVFANTEKLNDAV | 305 |
|
| Query: | 317 | EALAEHL--HYQWQ-------GFLWLSTNMRQEISALVILLQRTRRK | 354 |
| | E L + L H+ + G++WL+ ++ L L+ R RK | |
| Sbjct: | 306 | EELRQLLNNHHDLKVVETPIYGYVWLNMETAHQLELLSNLICRALRK | 352 |
[1352]On the basis of this analysis, including the identification of several transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 76
[1353]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 639>
[0000]| 1 | ..GCCGAAGACA CGCGCGTTAC CGCACAGCTT TTGAGCGCGT ACGGCATTCA | |
|
| 51 | GGGCAAACTC GTCAGTGTGC GCGAACACAA CGAACGGCAG ATGGCGGACA |
|
| 101 | AGATTGTCGG CTATCTTTCA GACGGCATGG TTGTGGCACA GGTTTCCGAT |
|
| 151 | GCGGGTACGC CGGCCGTGTG CGACCCGGGC GCGAAACTCG CCCGCCGCGT |
|
| 201 | GCGTGAGGCC GGGTTTAAAG TCGTTCCCGT CGTGGGCGCA AC.GCGGTGA |
|
| 251 | TGGCGGCTTT GAGCGTGGCC GGTGTGGAAG GATCCGATTT TTATTTCAAC |
|
| 301 | GGTTTTGTAC CGCCGAAATC GGGAGAACGC AGGAAACTGT TTGCCAAATG |
|
| 351 | GGTGCGGGCG GCGTTTCCTA TCGTCATGTT TGAAACGCCG CACCGCATCG |
|
| 401 | GTGCAGCGCT TGCCGATATG GCGGAACTGT TCCCCGAACG CCGATTAATG |
|
| 451 | CTGGCGCGCG AAATTACGAA AACGTTTGAA ACGTTCTTAA GCGGCACGGT |
|
| 501 | TGGGGAAATT CAGACGGCAT TGTCTGCCGA CGGCGACCAA TCGCGCGGCG |
|
| 551 | AGATGGTGTT GGTGCTTTAT CCGGCGCAGG ATGAAAAACA CGAAGGCTTG |
|
| 601 | TCCGAGTCCG CGCAAAACAT CATGAAAATC CTCACAGCCG AGCTGCCGAC |
|
| 651 | CAAACAGGCG GCGGAGCTTG CTGCCAAAAT CACGGGCGAG GGAAAGAAAG |
|
| 701 | CTTTGTACGA T.. |
[1354]This corresponds to the amino acid sequence <SEQ ID 640; ORF147>:
[0000]| 1 | ..AEDTRVTAQL LSAYGIQGKL VSVREHNERQ MADKIVGYLS DGMVVAQVSD | |
|
| 51 | AGTPAVCDPG AKLARRVREA GFKVVPVVGA XAVMAALSVA GVEGSDFYFN |
|
| 101 | GFVPPKSGER RKLFAKWVRA AFPIVMFETP HRIGAALADM AELFPERRLM |
|
| 151 | LAREITKTFE TFLSGTVGEI QTALSADGDQ SRGEMVLVLY PAQDEKHEGL |
|
| 201 | SESAQNIMKI LTAELPTKQA AELAAKITGE GKKALYD.. |
[1355]Further work revealed the complete nucleotide sequence <SEQ ID 641>:
[0000]| 1 | ATGTTTCAGA AACATTTGCA GAAAGCCTCC GACAGCGTCG TCGGAGGGAC | |
|
| 51 | ATTATACGTG GTTGCCACGC CCATCGGCAA TTTGGCGGAC ATTACCCTGC |
|
| 101 | GCGCTTTGGC GGTATTGCAA AAGGCGGACA TCATCTGTGC CGAAGACACG |
|
| 151 | CGCGTTACCG CACAGCTTTT GAGCGCGTAC GGCATTCAGG GCAAACTCGT |
|
| 201 | CAGTGTGCGC GAACACAACG AACGGCAGAT GGCGGACAAG ATTGTCGGCT |
|
| 251 | ATCTTTCAGA CGGCATGGTT GTGGCACAGG TTTCCGATGC GGGTACGCCG |
|
| 301 | GCCGTGTGCG ACCCGGGCGC GAAACTCGCC CGCCGCGTGC GTGAGGCCGG |
|
| 351 | GTTTAAAGTC GTTCCCGTCG TGGGCGCAAG CGCGGTGATG GCGGCTTTGA |
|
| 401 | GCGTGGCCGG TGTGGAAGGA TCCGATTTTT ATTTCAACGG TTTTGTACCG |
|
| 451 | CCGAAATCGG GAGAACGCAG GAAACTGTTT GCCAAATGGG TGCGGGCGGC |
|
| 501 | GTTTCCTATC GTCATGTTTG AAACGCCGCA CCGCATCGGT GCGACGCTTG |
|
| 551 | CCGATATGGC GGAACTGTTC CCCGAACGCC GATTAATGCT GGCGCGCGAA |
|
| 601 | ATTACGAAAA CGTTTGAAAC GTTCTTAAGC GGCACGGTTG GGGAAATTCA |
|
| 651 | GACGGCATTG TCTGCCGACG GCAACCAATC GCGCGGCGAG ATGGTGTTGG |
|
| 701 | TGCTTTATCC GGCGCAGGAT GAAAAACACG AAGGCTTGTC CGAGTCCGCG |
|
| 751 | CAAAACATCA TGAAAATCCT CACAGCCGAG CTGCCGACCA AACAGGCGGC |
|
| 801 | GGAGCTTGCT GCCAAAATCA CGGGCGAGGG AAAGAAAGCT TTGTACGATC |
|
| 851 | TGGCTCTGTC TTGGAAAAAC AAATAG |
[1356]This corresponds to the amino acid sequence <SEQ ID 642; ORF147-1>:
[0000]| 1 | MFQKHLQKAS DSVVGGTLYV VATPIGNLAD ITLRALAVLQ KADIICAEDT | |
|
| 51 | RVTAQLLSAY GIQGKLVSVR EHNERQMADK IVGYLSDGMV VAQVSDAGTP |
|
| 101 | AVCDPGAKLA RRVREAGFKV VPVVGASAVM AALSVAGVEG SDFYFNGFVP |
|
| 151 | PKSGERRKLF AKWVRAAFPI VMFETPHRIG ATLADMAELF PERRLMLARE |
|
| 201 | ITKTFETFLS GTVGEIQTAL SADGNQSRGE MVLVLYPAQD EKHEGLSESA |
|
| 251 | QNIMKILTAE LPTKQAAELA AKITGEGKKA LYDLALSWKN K* |
[1357]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical Protein ORF286 of E. coli (Accession Number U18997)
[1358]ORF147 and E. coli ORF286 protein show 36% aa identity in 237aa overlap:
[0000]| Orf147: | 1 | AEDTRVTAQLLSAYGIQGKLVSVREHNERQMADKIVGYLSDGMVVAQVSDAGTPAVCDPG | 60 | |
| | AEDTR T LL +GI +L ++ +HNE+Q A+ ++ L +G +A VSDAGTP + DPG | |
|
| Orf286: | 43 | AEDTRHTGLLLQHFGINARLFALHDHNEQQKAETLLAKLQEGQNIALVSDAGTPLINDPG | 102 |
|
| Orf147: | 61 | AKLARRVREXXXXXXXXXXXXXXXXXXXXXXXEGSDFYFNGFVPPKSGERRKLFAKWVRA | 120 |
| | L R RE F + GF+P KS RR | |
|
| Orf286: | 103 | YHLVRTCREAGIRVVPLPGPCAAITALSAAGLPSDRFCYEGFLPAKSKGRRDALKAIEAE | 162 |
|
| Orf147: | 121 | AFPIVMFETPHRIGAALADMAELFPERR-LMLAREITKTFETFLSGTVGEIQTALSADGD | 179 |
| | ++ +E+ HR+ +L D+ + E R ++LARE+TKT+ET VGE+ + D + | |
|
| Orf286: | 163 | PRTLIFYESTHRLLDSLEDIVAVLGESRYVVLARELTKTWETIHGAPVGELLAWVKEDEN | 222 |
|
| Orf147: | 180 | QSRGEMVLVLYPAQDEKHEGLSESAQNIMKILTAELPTKQAAELAAKITGEGKKALY | 236 |
| | + +GEMVL++ + E L A + +L AELP K+AA LAA+I G K ALY | |
|
| Orf286: | 223 | RRKGEMVLIV-EGHKAQEEDLPADALRTLALLQAELPLKKAAALAAEIHGVKKNALY | 278 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1359]ORF147 shows 96.6% identity over a237aa overlap with ORF75a from strain A of N. meningitidis:
[0000]
[1360]ORF147a is identical to ORF75a, which includes aa 56-292 of ORF75.
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1361]ORF147 shows 94.1% identity over a 237aa overlap with a predicted ORF (ORF147ng) from N. gonorrhoeae:
[0000]
[1362]An ORF147ng nucleotide sequence <SEQ ID 643> was predicted to encode a protein having amino acid sequence <SEQ ID 644>:
[0000]| 1 | MSVFQTAFFM FQKHLQKASD SVVGGTLYVV ATPIGNLADI TLRALAVLQK | |
|
| 51 | ADIICAEDTR VTAQLLSAYG IQGRLVSVRE HNERQMADKV IGFLSDGLVV |
|
| 101 | AQVSDAGTPA VCDPGAKLAR RVREAGFKVV PVVGASAVMA ALSVAGVAES |
|
| 151 | DFYFNGFVPP KSGERRKLFA KWVRAAFPVV MFETPHRIGA TLADMAELFP |
|
| 201 | ERRLMLAREI TKTFETFLSG TVGEIQTALA ADGNQSRGEM VLVLYPAQDE |
|
| 251 | KHEGLSESAQ NAMKILAAEL PTKQAAELAA KITGEGKKAL YDLALSWKNK |
|
| 301 | * |
[1363]Further work revealed the following gonococcal DNA sequence <SEQ ID 645>:
[0000]| 1 | ATGTTTCAGA AACACTTGCA GAAAGCCTCC GACAGCGTCG TCGGAGGGAC | |
|
| 51 | ATTATACGTG GTTGCCACGC CCATCGGCAA TTTGGCAGAC ATTACCCTGC |
|
| 101 | GCGCTTTGGC GGTATTGCAA AAGGCGGACA TCATTTGTGC CGAAGACACG |
|
| 151 | CGCGTTACTG CGCAGCTTTT GAGCGCGTAC GGCATTCAGG GCAGGTTGGT |
|
| 201 | CAGTGTGCGC GAACACAACG AGCGGCAGAT GGCGGACAAG GTAATCGGTT |
|
| 251 | TCCTTTCAGA CGGCCTGGTT GTGGCGCAGG TTTCCGATGC GGGTACGCCG |
|
| 301 | GCCGTGTGCG ACCCGGGCGC GAAACTCGCC CGCCGCGTGC GCGAAGCAGG |
|
| 351 | GTTCAAAGTC GTTCCCGTCG TGGGCGCAAG CGCGGTAATG GCGGCGTTGA |
|
| 401 | GTGTGGCCGG TGTGGCGGAA TCCGATTTTT ATTTCAACGG TTTTGTACCG |
|
| 451 | CCGAAATCGG GCGAACGTAG GAAATTGTTT GCCAAATGGG TGCGGGCGGC |
|
| 501 | ATTTCCTGTC GTCATGTTTG AAACGCCGCA CCGAATCGGG GCAACGCTTG |
|
| 551 | CCGATATGGC GGAATTGTTC CCCGAACGCC GTCTGATGCT GGCGCGCGAA |
|
| 601 | ATCACGAAAA CGTTTGAAAC GTTCTTAAGC GGCACGGTTG GGGAAATTCA |
|
| 651 | GACGGCATTG GCGGCGGACG GCAACCAATC GCGCGGCGAG ATGGTGTTGG |
|
| 701 | TGCTTTATCC GGCGCAGGAT GAAAAACACG AAGGCTTGTC CGAGTCTGCG |
|
| 751 | CAAAATGCGA TGAAAATCCT TGCGGCCGAG CTGCCGACCA AGCAGGCGGC |
|
| 801 | GGAGCTTGCC GCCAAGATTA CAGGTGAGGG CAAAAAGGCT TTGTACGATT |
|
| 851 | TGGCACTGTC GTGGAAAAAC AAATGA |
[1364]This corresponds to the amino acid sequence <SEQ ID 646; ORF147ng-1>:
[0000]| 1 | MFQKHLQKAS DSVVGGTLYV VATPIGNLAD ITLRALAVLQ KADIICAEDT | |
|
| 51 | RVTAQLLSAY GIQGRLVSVR EHNERQMADK VIGFLSDGLV VAQVSDAGTP |
|
| 101 | AVCDPGAKLA RRVREAGFKV VPVVGASAVM AALSVAGVAE SDFYFNGFVP |
|
| 151 | PKSGERRKLF AKWVRAAFPV VMFETPHRIG ATLADMAELF PERRLMLARE |
|
| 201 | ITKTFETFLS GTVGEIQTAL AADGNQSRGE MVLVLYPAQD EKHEGLSESA |
|
| 251 | QNAMKILAAE LPTKQAAELA AKITGEGKKA LYDLALSWKN K* |
[1365]ORF147ng shows homology to a hypothetical E. coli protein:
[0000]| sp|P45528|YRAL_ECOLI HYPOTHETICAL 31.3 KD | |
| PROTEIN IN AGAI-MTR INTERGENIC REGION (F286) |
| >gi|606086 (U18997) ORF_f286 [Escherichia coli] |
| >gi|1789535 (AE000395) hypothetical 31.3 kD protein in agai-mtr intergenic region |
| [Escherichia coli] Length = 286 |
| Score = 218 bits (550), Expect = 3e−56 |
| Identities = 128/284 (45%), Positives = 171/284 (60%), |
| Gaps = 4/284 (1%) |
|
| Query: | 4 | KHLQKASDSVVGGTLYVVATPIGNLADITLRALAVLQKADIICAEDTRVTAQLLSAYGIQ | 63 | |
| | K Q A +S G LY+V TPIGNLADIT RAL VLQ D+I AEDTR T LL +GI | |
| Sbjct: | 2 | KQHQSADNSQ--GQLYIVPTPIGNLADITQRALEVLQAVDLIAAEDTRHTGLLLQHFGIN | 59 |
|
| Query: | 64 | GRLVSVREHNERQMADKVIGFLSDGLVVAQVSDAGTPAVCDPGAKLARRVREAGFKVVPV | 123 |
| | RL ++ +HNE+Q A+ ++ L +G +A VSDAGTP + DPG L R REAG +VVP+ | |
| Sbjct: | 60 | ARLFALHDHNEQQKAETLLAKLQEGQNIALVSDAGTPLINDPGYHLVRTCREAGIRVVPL | 119 |
|
| Query: | 124 | VGASAVMAALSVAGVAESDFYFNGFVPPKSGERRKLFAKWVRAAFPVVMFETPHRIGATL | 183 |
| | G A + ALS AG+ F + GF+P KS RR ++ +E+ HR+ +L | |
| Sbjct: | 120 | PGPCAAITALSAAGLPSDRFCYEGFLPAKSKGRRDALKAIEAEPRTLIFYESTHRLLDSL | 179 |
|
| Query: | 184 | ADMAELFPERR-LMLAREITKTFETFLSGTVGEIQTALAADGNQSRGEMVLVLYPAQDEK | 242 |
| | D+ + E R ++LARE+TKT+ET VGE+ + D N+ +GEMVL++ + | |
| Sbjct: | 180 | EDIVAVLGESRYVVLARELTKTWETIHGAPVGELLAWVKEDENRRKGEMVLIV-EGHKAQ | 238 |
|
| Query: | 243 | HEGLSESAQNAMKILAAELPTKQAAELAAKITGEGKKALYDLAL | 286 |
| | E L A + +L AELP K+AA LAA+I G K ALY AL | |
| Sbjct: | 239 | EEDLPADALRTLALLQAELPLKKAAALAAEIHGVKKNALYKYAL | 282 |
[1366]Based on the computer analysis and the presence of a putative transmembrane domain in the gonococcal protein, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 77
[1367]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 647>
[0000]
[1368]This corresponds to the amino acid sequence <SEQ ID 648; ORF1>:
[0000]
[1369]Further sequencing analysis revealed the complete nucleotide sequence <SEQ ID 649>:
[0000]| 1 | ATGAAAACAA CCGACAAACG GACAACCGAA ACACACCGCA AAGCCCCGAA | |
|
| 51 | AACCGGCCGC ATCCGCTTCT CGCCTGCTTA CTTAGCCATA TGCCTGTCGT |
|
| 101 | TCGGCATTCT TCCCCAAGCC TGGGCGGGAC ACACTTATTT CGGCATCAAC |
|
| 151 | TACCAATACT ATCGCGACTT TGCCGAAAAT AAAGGCAAGT TTGCAGTCGG |
|
| 201 | GGCGAAAGAT ATTGAGGTTT ACAACAAAAA AGGGGAGTTG GTCGGCAAAT |
|
| 251 | CAATGACAAA AGCCCCGATG ATTGATTTTT CTGTGGTGTC GCGTAACGGC |
|
| 301 | GTGGCGGCAT TGGTGGGCGA TCAATATATT GTGAGCGTGG CACATAACGG |
|
| 351 | CGGCTATAAC AACGTTGATT TTGGTGCGGA AGGAAGAAAT CCCGATCAAC |
|
| 401 | ATCGTTTTAC TTATAAAATT GTGAAACGGA ATAATTATAA AGCAGGGACT |
|
| 451 | AAAGGCCATC CTTATGGCGG CGATTATCAT ATGCCGCGTT TGCATAAATT |
|
| 501 | TGTCACAGAT GCAGAACCTG TTGAAATGAC CAGTTATATG GATGGGCGGA |
|
| 551 | AATATATCGA TCAAAATAAT TACCCTGACC GTGTTCGTAT TGGGGCAGGC |
|
| 601 | AGGCAATATT GGCGATCTGA TGAAGATGAG CCCAATAACC GCGAAAGTTC |
|
| 651 | ATATCATATT GCAAGTGCGT ATTCTTGGCT CGTTGGTGGC AATACCTTTG |
|
| 701 | CACAAAATGG ATCAGGTGGT GGCACAGTCA ACTTAGGTAG TGAAAAAATT |
|
| 751 | AAACATAGCC CATATGGTTT TTTACCAACA GGAGGCTCAT TTGGCGACAG |
|
| 801 | TGGCTCACCA ATGTTTATCT ATGATGCCCA AAAGCAAAAG TGGTTAATTA |
|
| 851 | ATGGGGTATT GCAAACGGGC AACCCCTATA TAGGAAAAAG CAATGGCTTC |
|
| 901 | CAGCTGGTTC GTAAAGATTG GTTCTATGAT GAAATCTTTG CTGGAGATAC |
|
| 951 | CCATTCAGTA TTCTACGAAC CACGTCAAAA TGGGAAATAC TCTTTTAACG |
|
| 1001 | ACGATAATAA TGGCACAGGA AAAATCAATG CCAAACATGA ACACAATTCT |
|
| 1051 | CTGCCTAATA GATTAAAAAC ACGAACCGTT CAATTGTTTA ATGTTTCTTT |
|
| 1101 | ATCCGAGACA GCAAGAGAAC CTGTTTATCA TGCTGCAGGT GGTGTCAACA |
|
| 1151 | GTTATCGACC CAGACTGAAT AATGGAGAAA ATATTTCCTT TATTGACGAA |
|
| 1201 | GGAAAAGGCG AATTGATACT TACCAGCAAC ATCAATCAAG GTGCTGGAGG |
|
| 1251 | ATTATATTTC CAAGGAGATT TTACGGTCTC GCCTGAAAAT AACGAAACTT |
|
| 1301 | GGCAAGGCGC GGGCGTTCAT ATCAGTGAAG ACAGTACCGT TACTTGGAAA |
|
| 1351 | GTAAACGGCG TGGCAAACGA CCGCCTGTCC AAAATCGGCA AAGGCACGCT |
|
| 1401 | GCACGTTCAA GCCAAAGGGG AAAACCAAGG CTCGATCAGC GTGGGCGACG |
|
| 1451 | GTACAGTCAT TTTGGATCAG CAGGCAGACG ATAAAGGCAA AAAACAAGCC |
|
| 1501 | TTTAGTGAAA TCGGCTTGGT CAGCGGCAGG GGTACGGTGC AACTGAATGC |
|
| 1551 | CGATAATCAG TTCAACCCCG ACAAACTCTA TTTCGGCTTT CGCGGCGGAC |
|
| 1601 | GTTTGGATTT AAACGGGCAT TCGCTTTCGT TCCACCGTAT TCAAAATACC |
|
| 1651 | GATGAAGGGG CGATGATTGT CAACCACAAT CAAGACAAAG AATCCACCGT |
|
| 1701 | TACCATTACA GGCAATAAAG ATATTGCTAC AACCGGCAAT AACAACAGCT |
|
| 1751 | TGGATAGCAA AAAAGAAATT GCCTACAACG GTTGGTTTGG CGAGAAAGAT |
|
| 1801 | ACGACCAAAA CGAACGGGCG GCTCAACCTT GTTTACCAGC CCGCCGCAGA |
|
| 1851 | AGACCGCACC CTGCTGCTTT CCGGCGGAAC AAATTTAAAC GGCAACATCA |
|
| 1901 | CGCAAACAAA CGGCAAACTG TTTTTCAGCG GCAGACCAAC ACCGCACGCC |
|
| 1951 | TACAATCATT TAAACGACCA TTGGTCGCAA AAAGAGGGCA TTCCTCGCGG |
|
| 2001 | GGAAATCGTG TGGGACAACG ACTGGATCAA CCGCACATTT AAAGCGGAAA |
|
| 2051 | ACTTCCAAAT TAAAGGCGGA CAGGCGGTGG TTTCCCGCAA TGTTGCCAAA |
|
| 2101 | GTGAAAGGCG ATTGGCATTT GAGCAATCAC GCCCAAGCAG TTTTTGGTGT |
|
| 2151 | CGCACCGCAT CAAAGCCACA CAATCTGTAC ACGTTCGGAC TGGACGGGTC |
|
| 2201 | TGACAAATTG TGTCGAAAAA ACCATTACCG ACGATAAAGT GATTGCTTCA |
|
| 2251 | TTGACTAAGA CCGACATCAG CGGCAATGTC GATCTTGCCG ATCACGCTCA |
|
| 2301 | TTTAAATCTC ACAGGGCTTG CCACACTCAA CGGCAATCTT AGTGCAAATG |
|
| 2351 | GCGATACACG TTATACAGTC AGCCACAACG CCACCCAAAA CGGCAACCTT |
|
| 2401 | AGCCTCGTGG GCAATGCCCA AGCAACATTT AATCAAGCCA CATTAAACGG |
|
| 2451 | CAACACATCG GCTTCGGGCA ATGCTTCATT TAATCTAAGC GACCACGCCG |
|
| 2501 | TACAAAACGG CAGTCTGACG CTTTCCGGCA ACGCTAAGGC AAACGTAAGC |
|
| 2551 | CATTCCGCAC TCAACGGTAA TGTCTCCCTA GCCGATAAGG CAGTATTCCA |
|
| 2601 | TTTTGAAAGC AGCCGCTTTA CCGGACAAAT CAGCGGCGGC AAGGATACGG |
|
| 2651 | CATTACACTT AAAAGACAGC GAATGGACGC TGCCGTCAGG CACGGAATTA |
|
| 2701 | GGCAATTTAA ACCTTGACAA CGCCACCATT ACACTCAATT CCGCCTATCG |
|
| 2751 | CCACGATGCG GCAGGGGCGC AAACCGGCAG TGCGACAGAT GCGCCGCGCC |
|
| 2801 | GCCGTTCGCG CCGTTCGCGC CGTTCCCTAT TATCCGTTAC ACCGCCAACT |
|
| 2851 | TCGGTAGAAT CCCGTTTCAA CACGCTGACG GTAAACGGCA AATTGAACGG |
|
| 2901 | TCAGGGAACA TTCCGCTTTA TGTCGGAACT CTTCGGCTAC CGCAGCGACA |
|
| 2951 | AATTGAAGCT GGCGGAAAGT TCCGAAGGCA CTTACACCTT GGCGGTCAAC |
|
| 3001 | AATACCGGCA ACGAACCTGC AAGCCTCGAA CAATTGACGG TAGTGGAAGG |
|
| 3051 | AAAAGACAAC AAACCGCTGT CCGAAAACCT TAATTTCACC CTGCAAAACG |
|
| 3101 | AACACGTCGA TGCCGGCGCG TGGCGTTACC AACTCATCCG CAAAGACGGC |
|
| 3151 | GAGTTCCGCC TGCATAATCC GGTCAAAGAA CAAGAGCTTT CCGACAAACT |
|
| 3201 | CGGCAAGGCA GAAGCCAAAA AACAGGCGGA AAAAGACAAC GCGCAAAGCC |
|
| 3251 | TTGACGCGCT GATTGCGGCC GGGCGCGATG CCGTCGAAAA GACAGAAAGC |
|
| 3301 | GTTGCCGAAC CGGCCCGGCA GGCAGGCGGG GAAAATGTCG GCATTATGCA |
|
| 3351 | GGCGGAGGAA GAGAAAAAAC GGGTGCAGGC GGATAAAGAC ACCGCCTTGG |
|
| 3401 | CGAAACAGCG CGAAGCGGAA ACCCGGCCGG CTACCACCGC CTTCCCCCGC |
|
| 3451 | GCCCGCCGCG CCCGCCGGGA TTTGCCGCAA CTGCAACCCC AACCGCAGCC |
|
| 3501 | CCAACCGCAG CGCGACCTGA TCAGCCGTTA TGCCAATAGC GGTTTGAGTG |
|
| 3551 | AATTTTCCGC CACGCTCAAC AGCGTTTTCG CCGTACAGGA CGAATTAGAC |
|
| 3601 | CGCGTATTTG CCGAAGACCG CCGCAACGCC GTTTGGACAA GCGGCATCCG |
|
| 3651 | GGACACCAAA CACTACCGTT CGCAAGATTT CCGCGCCTAC CGCCAACAAA |
|
| 3701 | CCGACCTGCG CCAAATCGGT ATGCAGAAAA ACCTCGGCAG CGGGCGCGTC |
|
| 3751 | GGCATCCTGT TTTCGCACAA CCGGACCGAA AACACCTTCG ACGACGGCAT |
|
| 3801 | CGGCAACTCG GCACGGCTTG CCCACGGCGC CGTTTTCGGG CAATACGGCA |
|
| 3851 | TCGACAGGTT CTACATCGGC ATCAGCGCGG GCGCGGGTTT TAGCAGCGGC |
|
| 3901 | AGCCTTTCAG ACGGCATCGG AGGCAAAATC CGCCGCCGCG TGCTGCATTA |
|
| 3951 | CGGCATTCAG GCACGATACC GCGCCGGTTT CGGCGGATTC GGCATCGAAC |
|
| 4001 | CGCACATCGG CGCAACGCGC TATTTCGTCC AAAAAGCGGA TTACCGCTAC |
|
| 4051 | GAAAACGTCA ATATCGCCAC CCCCGGCCTT GCATTCAACC GCTACCGCGC |
|
| 4101 | GGGCATTAAG GCAGATTATT CATTCAAACC GGCGCAACAC ATTTCCATCA |
|
| 4151 | CGCCTTATTT GAGCCTGTCC TATACCGATG CCGCTTCGGG CAAAGTCCGA |
|
| 4201 | ACACGCGTCA ATACCGCCGT ATTGGCTCAG GATTTCGGCA AAACCCGCAG |
|
| 4251 | TGCGGAATGG GGCGTAAACG CCGAAATCAA AGGTTTCACG CTGTCCCTCC |
|
| 4301 | ACGCTGCCGC CGCCAAAGGC CCGCAACTGG AAGCGCAACA CAGCGCGGGC |
|
| 4351 | ATCAAATTAG GCTACCGCTG GTAA |
[1370]This corresponds to the amino acid sequence <SEQ ID 650; ORF1-1>:
[0000]| 1 | MKTTDKRTTE THRKAPKTGR IRFSPAYLAI CLSFGILPQA WAGHTYFGIN | |
|
| 51 | YQYYRDFAEN KGKFAVGAKD IEVYNKKGEL VGKSMTKAPM IDFSVVSRNG |
|
| 101 | VAALVGDQYI VSVAHNGGYN NVDFGAEGRN PDQHRFTYKI VKRNNYKAGT |
|
| 151 | KGHPYGGDYH MPRLHKFVTD AEPVEMTSYM DGRKYIDQNN YPDRVRIGAG |
|
| 201 | RQYWRSDEDE PNNRESSYHI ASAYSWLVGG NTFAQNGSGG GTVNLGSEKI |
|
| 251 | KHSPYGFLPT GGSFGDSGSP MFIYDAQKQK WLINGVLQTG NPYIGKSNGF |
|
| 301 | QLVRKDWFYD EIFAGDTHSV FYEPRQNGKY SFNDDNNGTG KINAKHEHNS |
|
| 351 | LPNRLKTRTV QLFNVSLSET AREPVYHAAG GVNSYRPRLN NGENISFIDE |
|
| 401 | GKGELILTSN INQGAGGLYF QGDFTVSPEN NETWQGAGVH ISEDSTVTWK |
|
| 451 | VNGVANDRLS KIGKGTLHVQ AKGENQGSIS VGDGTVILDQ QADDKGKKQA |
|
| 501 | FSEIGLVSGR GTVQLNADNQ FNPDKLYFGF RGGRLDLNGH SLSFHRIQNT |
|
| 551 | DEGAMIVNHN QDKESTVTIT GNKDIATTGN NNSLDSKKEI AYNGWFGEKD |
|
| 601 | TTKTNGRLNL VYQPAAEDRT LLLSGGTNLN GNITQTNGKL FFSGRPTPHA |
|
| 651 | YNHLNDHWSQ KEGIPRGEIV WDNDWINRTF KAENFQIKGG QAWSRNVAK |
|
| 701 | VKGDWHLSNH AQAVFGVAPH QSHTICTRSD WTGLTNCVEK TITDDKVIAS |
|
| 751 | LTKTDISGNV DLADHAHLNL TGLATLNGNL SANGDTRYTV SHNATQNGNL |
|
| 801 | SLVGNAQATF NQATLNGNTS ASGNASFNLS DHAVQNGSLT LSGNAKANVS |
|
| 851 | HSALNGNVSL ADKAVFHFES SRFTGQISGG KDTALHLKDS EWTLPSGTEL |
|
| 901 | GNLNLDNATI TLNSAYRHDA AGAQTGSATD APRRRSRRSR RSLLSVTPPT |
|
| 951 | SVESRFNTLT VNGKLNGQGT FRFMSELFGY RSDKLKLAES SEGTYTLAVN |
|
| 1001 | NTGNEPASLE QLTVVEGKDN KPLSENLNFT LQNEHVDAGA WRYQLIRKDG |
|
| 1051 | EFRLHNPVKE QELSDKLGKA EAKKQAEKDN AQSLDALIAA GRDAVEKTES |
|
| 1101 | VAEPARQAGG ENVGIMQAEE EKKRVQADKD TALAKQREAE TRPATTAFPR |
|
| 1151 | ARRARRDLPQ LQPQPQPQPQ RDLISRYANS GLSEFSATLN SVFAVQDELD |
|
| 1201 | RVFAEDRRNA VWTSGIRDTK HYRSQDFRAY RQQTDLRQIG MQKNLGSGRV |
|
| 1251 | GILFSHNRTE NTFDDGIGNS ARLAHGAVFG QYGIDRFYIG ISAGAGFSSG |
|
| 1301 | SLSDGIGGKI RRRVLHYGIQ ARYRAGFGGF GIEPHIGATR YFVQKADYRY |
|
| 1351 | ENVNIATPGL AFNRYRAGIK ADYSFKPAQH ISITPYLSLS YTDAASGKVR |
|
| 1401 | TRVNTAVLAQ DFGKTRSAEW GVNAEIKGFT LSLHAAAAKG PQLEAQHSAG |
|
| 1451 | IKLGYRW* |
[1371]Computer analysis of these sequences gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1372]ORF1 shows 57.8% identity over a 1456aa overlap with an ORF (ORF1a) from strain A of N. meningitidis:
[0000]

[1373]The complete length ORFla nucleotide sequence <SEQ ID 651> is:
[0000]| 1 | ATGAAAACAA CCGACAAACG GACAACCGAA ACACACCGCA AAGCCCCGAA | |
|
| 51 | AACCGGCCGC ATCCGCTTCT CGCCTGCTTA CTTAGCCATA TGCCTGTCGT |
|
| 101 | TCGGCATTCT TCCCCAAGCT TGGGCGGGAC ACACTTATTT CGGCATCAAC |
|
| 151 | TACCAATACT ATCGCGACTT TGCCGAAAAT AAAGGCAAGT TTGCAGTCGG |
|
| 201 | GGCGAAAGAT ATTGAGGTNT ACAACAAAAA AGGGGAGTTG GTCGGCAAAT |
|
| 251 | CAATGACAAA AGCCCCGATG ATTGATTTTT CTGTGGTGTC GCGTAACGGC |
|
| 301 | GTGGCGGCAT TGGTGGGCGA TCAATATATT GTGAGCGTGG CACATAACGG |
|
| 351 | CGGCTATAAC AACGTTGATT TTGGTGCGGA AGGAAGNAAT CCCGATCAGC |
|
| 401 | ACCGTTTTTC TTACCAAATT GTGAAAAGAA ATAATTATAA GCCTGACAAT |
|
| 451 | TCACACCCTT ACAACGGCGA TTANCATATG CCGCGTTTGC ATAAATTTGT |
|
| 501 | CACAGATGCA GAACCTGTCG AAATGACGAG TGACATGAGG GGGAATACCT |
|
| 551 | ATTCCGATAA AGAAAAATAT CCCGAGCGTG TCCGCATCGG CTCAGGACAC |
|
| 601 | CACTATTGGC GTTATGATGA TGACAAACAC GGCGATTTAT CCTACTCCGG |
|
| 651 | CGCATGGTTA ATTGGCGGCA ATACACATAT GCAGGGTTGG GGAAATAATG |
|
| 701 | GCGTANTTAG TTTGAGCGGC GATGTGCGCC ATGCCAACGA CTATGGCCCT |
|
| 751 | ATGCCGATTG CAGGTGCGGC AGGCGACAGC GGTTCGCCAA TGTTTATTTA |
|
| 801 | TGACAAAACA AACAATAAAT GGCTGCTCAA CGGAGTTTTA CAAACCGGCT |
|
| 851 | ACCCTTATTC CGGCAGGGAA AACGGTTTCC AGCTGATACG CAAAGATTGG |
|
| 901 | TTCTACGATG ACATTTACAG AGGCGATACA CATACCGTCT NTTTTGAACC |
|
| 951 | GCGCAGTAAC GGACATTTTT CCTTTACATC CAACAACAAC GGTACGGGTA |
|
| 1001 | CGGTAACAGA AACCAACGAA AAGGTNTCCA ATCCAAAGCT TAAAGTACAG |
|
| 1051 | ACAGTCCGAC TGTTTGACGA ATCTTTGAAT GAAACTGATA AAGAACCAGT |
|
| 1101 | TTACGCGGCA GGGGGTGTTA ATCAGTACCG TCCAAGGTTA AACAACGGTG |
|
| 1151 | AAAACCTTTC TTTTATCGAT TACGGCAACG GCAAACTCAT CTTATCAAAC |
|
| 1201 | AACATCAACC AAGGCGCGGG CGGTTTGTAT TTTGAAGGTG ATTTTACGGT |
|
| 1251 | CTCGCCTGAA AACAACGAAA CGTGGCAAGG CGCGGGCGTT CATATCAGTG |
|
| 1301 | AAGACAGTAC CGTTACTTGG AAAGTAAACG GCGTGGCAAA CGACCGCCTG |
|
| 1351 | TCCAAAATCG GCAAAGGCAC GCTGCACGTT CAAGCCAAAG GGGAAAACCA |
|
| 1401 | AGGCTCGATC AGCGTGGGCG ACGGTACAGT CATTTTGGAT CAGCAGGCAG |
|
| 1451 | ACGATAAAGG CAAAAAACAA GCCTTTAGTG AAATCGGCTT GNTCAGCGGC |
|
| 1501 | AGGGGTACGG TGCAACTGAA TGCCGATAAT CAGTTCAACC CCGACAAACT |
|
| 1551 | CTATTTCGGC TTTCGCGGCG GACGTTTGGA TTTAAACGGG CATTCGCTTT |
|
| 1601 | CGTTCCACCG TATTCAAAAT ACCGATGAAG GGGCGATGAT TGNCNATCAT |
|
| 1651 | AATGCCACAA CAACATCCAC CGTTACCATT ACAGGGAATG AAAGTATTAC |
|
| 1701 | ACAACCGAGT GGTAAGAATA TCAATAGACT TAATTACAGC AAAGAAATTG |
|
| 1751 | CCTACAACGG TTGGTTTGGC GAGAAAGATA CGACCAAAAC GAACGGGCGG |
|
| 1801 | CTCAACCTTG TTTACCAGCC CGCCGCAGAA GACCGCACCC NGCTGCTTTC |
|
| 1851 | CGGCGGAACA AATTTAAACG GCAACATCAC GCAAACAAAC GGCAAACTGT |
|
| 1901 | TTTTCAGCGG CAGACCGACA CCGCACGCCT ACAATCATTT AGGAAGCGGG |
|
| 1951 | TGGTCAAAAA TGGAAGGTAT CCCACAAGGA GAAATCGTGT GGGACAACGA |
|
| 2001 | CTGGATCNAC CGCACGTTTA AAGCGGAAAA TTTCCATATT CAGGGCGGGC |
|
| 2051 | AGGCGGTGAT TTCCCGCAAT GTTGCCAAAG TGGAAGGCGA TTGNCATTTG |
|
| 2101 | AGCAATCACG CCCAAGCAGT TTTTGGTGTC GCACCGCATC AAAGCCATAC |
|
| 2151 | AATCTGTACA CGTTCGGACT GGACNGGTCT GACAAATTGT GTCGAANAAA |
|
| 2201 | NCATTACCGA CGATAAAGTG ATTGCTTCAT TGACTAAGAC NGACNTNAGC |
|
| 2251 | GGCANTGTNA GNCTNNCCNA TNACGNTNNT TNAAANCTCN CNGGGCNTGC |
|
| 2301 | NNCACTNAAN GGCAATCTTA GTGCAAATGG CGATACACGT TATACAGTCA |
|
| 2351 | GCCACAACGC CACCCAAAAC GGCAACCTTA GCCTCGTGGG CAATGCCCAA |
|
| 2401 | GCAACATTTA ATCAAGCCAC ATTAAACGGC AACNCATCGG NTTCGGGCAA |
|
| 2451 | TGCTTCATTT AATCTAAGCA ACAACGCCGC ACAAAACGGC AGTCTGACGC |
|
| 2501 | TTTCCGACAA CGCTAAGGCA AACGTAAGCC ATTCCGCACT CAACGGCAAT |
|
| 2551 | GTCTCCCTAG CCGATAAGGC AGTATTCCAT TTTGAAAACA GCCGCTTTAC |
|
| 2601 | CGGACAACTC AGCGGCAGCA AGGANACAGC ATTACACTTA AAAGACAGCG |
|
| 2651 | AATGGACGCT GCCGTCAGGC ACGGAATTAG GCAATTTAAA CCTTGACAAC |
|
| 2701 | GCCACCATTA CACTCAATTC CGCCTATCGC CACGATGCTG CAGGCGCGCA |
|
| 2751 | AACCGGCAGN GTGTCAGACA CGCCGCGCCG CCGTTCGCGC CGTTCCCTAT |
|
| 2801 | TATCCGTTAC ACCGCCAACT TCGGTAGAAT CCCGTTTCAA CACGCTGACG |
|
| 2851 | GTAAACGGCA AATTGAACNG TCAAGGAACA TTCCGCTTTA TGTCGGAACT |
|
| 2901 | CTTCGGCTAC CGAAGCGACA AATTGAAGCT GGCGGAAAGT TCCGAAGGNA |
|
| 2951 | CTTACACCTT GGCGGTCAAC AATACCGGCA ACGAACCCGT AAGCCTCGAT |
|
| 3001 | CAATTGACGG TAGTGGAAGG GAAAGACAAC AAACCGCTGT CCGAAAACCT |
|
| 3051 | TAATTTCACC CTGCAAAACG AACACGTCGA TGCCGGCGCG TGGCGTTACC |
|
| 3101 | AACTCATCCG CAAAGACGGC GAGTTCCGCC TGCATAATCC GGTCAAAGAA |
|
| 3151 | CAAGAGCTTT CCGACAAACT CGGCAAGGCA GAAGCCAAAA AACAGGCGGA |
|
| 3201 | AAAAGACAAC GCGCAAAGCC TTGACGCGCT GATTGCGGCC GGGCGCGATG |
|
| 3251 | CCGCCGAAAA GACAGAAAGC GTTGCCGAAC CGGCCCGGCN GGCAGGCGGG |
|
| 3301 | GAAAATGTCG GCATTATGCA GGCGGAGGAA GAGAAAAAAC GGGTGCAGGC |
|
| 3351 | GGATAAAGAC AGCGCNTTGG CGAAACAGCG CGAAGCGGAA ACCCGGCCGG |
|
| 3401 | NTACCACCGC CTTCCCCCGC GCCCGCNGCG CCCGCCGGGA TTTGCCGCAA |
|
| 3451 | CCGCAGCCCC AACCGCAACC TCAACCCCAA CCGCAGCGCG ACCTGATNAG |
|
| 3501 | CCGTTATGCC AATAGCGGTT TGAGTGAATT TTCCGCCACG CTCAACAGCG |
|
| 3551 | TTTTCGCCGT ACAGGACGAA TTGGACCGCG TGTTTGCCGA AGACCGCCGC |
|
| 3601 | AACGCNGTTT GGACAAGCNG CATCCGGNAC ACCAAACACT ACCGTTCGCA |
|
| 3651 | AGATTTCCGC GCCTACCGCC AACAAACCGA CCTGCGCCAA ATCGGTATGC |
|
| 3701 | AGAAAAACCT CGGCAGCGGG CGCGTCGGCA TCCTGTTTTC GCACAACCGG |
|
| 3751 | ACCGAAAACA NCTTCGACGA CGGCATCGGC AACTCGGCAC GGCTTGCCCA |
|
| 3801 | CGGCGCCGTT TTCGGGCAAT ACGGCATCGG CAGGTTCGAC ATCGGCATCA |
|
| 3851 | GCACGGGCGC GGGTTTTAGC AGCGGCANTC TNTCAGACGG CATCGGAGGC |
|
| 3901 | AAAATCCGCC GCCGCGTGCT GCATTACGGC ATTCAGGCAC GATACCGCGC |
|
| 3951 | CGGTTTCGGC GGATTCGGCA TCGAACCGTA CATCGGCGCA ACGCGCTATT |
|
| 4001 | TCGTCCAAAA AGCGGATTAC CGCTACGAAA ACGTCAATAT CGCCACCCCC |
|
| 4051 | GGTCTTGCGT TCAACCGNTA CCGNGCGGGC ATTAAGGCAG ATTATTCATT |
|
| 4101 | CAAACCGGCG CAACACATNT CCATCACNCC TTATTTNAGC CTGTCCTATA |
|
| 4151 | CCGATGCCGC TTCGGGCAAA GTCCGAACAC GCGTCAATAC CGCNGTATTG |
|
| 4201 | GCTCAGGATT TCGGCAAAAC CCGCAGTGCG GAATGGGGCG TAAACGCCGA |
|
| 4251 | AATCAAAGGT TTCACGCTGT CCNTCCACGC TGCCGCCGCC AAAGGNCCGC |
|
| 4301 | AACTGGAAGC GCAACACAGC GCGGGCATCA AATTAGGCTA CCGCTGGTAA |
[1374]This encodes a protein having amino acid sequence <SEQ ID 652>:
[0000]| 1 | MKTTDKRTTE THRKAPKTGR IRFSPAYLAI CLSFGILPQA WAGHTYFGIN | |
|
| 51 | YQYYRDFAEN KGKFAVGAKD IEVYNKKGEL VGKSMTKAPM IDFSVVSRNG |
|
| 101 | VAALVGDQYI VSVAHNGGYN NVDFGAEGXN PDQHRFSYQI VKRNNYKPDN |
|
| 151 | SHPYNGDXHM PRLHKFVTDA EPVEMTSDMR GNTYSDKEKY PERVRIGSGH |
|
| 201 | HYWRYDDDKH GDLSYSGAWL IGGNTHMQGW GNNGVXSLSG DVRHANDYGP |
|
| 251 | MPIAGAAGDS GSPMFIYDKT NNKWLLNGVL QTGYPYSGRE NGFQLIRKDW |
|
| 301 | FYDDIYRGDT HTVXFEPRSN GHFSFTSNNN GTGTVTETNE KVSNPKLKVQ |
|
| 351 | TVRLFDESLN ETDKEPVYAA GGVNQYRPRL NNGENLSFID YGNGKLILSN |
|
| 401 | NINQGAGGLY FEGDFTVSPE NNETWQGAGV HISEDSTVTW KVNGVANDRL |
|
| 451 | SKIGKGTLHV QAKGENQGSI SVGDGTVILD QQADDKGKKQ AFSEIGLXSG |
|
| 501 | RGTVQLNADN QFNPDKLYFG FRGGRLDLNG HSLSFHRIQN TDEGAMIXXH |
|
| 551 | NATTTSTVTI TGNESITQPS GKNINRLNYS KEIAYNGWFG EKDTTKTNGR |
|
| 601 | LNLVYQPAAE DRTXLLSGGT NLNGNITQTN GKLFFSGRPT PHAYNHLGSG |
|
| 651 | WSKMEGIPQG EIVWDNDWIX RTFKAENFHI QGGQAVISRN VAKVEGDXHL |
|
| 701 | SNHAQAVFGV APHQSHTICT RSDWTGLTNC VEXXITDDKV IASLTKTDXS |
|
| 751 | GXVXLXXXXX XXLXGXAXLX GNLSANGDTR YTVSHNATQN GNLSLVGNAQ |
|
| 801 | ATFNQATLNG NXSXSGNASF NLSNNAAQNG SLTLSDNAKA NVSHSALNGN |
|
| 851 | VSLADKAVFH FENSRFTGQL SGSKXTALHL KDSEWTLPSG TELGNLNLDN |
|
| 901 | ATITLNSAYR HDAAGAQTGX VSDTPRRRSR RSLLSVTPPT SVESRFNTLT |
|
| 951 | VNGKLNXQGT FRFMSELFGY RSDKLKLAES SEGTYTLAVN NTGNEPVSLD |
|
| 1001 | QLTVVEGKDN KPLSENLNFT LQNEHVDAGA WRYQLIRKDG EFRLHNPVKE |
|
| 1051 | QELSDKLGKA EAKKQAEKDN AQSLDALIAA GRDAAEKTES VAEPARXAGG |
|
| 1101 | ENVGIMQAEE EKKRVQADKD SALAKQREAE TRPXTTAFPR ARXARRDLPQ |
|
| 1151 | PQPQPQPQPQ PQRDLXSRYA NSGLSEFSAT LNSVFAVQDE LDRVFAEDRR |
|
| 1201 | NAVWTSXIRX TKHYRSQDFR AYRQQTDLRQ IGMQKNLGSG RVGILFSHNR |
|
| 1251 | TENXFDDGIG NSARLAHGAV FGQYGIGRFD IGISTGAGFS SGXLSDGIGG |
|
| 1301 | KIRRRVLHYG IQARYRAGFG GFGIEPYIGA TRYFVQKADY RYENVNIATP |
|
| 1351 | GLAFNRYRAG IKADYSFKPA QHXSITPYXS LSYTDAASGK VRTRVNTAVL |
|
| 1401 | AQDFGKTRSA EWGVNAEIKG FTLSXHAAAA KGPQLEAQHS AGIKLGYRW* |
[1375]A transmembrane region is underlined.
[1376]ORF1-1 shows 86.3% identity over a 1462aa overlap with ORF1a:
[0000]

[0000]Homology with Adhesion and Penetration Protein Hap Precursor of H. influenzae (Accession Number P45387)
[1377]Amino acids 23-423 of ORF1 show 59% aa identity with hap protein in 450aa overlap:
[0000]| orf1 | 23 | FXAAYLAICLSFGILPQAWAGHTYFGINYQYYRDFAENKGKFAVGAKDIEVYNKKGELVG | 82 | |
| | F +L C+S GI QAWAGHTYFGI+YQYYRDFAENKGKF VGAK+IEVYNK+G+LVG | |
| hap | 6 | FRLNFLTACVSLGIASQAWAGHTYFGIDYQYYRDFAENKGKFTVGAKNIEVYNKEGQLVG | 65 |
|
| orf1 | 83 | KSMTKAPMIDFSVVSRNGVAALVGVQYIVSVAHNGGYNNVDFGAEGXNIXDQXRXTYKIV | 142 |
| | SMTKAPMIDFSVVSRNGVAALVG QYIVSVAHNGGYN+VDFGAEG N DQ R TY+IV | |
| hap | 66 | TSMTKAPMIDFSVVSRNGVAALVGDQYIVSVAHNGGYNDVDFGAEGRN-PDQHRFTYQIV | 124 |
|
| orf1 | 143 | KRNNYKAGTKGHPYGGDYHMPRLHKXVTDAEPVEMTSYMDGRKYIDQNNYPDRVRIGAGR | 202 |
| | KRNNY+A + HPY GDYHMPRLHK VT+AEPV MT+ MDG+ Y D+ NYP+RVRIG+GR | |
| hap | 125 | KRNNYQAWERKHPYDGDYHMPRLHKFVTEAEPVGMTTNMDGKVYADRENYPERVRIGSGR | 184 |
|
| orf1 | 203 | QYWRSDEDEPNNRESSYHIA---------------------------------------- | 222 |
| | QYWR+D+DE N SSY+++ | |
| hap | 185 | QYWRTDKDEETNVHSSYYVSGAYRYLTAGNTHTQSGNGNGTVNLSGNVVSPNHYGPLPTG | 244 |
|
| orf1 | 223 | -----SGSPMFIYDAQKQKWLINGVLQTGNPYIGKSNGFQLVRKDWFYDEIFAGDTHSVF | 277 |
| | SGSPMFIYDA+K++WLIN VLQTG+P+ G+ NGFQL+R++WFY+E+ A DT SVF | |
| hap | 245 | GSKGDSGSPMFIYDAKKKQWLINAVLQTGHPFFGRGNGFQLIREEWFYNEVLAVDTPSVF | 304 |
|
| orf1 | 278 | --YEPRQNGKYSFNDDNNGTGKIN-AKHEHNSLPNRLKTRTVQLFNVSLSETAREPVYHA | 334 |
| | Y P NG YSF +N+GTGK+ + + + + TV+LFN SL++TA+E V A | |
| hap | 305 | QRYIPPINGHYSFVSNNDGTGKLTLTRPSKDGSKAKSEVGTVKLFNPSLNQTAKEHV-KA | 363 |
|
| orf1 | 335 | AGGVNSYRPRLNNGENISFIDEGKGELILTSNINQGAGGLYFQGDFTV-SPENNETWQGA | 393 |
| | A G N Y+PR+ G+NI D+GKG L + +NINQGAGGLYF+G+F V +NN TWQGA | |
| hap | 364 | AAGYNIYQPRMEYGKNIYLGDQGKGTLTIENNINQGAGGLYFEGNFVVKGKQNNITWQGA | 423 |
|
| orf1 | 394 | GVHISEDSTVTWKVNGVANDRLSKIGKGTL | 423 |
| | GV I +D+TV WKV+ NDRLSKIG GTL | |
| hap | 424 | GVSIGQDATVEWKVHNPENDRLSKIGIGTL | 453 |
[1378]Amino acids 715-1011 of ORF1 show 50% aa identity with hap protein in 258aa overlap:
[0000]| Orf1 | 41 | DTRYTVSHNATQ-NGNXSLVXNAQATFNQ-ATLNGNTSASGNASFNLSDHAVQNGSLTLS | 98 | |
| | DT+ S TQ NG+ +L NA + A LNGN + ++ F LS++A Q G++ LS | |
| hap | 733 | DTKVINSIPITQINGSINLTNNATVNIHGLAKLNGNVTLIDHSQFTLSNNATQTGNIKLS | 792 |
|
| orf1 | 99 | GNAKANVSHSALNGNVSLADKAVFHFESSRFTGQISGGKDTALHLKDSEWTLPSGXELGN | 158 |
| | +A A V+++ LNGNV L D A F ++S F QI G KDT + L+++ WT+PS L N | |
| hap | 793 | NHANATVNNATLNGNVHLTDSAQFSLKNSHFWHQIQGDKDTTVTLENATWTMPSDTTLQN | 852 |
|
| orf1 | 159 | LNLDNATITLNSAYRHDAAGAQTGSATDAPXXXXXXXXXXLLXVTPPTSVESRFNTLTVN | 218 |
| | L L+N+T+TLNSAY + S+ +AP L T PTS E RFNTLTVN | |
| hap | 853 | LTLNNSTVTLNSAY--------SASSNNAPRHRRS-----LETETTPTSAEHRFNTLTVN | 899 |
|
| orf1 | 219 | GKLNGQGTFRFMSELFGYRSDKLKLAESSEGTYTLAVNNTGNEPASLEQLTVVEGKDNKP | 278 |
| | GKL+GQGTF+F S LFGY+SDKLKL+ +EG YTL+V NTG EP +LEQLT++E DNKP | |
| hap | 900 | GKLSGQGTFQFTSSLFGYKSDKLKLSNDAEGDYTLSVRNTGKEPVTLEQLTLIESLDNKP | 959 |
|
| orf1 | 279 | LSENLNFTLQNEHVDAGA | 296 |
| | LS+ L FTL+N+HVDAGA | |
| hap | 960 | LSDKLKFTLENDHVDAGA | 977 |
[1379]Amino acids 1192-1450 of ORF1 show 41% aa identity with hap protein in 259aa overlap:
[0000]| Orf1 | 1 | LDRVFAEDRRNAVWTSGIRDTKHYRSQDFRAYRQQTDLRQIGMQKNLGSGRVGILFSHNR | 60 | |
| | LDR+F + ++AVWT+ +D + Y S FRAY+Q+T+LRQIG+QK L +GR+G +FSH+R | |
| hap | 1135 | LDRLFVDQAQSAVWTNIAQDKRRYDSDAFRAYQQKTNLRQIGVQKALANGRIGAVFSHSR | 1194 |
|
| orf1 | 61 | TENTFDDGIGNSARLAHGAVFGQYGIDRFYXXXXXXXXXXXXXXXXXIGXKXRRRVLHYG | 120 |
| | ++NTFD+ + N A L + F QY K R+ ++YG | |
| hap | 1195 | SDNTFDEQVKNHATLTMMSGFAQYQWGDLQFGVNVGTGISASKMAEEQSRKIHRKAINYG | 1254 |
|
| orf1 | 121 | IQARYRAGFGGFGIEPHIGATRYFVQKADYRYENVNIATPGLAFNRYRAGIKADYSFKPA | 180 |
| | + A Y+ G GI+P+ G RYF+++ +Y+ E V + TP LAFNRY AGI+ DY+F P | |
| hap | 1255 | VNASYQFRLGQLGIQPYFGVNRYFIERENYQSEEVRVKTPSLAFNRYNAGIRVDYTFTPT | 1314 |
|
| orf1 | 181 | QHISITPYLSLSYTDAASGKVRTRVNTAVLAQDFGKTRSAEWGVNAEIKGFTLSLHAAAA | 240 |
| | +IS+ PY ++Y D ++ V+T VN VL Q FG+ E G+ AEI F +S + + | |
| hap | 1315 | DNISVKPYFFVNYVDVSNANVQTTVNLTVLQQPFGRYWQKEVGLKAEILHFQISAFISKS | 1374 |
|
| orf1 | 241 | KGPQLEAQHSAGIKLGYRW | 259 |
| | +G QL Q + G+KLGYRW | |
| hap | 1375 | QGSQLGKQQNVGVKLGYRW | 1393 |
Homology with a Predicted ORF from
N. gonorrhoeae [1380]The blocks of ORF1 show 83.5%, 88.3%, and 97.7% identities in 467, 298, and 259 aa overlap, respectively with a predicted ORF (ORF1ng) from N. gonorrhoeae:
[0000]

[1381]The complete length ORF1ng nucleotide sequence was identified <SEQ ID 653>:
[0000]| 1 | ATGAAAACAA CCGACAAACG GACAACCGAA ACACACCGCA AAGCCCCTAA | |
|
| 51 | AACCGGCCGC ATCCGCTTCT CGCCCGCTTA CTTAGCCATA TGCCTGTCGT |
|
| 101 | TCGGCATTCT GCCCCAAGCC CGGGCGGGAC ACACTTATTT CGGCATCAAC |
|
| 151 | TACCAATACT ATCGCGACTT TGCCGAAAAT AAAGGCAAGT TTGCAGTCGG |
|
| 201 | GGCGAAAGAT ATTGAGGTTT ACAACAAAAA AGGGGAGTTG GTCGGCAAAT |
|
| 251 | CGATGACGAA AGCCCCGATG ATTGATTTTT CTGTGGTATC GCGTAACGGC |
|
| 301 | GTGGCGGCAT TGGCGGGCGA TCAATATATT GTGAGCGTGG CACATAACGG |
|
| 351 | CGGCTATAAC AATGTTGATT TTGGTGCGGA GGGAAGCAAT CCCGATCAGC |
|
| 401 | ACCGCTTTTC TTACCAAATT GTGAAAAGAA ATAATTATAA AGCAGGGACT |
|
| 451 | AACGGCCATC CTTATGGCGG CGATTATCAT ATGCCGCGTT TGCACAAATT |
|
| 501 | TGTCACAGAT GCAGAACCTG TTGAGATGAC CAGTTATATG GATGGGTGGA |
|
| 551 | AATACGCTGA TTTAAATAAA TACCCTGATC GTGTTCGAAT CGGAGCAGGC |
|
| 601 | AGACAATATT GGCGGTCTGA TGAAGACGAA CCCAATAACC GCGAAAGTTC |
|
| 651 | ATATCATATT GCAAGCGCAT ATTCTTGGCT CGTCGGTGGC AATACCTTTG |
|
| 701 | CACAAAATGG ATCAGGTGGT GGCACAGTCA ACTTAGGTAG CGAAAAAATT |
|
| 751 | AAACATAGCC CATATGGTTT TTTACCAACA GGAGGCTCAT TTGGCGACAG |
|
| 801 | TGGCTCACCA ATGTTTATCT ATGATGCCCA AAAGCAAAAG TGGTTAATTA |
|
| 851 | ATGGGGTATT GCAAACAGGC AACCCCTATA TAGGAAAAAG CAATGGCTTC |
|
| 901 | CAGCTAGTTC GTAAAGATTG GTTCTATGAT GAAATCTTTG CTGGAGATAC |
|
| 951 | CCATTCAGTA TTCTACGAAC CACATCAAAA TGGGAAATAC TTTTTTAACG |
|
| 1001 | ACAATAATAA TGGCGCAGGA AAAATCGATG CCAAACATAA ACACTATTCT |
|
| 1051 | CTACCTTATA GATTAAAAAC ACGAACCGTT CAATTGTTTA ATGTTTCTTT |
|
| 1101 | ATCCGAGACA GCAAGAGAAC CTGTTTATCA TGCTGCAGGT GGGGTCAACA |
|
| 1151 | GTTATCGACC CAGACTGAAT AATGGAGAAA ATATTTCCTT TATTGACAAA |
|
| 1201 | GGAAAAGGTG AATTGATACT TACCAGCAAC ATCAACCAAG GCGCGGGCGG |
|
| 1251 | TTTGTATTTT GAGGGTAATT TTACGGTCTC GCCTAAAAAC AACGAAACGT |
|
| 1301 | GGCAAGGCGC GGGCGTTCAT ATCAGTGATG GCAGTACCGT TACTTGGAAA |
|
| 1351 | GTAAACGGCG TGGCAAACGA CCGCCTGTCC AAAATCGGCA AAGGCACGCT |
|
| 1401 | GCTGGTTCAA GCCAAAGGGG AAAACCAAGG CTCGGTCAGC GTGGGCGACG |
|
| 1451 | GTAAAGTCAT CTTAGATCAG CAGGCGGACG ATCAAGGCAA AAAACAAGCC |
|
| 1501 | TTTAGTGAAA TCGGCTTGGT CAGCGGCAGG GGGACGGTGC AACTGAATGC |
|
| 1551 | CGATAATCAG TTCAACCCCG ACAAACTCTA TTTCGGCTTT CGCGGCGGAC |
|
| 1601 | GTTTGGATTT GAACGGGCAT TCGCTTTCGT TCCACCGCAT TCAAAATACC |
|
| 1651 | GATGAAGGGG CGATGATTGT CAACCACAAT CAAGACAAAG AATCCACCGT |
|
| 1701 | TACCATTACA GGCAATAAAG ATATTACTAC AACCGGCAAT AACAACAACT |
|
| 1751 | TGGATAGCAA AAAAGAAATT GCCTACAACG GTTGGTTTGG CGAGAAAGAT |
|
| 1801 | GCAACCAAAA CGAACGGGCG GCTCAATCTG AATTACCAAC CGGAAGAAGC |
|
| 1851 | GGATCGCACT TTACTGCTTT CCGGCGGAAC AAATTTAAAC GGCAATATCA |
|
| 1901 | CGCAAACAAA CGGCAAACTG TTTTTCAGCG GCAGACCGAC ACCGCACGCC |
|
| 1951 | TACAATCATT TAGGAAGCGG GTGGTCAAAA ATGGAAGGTA TCCCACAAGG |
|
| 2001 | AGAAATCGTG TGGGACAACG ATTGGATCGA CCGCACATTT AAAGCGGAAA |
|
| 2051 | ACTTCCATAT TCAGGGCGGA CAAGCGGTGG TTTCCCGCAA TGTTGCCAAA |
|
| 2101 | GTGGAAGGCG ATTGGCATTT AAGCAATCAC GCCCAAGCAG TTTTCGGTGT |
|
| 2151 | CGCACCGCAT CAAAGCCACA CAATCTGTAC ACGTTCGGAC TGGACGGGTC |
|
| 2201 | TGACAAGTTG TACCGAAAAA ACCATTACCG ACGATAAAGT GATTGCTTCA |
|
| 2251 | TTGAGCAAGA CCGACATCAG AGGCAATGTC AGCCTTGCCG ATCACGCTCA |
|
| 2301 | TTTAAATCTC ACAGGACTTG CCACACTCAA CGGCAATCTT AGTGCAGGCG |
|
| 2351 | GAGACACGCA CTATACGGTT ACGCGCAACG CCACCCAAAA CGGCAACCTC |
|
| 2401 | AGCCTCGTGG GCAATGCCCA AGCAACATTT AATCAAGCCA CATTAAACGG |
|
| 2451 | CAACACATCG GCTTCGGACA ATGCTTCATT TAATCTAAGC AACAACGCCG |
|
| 2501 | TACAAAACGG CAGTCTGACG CTTTCCGACA ACGCTAAGGC AAACGTAAGC |
|
| 2551 | CATTCCGCAC TCAACGGCAA TGTCTCCCTA GCCGATAAGG CAGTATTCCA |
|
| 2601 | TTTTGAAAAC AGCCGCTTTA CCGGAAAAAT CAGCGGCGGC AAGGATACGG |
|
| 2651 | CATTACACTT AAAAGACAGC GAATGGACGC TGCCGTCGGG CACGGAATTA |
|
| 2701 | GGCAATTTAA ACCTTGACAA CGCCACCATT ACACTCAATT CCGCCTATCG |
|
| 2751 | ACACGATGCG GCAGGCGCGC AAACCGGCAG TGCGGCAGAT GCGCCGCGCC |
|
| 2801 | GCCGTTCGCG CCGTTCCCTA TTATCCGTTA CGCCGCCAAC TTCGGCAGAA |
|
| 2851 | TCCCGTTTCA ACACGCTGAC GGTAAACGGC AAATTGAACG GTCAGGGAAC |
|
| 2901 | ATTCCGCTTT ATGTCGGAAC TCTTCGGCTA CCGCAGCGGC AAATTGAAGC |
|
| 2951 | TGGCGGAAAG TTCCGAAGGC ACTTACACCT TGGCTGTCAA CAATACCGGC |
|
| 3001 | AACGAACCCG TAAGTCTCGA GCAATTGACG GTAGTGGAAG GAAAAGACAA |
|
| 3051 | CACACCGCTG TCCGAAAATC TTAATTTCAC CCTGCaaaAc gaacacgtcg |
|
| 3101 | atgccggcgc atggCGTTAT CAGCTTATCC gcaaagacgG CGAGTTCCgc |
|
| 3151 | CTGCATAATC CGGTCAAAGA ACAAGAGCTT TCCGACAAAC TCGGCAAGgc |
|
| 3201 | gggagaaACA GAggccgccT TGACGGCAAA ACAGGCacaA CTTGCCGCCA |
|
| 3251 | AAcaacaggc ggaaaAAGAC AACgcgcaaa gccttgAcgc gctgattgcg |
|
| 3301 | gCcgggcgca atgccaccga AAAGGCAgaa agtgttgccg aaccgGCCCG |
|
| 3351 | GCAGGCAGGC GGGGAAAAtg ccgGCATTAT GCAGGCGGAG GAAGAGAAAA |
|
| 3401 | AACGGGTGCA GGCGGATAAA GACACCGCCT TGGCGAAACA GCGCGAAGCG |
|
| 3451 | GAAACCCGGC CGGCTACCAC CGCCTTCCCC CGCGCCCGCC GCGCCCGCCG |
|
| 3501 | GGATTTGCCG CAACCGCAGC CCCAACCGCA ACCCCAACCG CAGCGCGACC |
|
| 3551 | TGATCAGCCG TTATGCCAAT AGCGGTTTGA GTGAATTTTC CGCCACGCTC |
|
| 3601 | AACAGCGTTT TCGCCGTACA GGACGAATTG GACCGCGTGT TTGCCGAAGA |
|
| 3651 | CCGCCGCAAC GCCGTTTGGA CAAGCGGCAT CCGGGACACC AAACACTACC |
|
| 3701 | GTTCGCAAGA TTTCCGCGCC TACCGCCAAC AAACCGACCT GCGCCAAATC |
|
| 3751 | GGTATGCAGA AAAACCTCGG CAGCGGGCGC GTCGGCATCC TGTTTTCGCA |
|
| 3801 | CAACCGGACC GGAAACACCT TCGACGACGG CATCGGCAAC TCGGCACGGC |
|
| 3851 | TTGCCCACGG TGCCGTTTTC GGGCAATACG GCATCGGCAG GTTCGACATC |
|
| 3901 | GGCATCAGCG CGGGCGCGGG TTTTAGTAGC GGCAGCCTTT CAGACGGCAT |
|
| 3951 | CAGAGGCAAA ATCCGCCGCC GCGTGCTGCA TTACGGCATT CAGGCAAGAT |
|
| 4001 | ACCGCGCAGG TTTCGGCGGA TTCGGCATCG AACCGCACAT CGGCGCAACG |
|
| 4051 | CGCTATTTCG TCCAAAAAGC GGATTACCGA TACGAAAACG TCAATATCGC |
|
| 4101 | CACCCCGGGC CTTGCATTCA ACCGCTACCG CGCGGGCATT AAGGCAGATT |
|
| 4151 | ATTCATTCAA ACCGGCGCAA CACATTTCCA TCACGCCTTA TTTGAGCCTG |
|
| 4201 | TCCTATACCG ATGCCGCTTC CGGCAAAGTC CGAACGCGCG TCAATACCGC |
|
| 4251 | CGTATTGGCG CAGGATTTCG GCAAAACCCG CAGTGCGGAA TGGGGCGTAA |
|
| 4301 | ACGCCGAAAT CAAAGGTTTC ACGCTGTCCC TCCACGCTGC CGCCGCCAAG |
|
| 4351 | GGGCCGCAAT TGGAAGCGCA GCACAGCGCG GGCATCAAAT TAGGCTACCG |
|
| 4401 | CTGGTAA |
[1382]This is predicted to encode a protein having amino acid sequence <SEQ ID 654>:
[0000]| 1 | MKTTDKRTTE THRKAPKTGR IRFSPAYLAI CLSFGILPQA RAGHTYFGIN | |
|
| 51 | YQYYRDFAEN KGKFAVGAKD IEVYNKKGEL VGKSMTKAPM IDFSVVSRNG |
|
| 101 | VAALAGDQYI VSVAHNGGYN NVDFGAEGSN PDQHRFSYQI VKRNNYKAGT |
|
| 151 | NGHPYGGDYH MPRLHKFVTD AEPVEMTSYM DGWKYADLNK YPDRVRIGAG |
|
| 201 | RQYWRSDEDE PNNRESSYHI ASAYSWLVGG NTFAQNGSGG GTVNLGSEKI |
|
| 251 | KHSPYGFLPT GGSFGDSGSP MFIYDAQKQK WLINGVLQTG NPYIGKSNGF |
|
| 301 | QLVRKDWFYD EIFAGDTHSV FYEPHQNGKY FFNDNNNGAG KIDAKHKHYS |
|
| 351 | LPYRLKTRTV QLFNVSLSET AREPVYHAAG GVNSYRPRLN NGENISFIDK |
|
| 401 | GKGELILTSN INQGAGGLYF EGNFTVSPKN NETWQGAGVH ISDGSTVTWK |
|
| 451 | VNGVANDRLS KIGKGTLLVQ AKGENQGSVS VGDGKVILDQ QADDQGKKQA |
|
| 501 | FSEIGLVSGR GTVQLNADNQ FNPDKLYFGF RGGRLDLNGH SLSFHRIQNT |
|
| 551 | DEGAMIVNHN QDKESTVTIT GNKDITTTGN NNNLDSKKEI AYNGWFGEKD |
|
| 601 | ATKTNGGLNL NYPPEEADRT LLLSGGTNLN GNITQTNGKL FFSGRPTPHA |
|
| 651 | YNHLGSGWSK MEGIPQGEIV WDNDWIDRTF KAENFHIQGG QAVVSRNVAK |
|
| 701 | VEGDWHLSNH AQAVFGVAPH QSHTICTRSD WTGLTSCTEK TITDDKVIAS |
|
| 751 | LSKTDVRGNV SLADHAHLNL TGLATFNGNL VQAETRTIRL RANATQNGNL |
|
| 801 | SLVGNAQATF NQATLNGNTS ASDNASFNLS NNAVQNGSLT LSDNAKANVS |
|
| 851 | HSALNGNVSL ADKAVFHFEN SRFTGKISGG KDTALHLKDS EWTLPSGTEL |
|
| 901 | GNLNLDNATI TLNSAYRHDA AGAQTGSAAD APRRRSRRSL LSVTPPTSAE |
|
| 951 | SRFNTLTVNG KLNGQGTFRF MSELFGYRSG KLKLAESSEG TYTLAVNNTG |
|
| 1001 | NEPVSLEQLT VVEGKDNTPL SENLNFTLQN EHVDAGAWRY QLIRKDGEFR |
|
| 1051 | LHNPVKEQEL SDKLGKAGET EAALTAKQAQ LAAKQQAEKD NAQSLDALIA |
|
| 1101 | AGRNATEKAE SVAEPARQAG GENAGIMQAE EEKKRVQADK DTALAKQREA |
|
| 1151 | ETRPATTAFP RARRARRDLP QPQPQPQPQP QRDLISRYAN SGLSEFSATL |
|
| 1201 | NSVFAVQDEL DRVFAEDRRN AVWTSGIRDT KHYRSQDFRA YRQQTDLRQI |
|
| 1251 | GMQKNLGSGR VGILFSHNRT GNTFDDGIGN SARLAHGAVF GQYGIGRFDI |
|
| 1301 | GISAGAGFSS GSLSDGIRGK IRRRVLHYGI QARYRAGFGG FGIEPHIGAT |
|
| 1351 | RYFVQKADYR YENVNIATPG LAFNRYRAGI KADYSFKPAQ HISITPYLSL |
|
| 1401 | SYTDAASGKV RTRVNTAVLA QDFGKTRSAE WGVNAEIKGF TLSLHAAAAK |
|
| 1451 | GPQLEAQHSA GIKLGYRW* |
[1383]Underlined and double-underlined sequences represent the active site of a serine protease (trypsin family) and an ATP/GTP-binding site motif A (P-loop).
[1384]ORF1-1 and ORF1 ng show 93.7% identity in 1471 aa overlap:
[0000]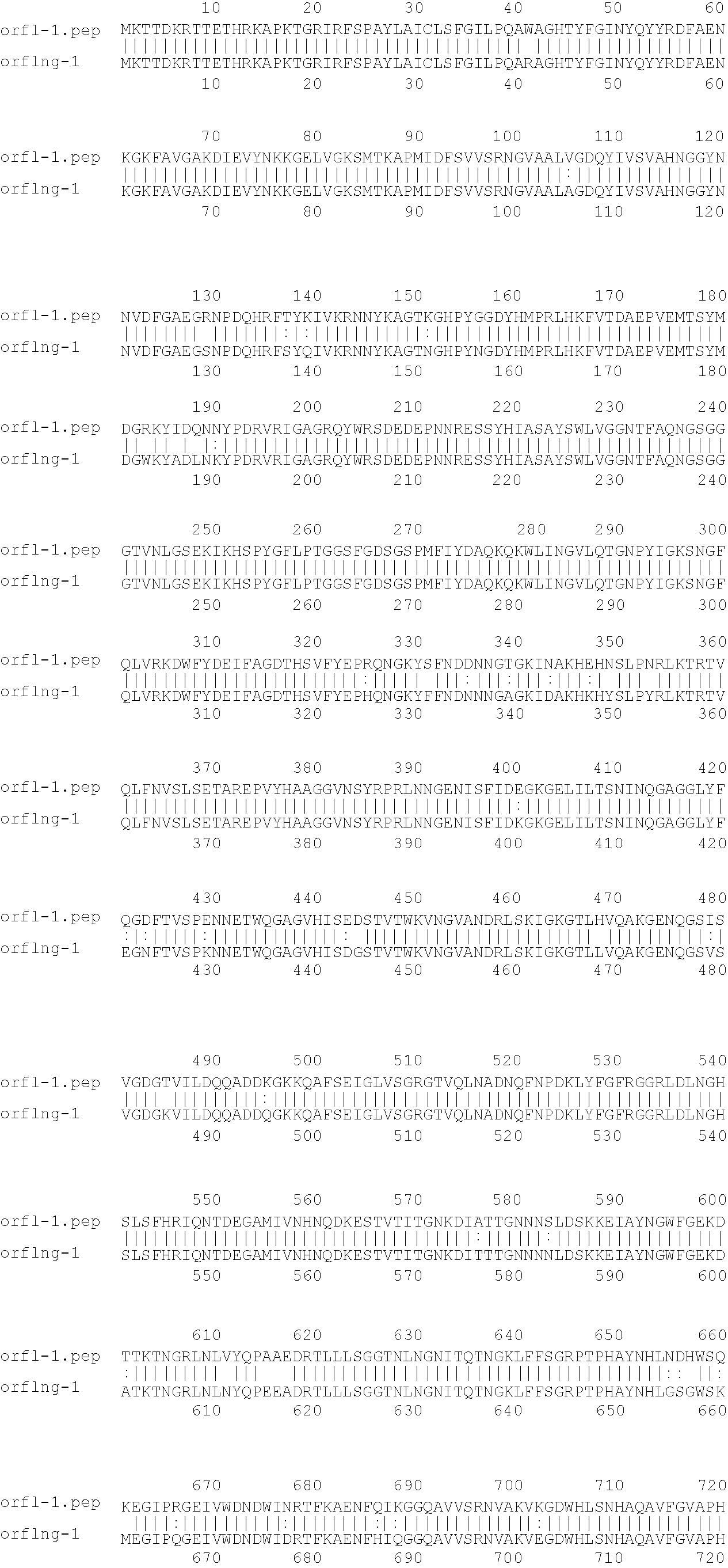
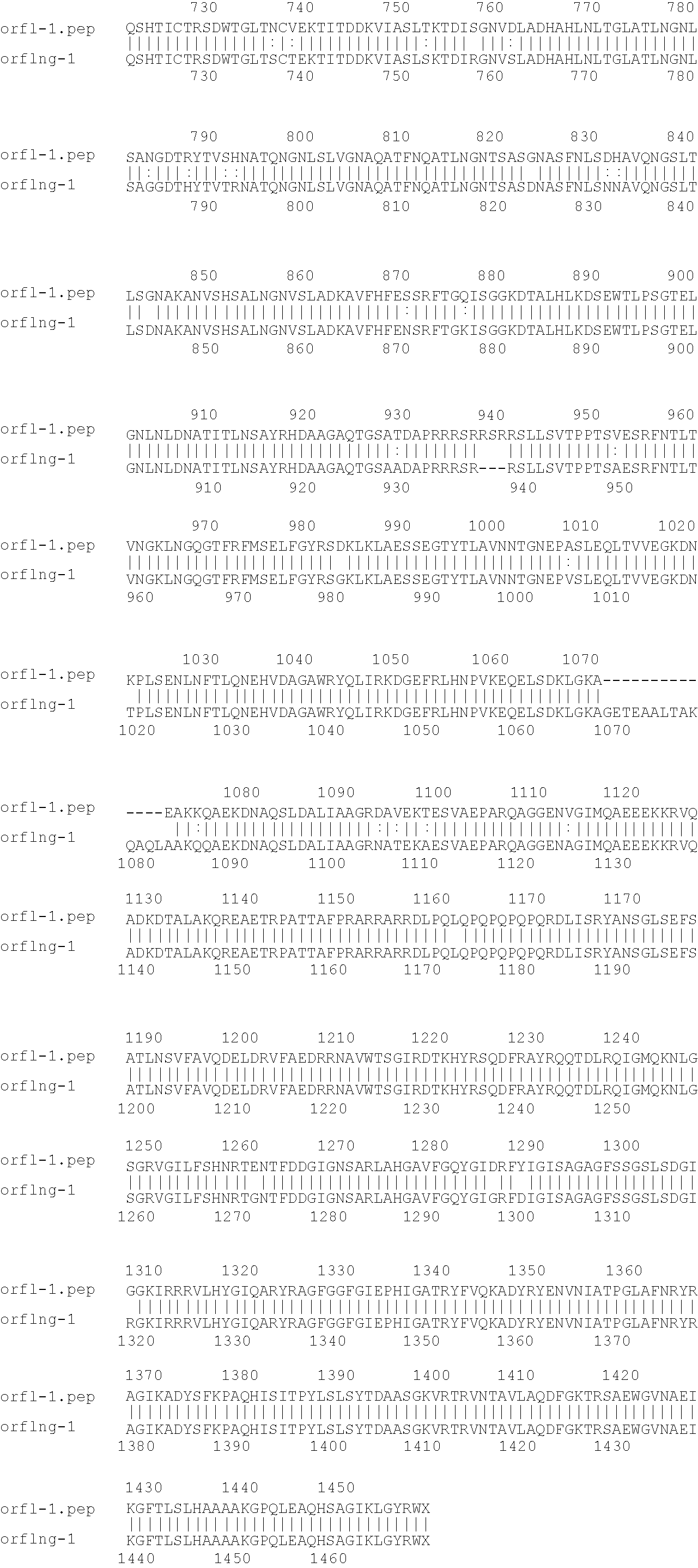
[1385]In addition, ORF1ng shows 55.7% identity with hap protein (P45387) over a 1455aa overlap:
[1386]Based on this analysis, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 78
[1387]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 655>:
[0000]| 1 | ..AAGGTGTGGC AATTTGTCGA AGA.CCGCTG CGTGCCGTCG TGCCTGCCGA | |
|
| 51 | CAGTTTTGAA CCGACCGCGC AAAAATTGAA CCTGTTTAAG GCGGGTGCGG |
|
| 101 | CAACCATTTT GTTTTATGAA GATCAAAATG TCGTCAAAGG TTTGCAGGAG |
|
| 151 | CAGTTCCCTG CTTATGCCGC TAACTTCCCC GTTTGGGCGg ATCAGGCAAA |
|
| 201 | CGCGATGGTG CAGTATGCCG TTTGGACGAC ACTTGCCGCG GTCGGCGTAG |
|
| 251 | GTGCAAACCT GCAACATTAC AATCCCTTGC CCGATGCGGC GATTGCCAAA |
|
| 301 | GCGTGGAATA TCCCCGAAAA CTGGTTGTTG CGCGCACAAA TGGTTATCGG |
|
| 351 | CGGTATTGAA GGGGCGGCAG GTGAAAAGAC CTTTGAACCC GTTGCAGAAC |
|
| 401 | GTTTGAAAGT GTTCGGCGCA TAA |
[1388]This corresponds to the amino acid sequence <SEQ ID 656; ORF6>:
[0000]| 1 | ..KVWQFVEXPL RAVVPADSFE PTAQKLNLFK AGAATILFYE DQNVVKGLQE | |
|
| 51 | QFPAYAANFP VWADQANAMV QYAVWTTLAA VGVGANLQHY NPLPDAAIAK |
|
| 101 | AWNIPENWLL RAQMVIGGIE GAAGEKTFEP VAERLKVFGA * |
[1389]Further sequence analysis revealed a further partial DNA sequence <SEQ ID 657>:
[0000]| 1 | ..CTGCGTGCCG TCGTGCCTGC CGACAGTTTT GAACCGACCG CGCAAAAATT | |
|
| 51 | GAACCTGTTT AAGGCGGGTG CGGCAACCAT TTTGTTTTAT GAAGATCAAA |
|
| 101 | ATGTCGTCAA AGGTTTGCAG GAGCAGTTCC CTGCTTATGC CGCTAACTTC |
|
| 151 | CCCGTTTGGG CGGATCAGGC AAACGCGATG GTGCAGTATG CCGTTTGGAC |
|
| 201 | GACACTTGCC GCGGTCGGCG TAGGTGCAAA CCTGCAACAT TACAATCCCT |
|
| 251 | TGCCCGATGC GGCGATTGCC AAAGCGTGGA ATATCCCCGA AAACTGGTTG |
|
| 301 | TTGCGCGCAC AAATGGTTAT CGGCGGTATT GAAGGGGCGG CAGGTGAAAA |
|
| 351 | GACCTTTGAA CCCGTTGCAG AACGTTTGAA AGTGTTCGGC GCATAA |
[1390]This corresponds to the amino acid sequence <SEQ ID 658; ORF6-1>:
[0000]| 1 | ..LRAVVPADSF EPTAQKLNLF KAGAATILFY EDQNVVKGLQ EQFPAYAANF | |
|
| 51 | PVWADQANAM VQYAVWTTLA AVGVGANLQH YNPLPDAAIA KAWNIPENWL |
|
| 101 | LRAQMVIGGI EGAAGEKTFE PVAERLKVFG A* |
[1391]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1392]ORF6 shows 98.6% identity over a 140aa overlap with an ORF (ORF6a) from strain A of N. meningitidis.
[0000]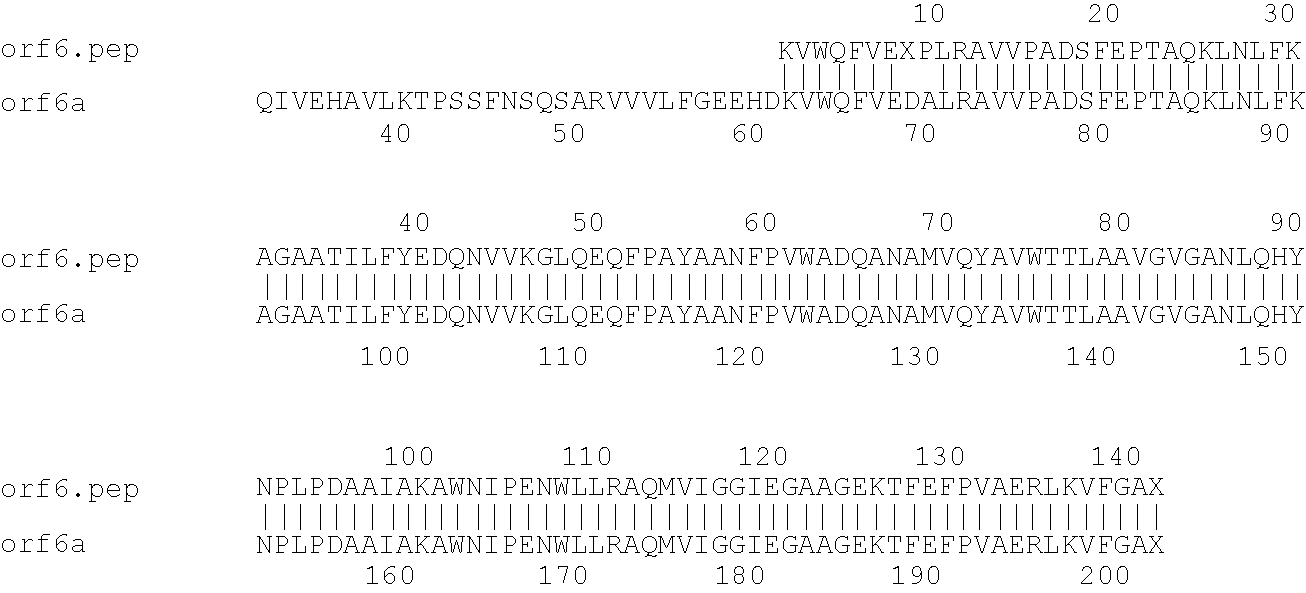
[1393]The complete length ORF6a nucleotide sequence <SEQ ID 659> is:
[0000]| 1 | ATGACCCGTC AATCTCTGCA ACAGGCTGCC GAAAGCCGCC GTTCCATTTA | |
|
| 51 | TTCGTTAAAT AAAAATCTGC CCGTCGGCAA AGATGAAATC GTCCAAATCG |
|
| 101 | TCGAACACGC CGTTTTGCAC ACACCTTCTT CGTTCAATTC CCAATCTGCC |
|
| 151 | CGTGTGGTCG TGCTGTTTGG CGAAGAGCAT GATAAGGTGT GGCAATTTGT |
|
| 201 | CGAAGACGCG CTGCGTGCCG TCGTGCCTGC CGACAGTTTT GAACCGACCG |
|
| 251 | CGCAAAAATT GAACCTGTTT AAGGCGGGTG CGGCAACTAT TTTGTTTTAT |
|
| 301 | GAAGATCAAA ATGTCGTCAA AGGTTTGCAG GAGCAGTTCC CTGCTTATGC |
|
| 351 | CGCCAACTTT CCCGTTTGGG CGGACCAGGC GAACGCGATG GTGCAGTATG |
|
| 401 | CCGTTTGGAC GACACTTGCC GCGGTCGGCG TAGGTGCAAA CCTGCAACAT |
|
| 451 | TACAATCCCT TGCCCGATGC GGCGATTGCC AAAGCGTGGA ATATCCCCGA |
|
| 501 | AAACTGGTTG TTGCGCGCAC AAATGGTTAT CGGCGGTATT GAAGGGGCGG |
|
| 551 | CAGGTGAAAA GACCTTTGAA CCAGTTGCAG AACGTTTGAA AGTGTTCGGC |
|
| 601 | GCATAA |
[1394]This is predicted to encode a protein having amino acid sequence <SEQ ID 660>:
[0000]| 1 | MTRQSLQQAA ESRRSIYSLN KNLPVGKDEI VQIVEHAVLH TPSSFNSQSA | |
|
| 51 | RVVVLFGEEH DKVWQFVEDA LRAVVPADSF EPTAQKLNLF KAGAATILFY |
|
| 101 | EDQNVVKGLQ EQFPAYAANF PVWADQANAM VQYAVWTTLA AVGVGANLQH |
|
| 151 | YNPLPDAAIA KAWNIPENWL LRAQMVIGGI EGAAGEKTFE PVAERLKVFG |
|
| 201 | A* |
[1395]ORF6a and ORF6-1 show 100.0% identity in 131 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1396]ORF6 shows 95.7% identity over a 140aa overlap with a predicted ORF (ORF6ng) from N. gonorrhoeae:
[0000]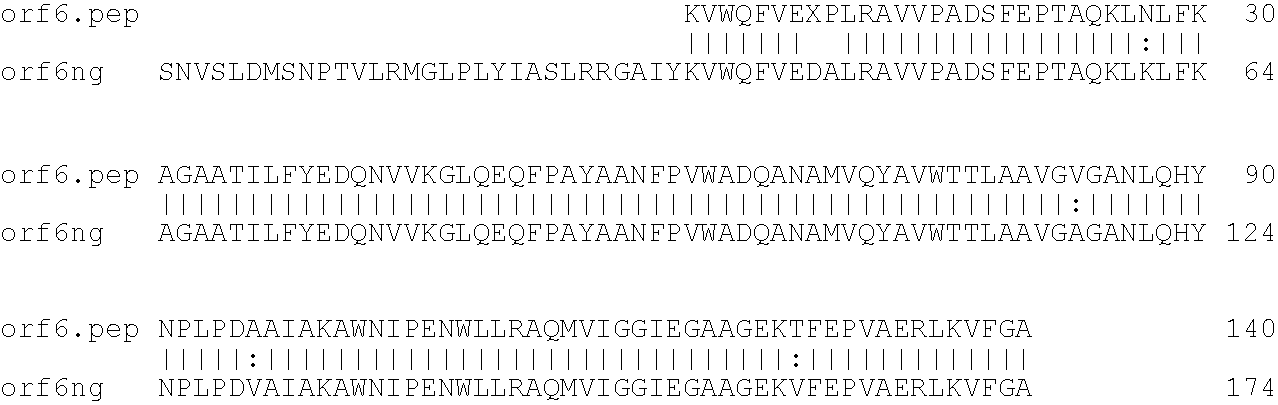
[1397]The complete length ORF6ng nucleotide sequence <SEQ ID 661> was identified as:
[0000]| 1 | ATGGCCGTTG CGTCAAATGT CAGCTTGGAT ATGTCCAATC CTACGGTGTT | |
|
| 51 | ACGCATGGGA TTACCCTTAT ATATTGCGTC CCTAAGAAGG GGCGCAATAT |
|
| 101 | ATAAGGTGTG GCAATTTGTC GAAGACGCGC TGCGTGCCGT CGTGCCTGCC |
|
| 151 | GACAGTTTTG AACCGACCGC GCAAAAATTG AAGCTGTTTA AGGCGGGCGC |
|
| 201 | GGCAACCATT TTGTTTTATG AAGATCAAAA TGTCGTCAAA GGTTTGCAGG |
|
| 251 | AGCAGTTCCC TGCTTATGCC GCCAACTTTC CCGTTTGGGC GGACCAGGCG |
|
| 301 | AACGCTATGG TACAGTATGC CGTCTGGACG ACACTTGCCG CGGTCGGTGC |
|
| 351 | AGGTGCAAAT CTGCAACATT ACAACCCCTT GCCCGATGTG GCGATTGCTA |
|
| 401 | AAGCGTGGAA TATTCCCGAA AACTGGCTGT TGCGCGCGCA AATGGTTATC |
|
| 451 | GGTGGTATTG AAGGGGcggc aggtgaaaaa gtctttgaac CCGTTGCgga |
|
| 501 | acgtttgAAA GTGTTCGGCG CATAA |
[1398]This encodes a protein having amino acid sequence <SEQ ID 662>:
[0000]| 1 | MAVASNVSLD MSNPTVLRMG LPLYIASLRR GAIYKVWQFV EDALRAVVPA | |
|
| 51 | DSFEPTAQKL KLFKAGAATI LFYEDQNVVK GLQEQFPAYA ANFPVWADQA |
|
| 101 | NAMVQYAVWT TLAAVGAGAN LQHYNPLPDV AIAKAWNIPE NWLLRAQMVI |
|
| 151 | GGIEGAAGEK VFEPVAERLK VFGA* |
[1399]ORF6ng and ORF6-1 show 96.9% identity in 131 aa overlap:
[0000]
[1400]It is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 79
[1401]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 663>
[0000]| 1 | ..GGCTACAACT ACCTGTTCGC GCGCGGCAGC CGCATCGCCA ACTACCAAAT | |
|
| 51 | CAACGGCATC CCCGTTGCCG ACGCGCTGGC CGATACGGGt CAATGCCAAC |
|
| 101 | ACCGCCGCCT ATGAGCGCGT AGAAGTCGTG CGCGGCGTGG CGGGGCTGCT |
|
| 151 | GGACGGCACG GGCGAGCCTT CCGCCACCGT CAATCTGGTG CGCAAACGCC |
|
| 201 | TGACCCGCAA GCCATTGTTT GAAGTCCGCG CCGAAGCgGG CAACCGcAAA |
|
| 251 | CATTTCGGGC TGGACGCGGA CGTATCGGGC AGCCTGAACA CCGAAG.crC |
|
| 301 | rCTGCGCgGC CGCCTGGTTT CCAcCTTCGG ACGCGGCGAC TCGTGGCGGC |
|
| 351 | GGCGCGAACG CAGCCGskAT GCCGAACTCT ACGGCATTTT GGAATACGAC |
|
| 401 | ATCGCACCGC AAACCCGCGT CCACGCArGC ATGGACTACC AGCAGGCGAA |
|
| 451 | AGAAACCGCC GACGCGCCGC TCAGcTACGC CGTGTACGAC AGCCAAGGTT |
|
| 501 | ATGCCACCGC CTTCGGCCCG AAAGACAACC CCGCCACAAA TTGGGCGAAC |
|
| 551 | AGCCACCACC GTGCGCTCAA CCTGTTCGCC GGCATCGAAC ACCGCTTCAA |
|
| 601 | CCAAGACTGG AAACTCAAAG CCGAATACGA CTAC.. |
[1402]This corresponds to the amino acid sequence <SEQ ID 664; ORF23>:
[0000]| 1 | ..GYNYLFARGS RIANYQINGI PVADALADTG NANTAAYERV EVVRGVAGLL | |
|
| 51 | DGTGEPSATV NLVRKRLTRK PLFEVRAEAG NRKHFGLDAD VSGSLNTEXX |
|
| 101 | LRGRLVSTFG RGDSWRRRER SRXAELYGIL EYDIAPQTRV HAXMDYQQAK |
|
| 151 | ETADAPLSYA VYDSQGYATA FGPKDNPATN WANSHHRALN LFAGIEHRFN |
|
| 201 | QDWKLKAEYD Y.. |
[1403]Further work revealed the complete nucleotide sequence <SEQ ID 665>:
[0000]| 1 | ATGACACGCT TCAAATATTC CCTGCTGTTT GCCGCCCTGT TGCCCGTGTA | |
|
| 51 | CGCGCAGGCC GATGTTTCTG TTTCAGACGA CCCCAAACCG CAGGAAAGCA |
|
| 101 | CTGAATTGCC GACCATCACC GTTACCGCCG ACCGCACCGC GAGTTCCAAC |
|
| 151 | GACGGCTACA CTGTTTCCGG CACGCACACC CCGCTCGGGC TGCCCATGAC |
|
| 201 | CCTGCGCGAA ATCCCGCAGA GCGTCAGCGT CATCACATCG CAACAAATGC |
|
| 251 | GCGACCAAAA CATCAAAACG CTCGACCGCG CCCTGTTGCA GGCGACCGGC |
|
| 301 | ACCAGCCGCC AGATTTACGG CTCCGACCGC GCGGGCTACA ACTACCTGTT |
|
| 351 | CGCGCGCGGC AGCCGCATCG CCAACTACCA AATCAACGGC ATCCCCGTTG |
|
| 401 | CCGACGCGCT GGCCGATACG GGCAATGCCA ACACCGCCGC CTATGAGCGC |
|
| 451 | GTAGAAGTCG TGCGCGGCGT GGCGGGGCTG CTGGACGGCA CGGGCGAGCC |
|
| 501 | TTCCGCCACC GTCAATCTGG TGCGCAAACG CCTGACCCGC AAGCCATTGT |
|
| 551 | TTGAAGTCCG CGCCGAAGCG GGCAACCGCA AACATTTCGG GCTGGACGCG |
|
| 601 | GACGTATCGG GCAGCCTGAA CACCGAAGGC ACGCTGCGCG GCCGCCTGGT |
|
| 651 | TTCCACCTTC GGACGCGGCG ACTCGTGGCG GCGGCGCGAA CGCAGCCGCG |
|
| 701 | ATGCCGAACT CTACGGCATT TTGGAATACG ACATCGCACC GCAAACCCGC |
|
| 751 | GTCCACGCAG GCATGGACTA CCAGCAGGCG AAAGAAACCG CCGACGCGCC |
|
| 801 | GCTCAGCTAC GCCGTGTACG ACAGCCAAGG TTATGCCACC GCCTTCGGCC |
|
| 851 | CGAAAGACAA CCCCGCCACA AATTGGGCGA ACAGCCGCCA CCGTGCGCTC |
|
| 901 | AACCTGTTCG CCGGCATCGA ACACCGCTTC AACCAAGACT GGAAACTCAA |
|
| 951 | AGCCGAATAC GACTACACCC GCAGCCGCTT CCGCCAGCCC TACGGCGTAG |
|
| 1001 | CAGGCGTGCT TTCCATCGAC CACAACACCG CCGCCACCGA CCTGATTCCC |
|
| 1051 | GGTTATTGGC ACGCCGACCC GCGCACCCAC AGCGCCAGCG TGTCATTGAT |
|
| 1101 | CGGCAAATAC CGCCTGTTCG GCCGCGAACA CGATTTAATC GCGGGTATCA |
|
| 1151 | ACGGTTACAA ATACGCCAGC AACAAATACG GCGAACGCAG CATCATCCCC |
|
| 1201 | AACGCCATTC CCAACGCCTA CGAATTTTCC CGCACGGGTG CCTACCCGCA |
|
| 1251 | GCCTGCATCG TTTGCCCAAA CCATCCCGCA ATACGGCACC AGGCGGCAAA |
|
| 1301 | TCGGCGGCTA TCTCGCCACC CGTTTCCGCG CCGCCGACAA CCTTTCGCTG |
|
| 1351 | ATTTTGGGCG GACGATACAC CCGTTACCGC ACCGGCAGCT ACGACAGCCG |
|
| 1401 | CACACAAGGC ATGACCTATG TGTCCGCCAA CCGTTTCACC CCCTACACAG |
|
| 1451 | GCATCGTGTT CGACCTGACC GGCAACCTGT CTCTTTACGG CTCGTACAGC |
|
| 1501 | AGCCTGTTCG TCCCGCAATC GCAAAAAGAC GAACACGGCA GCTACCTGAA |
|
| 1551 | ACCCGTAACC GGCAACAATC TGGAAGCCGG CATCAAAGGC GAATGGCTTG |
|
| 1601 | AAGGCCGTCT GAACGCATCC GCCGCCGTGT ACCGCGCCCG TAAAAACAAC |
|
| 1651 | CTCGCCACCG CAGCAGGACG CGACCCGAGC GGCAACACCT ACTACCGCGC |
|
| 1701 | CGCCAACCAA GCCAAAACCC ACGGCTGGGA AATCGAAGTC GGCGGCCGCA |
|
| 1751 | TCACGCCCGA ATGGCAGATA CAGGCAGGTT ACAGCCAAAG CAAAACCCGC |
|
| 1801 | GACCAAGACG GCAGCCGCCT GAACCCCGAC AGCGTACCCG AACGCAGCTT |
|
| 1851 | CAAACTCTTC ACTGCCTACC ACTTTGCCCC CGAAGCCCCC AGCGGCTGGA |
|
| 1901 | CCATCGGCGC AGGCGTGCGC TGGCAGAGCG AAACCCACAC CGACCCTGCC |
|
| 1951 | ACGCTCCGCA TCCCCAACCC CGCCGCCAAA GCCCGCGCCG CCGACAACAG |
|
| 2001 | CCGCCAAAAA GCCTACGCCG TCGCCGACAT CATGGCGCGT TACCGCTTCA |
|
| 2051 | ATCCGCGCGC CGAACTGTCG CTGAACGTGG ACAATCTGTT CAACAAACAC |
|
| 2101 | TACCGCACCC AGCCCGACCG CCACAGCTAC GGCGCACTGC GGACAGTGAA |
|
| 2151 | CGCGGCGTTT ACCTATCGGT TTAAATAA |
[1404]This corresponds to the amino acid sequence <SEQ ID 666; ORF23-1>:
[0000]| 1 | MTRFKYSLLF AALLPVYAQA DVSVSDDPKP QESTELPTIT VTADRTASSN | |
|
| 51 | DGYTVSGTHT PLGLPMTLRE IPQSVSVITS QQMRDQNIKT LDRALLQATG |
|
| 101 | TSRQIYGSDR AGYNYLFARG SRIANYQING IPVADALADT GNANTAAYER |
|
| 151 | VEVVRGVAGL LDGTGEPSAT VNLVRKRLTR KPLFEVRAEA GNRKHFGLDA |
|
| 201 | DVSGSLNTEG TLRGRLVSTF GRGDSWRRRE RSRDAELYGI LEYDIAPQTR |
|
| 251 | VHAGMDYQQA KETADAPLSY AVYDSQGYAT AFGPKDNPAT NWANSRHRAL |
|
| 301 | NLFAGIEHRF NQDWKLKAEY DYTRSRFRQP YGVAGVLSID HNTAATDLIP |
|
| 351 | GYWHADPRTH SASVSLIGKY RLFGREHDLI AGINGYKYAS NKYGERSIIP |
|
| 401 | NAIPNAYEFS RTGAYPQPAS FAQTIPQYGT RRQIGGYLAT RFRAADNLSL |
|
| 451 | ILGGRYTRYR TGSYDSRTQG MTYVSANRFT PYTGIVFDLT GNLSLYGSYS |
|
| 501 | SLFVPQSQKD EHGSYLKPVT GNNLEAGIKG EWLEGRLNAS AAVYRARKNN |
|
| 551 | LATAAGRDPS GNTYYRAANQ AKTHGWEIEV GGRITPEWQI QAGYSQSKTR |
|
| 601 | DQDGSRLNPD SVPERSFKLF TAYHFAPEAP SGWTIGAGVR WQSETHTDPA |
|
| 651 | TLRIPNPAAK ARAADNSRQK AYAVADIMAR YRFNPRAELS LNVDNLFNKH |
|
| 701 | YRTQPDRHSY GALRTVNAAF TYRFK* |
[1405]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the Ferric-Pseudobactin Receptor PupB of Pseudomonas putida (Accession Number P38047)
[1406]ORF23 and PupB protein show 32% aa identity in 205aa overlap:
[0000]| Orf23 | 6 | FARGSRIANYQINGIPVADALADTGNANTAAYERVEVVRGVAGLLDGTGEPSATVNLVRK | 65 | |
| | ++RG I NY+++G+P + L D + + A ++RVE+VRG GL+ G G PSAT+NL+RK |
| PupB | 215 | WSRGFAIQNYEVDGVPTSTRL-DNYSQSMAMFDRVEIVRGATGLISGMGNPSATINLIRK | 273 |
|
| Orf23 | 66 | RLTRKPLFEVRAEAGNRKHFGLDADVSGSLNTEXXLRGRLVSTFXXXXXXXXXXXXXXAE | 125 |
| | R T + + EAGN +G DVSG L +RGR V+ + |
| PupB | 274 | RPTAEAQASITGEAGNWDRYGTGFDVSGPLTETGNIRGRFVADYKTEKAWIDRYNQQSQL | 333 |
|
| Orf23 | 126 | LYGILEYDIAPQTRVHAXMDYQQAKETADAPLSYAVYD--SQGYATAFGPKDNPATNWAN | 183 |
| | +YGI E+D++ T + Y + D+PL + S G T N A +W+ |
| PupB | 334 | MYGITEFDLSEDTLLTVGFSY--LRSDIDSPLRSGLPTRFSTGERTNLKRSLNAAPDWSY | 391 |
|
| Orf23 | 184 | SHHRALNLFAGIEHRFNQDWKLKAE | 208 |
| | + H + F IE + W K E |
| PupB | 392 | NDHEQTSFFTSIEQQLGNGWSGKIE | 416 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1407]ORF23 shows 95.7% identity over a 211aa overlap with an ORF (ORF23a) from strain A of N. meningitidis:
[0000]
[1408]The complete length ORF23a nucleotide sequence <SEQ ID 667> is:
[0000]| 1 | ATGACACGCT TCAAATATTC CCTGCTGTTT GCCGCCCTGT TGCCCGTGTA | |
|
| 51 | CGCGCAGGCC GATGTTTCTG TTTCAGACGA CCCAAAACCG CAGGAAAGCA |
|
| 101 | CTGAATTGCC GACCATCACC GTTACCGCCG ACCGCACCGC GAGTTCCAAC |
|
| 151 | GACGGCTACA CTGTTTCCGG CACGCACACC CCGCTCGGGC TGCCCATGAC |
|
| 201 | CCTGCGCGAA ATCCCGCAGA GCGTCAGCGT CATCACATCG CAACAAATGC |
|
| 251 | GCGACCAAAA CATCAAAGCG CTCGACCGCG CCCTGTTGCA GGCGACCGGC |
|
| 301 | ACCAGCCGCC AGATTTACGG CTCCGACCGC GCGGGCTACA ACTACCTGTT |
|
| 351 | CGCGCGCGGC AGCCGCATCG CCAACTACCA AATCAACGGC ATCCCCGTTG |
|
| 401 | CCGACGCGCT GGCCGATACG GGCAATGCCA ACACCGCCGC CTATGAGCGC |
|
| 451 | GTAGAAGTCG TGCGCGGCGT GGCGGGGCTG CTGGACGGCA CGGGCGAGCC |
|
| 501 | TTCCGCCACC GTCAATCTGG TGCGCAAACG CCCGACCCGC AAGCCATTGT |
|
| 551 | TTGAAGTCCG CGCCGAAGCG GGCAACCGCA AACATTTCGG GCTGGGCGCG |
|
| 601 | GACGTATCGG GCAGCCTGAA TGCCGAAGGC ACGCTGCGCG GCCGCCTGGT |
|
| 651 | TTCCACCTTC GGACGCGGCG ACTCGTGGCG GCAGCGCGAA CGCAGCCGCG |
|
| 701 | ATGCCGAACT CTACGGCATT TTGGAATACG ACATCGCACC GCAAACCCGC |
|
| 751 | GTCCACGCAG GCATGGACTA CCAGCAGGCG AAAGAAACCG CCGACGCGCC |
|
| 801 | GCTCAGCTAC GCCGTGTACG ACAGCCAAGG TTATGCCACC GCCTTCGGCC |
|
| 851 | CGAAAGACAA CCCCGCCACA AATTGGGCGA ACAGCCGCCA CCGTGCGCTC |
|
| 901 | AACCTGTTCG CCGGCATCGA ACACCGCTTC AACCAAGACT GGAAACTCAA |
|
| 951 | AGCCGAATAC GACTACACCC GCAGCCGCTT CCGCCAGCCC TACGGCGTAG |
|
| 1001 | CAGGCGTGCT TTCCATCGAC CACAACACCG CCGCCACCGA CCTGATTCCC |
|
| 1051 | GGTTATTGGC ACGCCGACCC GCGCACCCAC AGCGCCAGCG TGTCATTAAT |
|
| 1101 | CGGCAAATAC CGCCTGTTCG GCCGCGAACA CGATTTAATC GCGGGTATCA |
|
| 1151 | ACGGTTACAA ATACGCCAGC AACAAATACG GCGAACGCAG CATCATCCCC |
|
| 1201 | AACGCCATTC CCAACGCCTA CGAATTTTCC CGCACGGGTG CCTACCCGCA |
|
| 1251 | GCCTGCATCG TTTGCCCAAA CCATCCCGCA ATACGGCACC AGGCGGCAAA |
|
| 1301 | TCGGCGGCTA TCTCGCCACC CGTTTCCGCG CCGCCGACAA CCTTTCGCTG |
|
| 1351 | ATACTCGGCG GCAGATACAG CCGTTACCGC ACCGGCAGCT ACGACAGCCG |
|
| 1401 | CACACAAGGC ATGACCTATG TGTCCGCCAA CCGTTTCACC CCCTACACAG |
|
| 1451 | GCATCGTGTT CGACCTGACC GGCAACCTGT CGCTTTACGG CTCGTACAGC |
|
| 1501 | AGCCTGTTCG TCCCGCAATC GCAAAAAGAC GAACACGGCA GCTACCTGAA |
|
| 1551 | ACCCGTAACC GGCAACAATC TGGAAGCCGG CATCAAAGGC GAATGGCTTG |
|
| 1601 | AAGGCCGTCT GAACGCATCC GCCGCCGTGT ACCGCGCCCG TAAAAACAAC |
|
| 1651 | CTCGCCACCG CAGCAGGACG CGACCCGAGC GGCAACACCT ACTACCGCGC |
|
| 1701 | CGCCAACCAA GCCAAAACCC ACGGCTGGGA AATCGAAGTC GGCGGCCGCA |
|
| 1751 | TCACGCCCGA ATGGCAGATA CAGGCAGGTT ACAGCCAAGG CAAAACCCGC |
|
| 1801 | GACCAAGACG GCAGCCGCCT GAACCCCGAC AGCGTACCCG AACGCAGCTT |
|
| 1851 | CAAACTCTTC ACTGCCTACC ACTTTGCCCC CGAAGCCCCC AGCGGCTGGA |
|
| 1901 | CCATCGGCGC AGGCGTGCGC TGGCAGAGCG AAACCCACAC CGACCCTGCC |
|
| 1951 | ACGCTCCGCA TCCCCAACCC CGCCGCCAAA GCCCGCGCCG CCGACAACAG |
|
| 2001 | CCGCCAAAAA GCCTACGCCG TCGCCGACAT CATGGCGCGT TACCGCTTCA |
|
| 2051 | ATCCGCGCGC CGAACTGTCG CTGAACGTGG ACAATCTGTT CAACAAACAC |
|
| 2101 | TACCGCACCC AGCCCGACCG CCACAGCTAC GGCGCACTGC GGACAGTGAA |
|
| 2151 | CGCGGCGTTT ACCTATCGGT TTAAATAA |
[1409]This encodes a protein having amino acid sequence <SEQ ID 668>:
[0000]| 1 | MTRFKYSLLF AALLPVYAQA DVSVSDDPKP QESTELPTIT VTADRTASSN | |
|
| 51 | DGYTVSGTHT PLGLPMTLRE IPQSVSVITS QQMRDQNIKA LDRALLQATG |
|
| 101 | TSRQIYGSDR AGYNYLFARG SRIANYQING IPVADALADT GNANTAAYER |
|
| 151 | VEVVRGVAGL LDGTGEPSAT VNLVRKRPTR KPLFEVRAEA GNRKHFGLGA |
|
| 201 | DVSGSLNAEG TLRGRLVSTF GRGDSWRQRE RSRDAELYGI LEYDIAPQTR |
|
| 251 | VHAGMDYQQA KETADAPLSY AVYDSQGYAT AFGPKDNPAT NWANSRHRAL |
|
| 301 | NLFAGIEHRF NQDWKLKAEY DYTRSRFRQP YGVAGVLSID HNTAATDLIP |
|
| 351 | GYWHADPRTH SASVSLIGKY RLFGREHDLI AGINGYKYAS NKYGERSIIP |
|
| 401 | NAIPNAYEFS RTGAYPQPAS FAQTIPQYGT RRQIGGYLAT RFRAADNLSL |
|
| 451 | ILGGRYSRYR TGSYDSRTQG MTYVSANRFT PYTGIVFDLT GNLSLYGSYS |
|
| 501 | SLFVPQSQKD EHGSYLKPVT GNNLEAGIKG EWLEGRLNAS AAVYRARKNN |
|
| 551 | LATAAGRDPS GNTYYRAANQ AKTHGWEIEV GGRITPEWQI QAGYSQSKTR |
|
| 601 | DQDGSRLNPD SVPERSFKLF TAYHFAPEAP SGWTIGAGVR WQSETHTDPA |
|
| 651 | TLRIPNPAAK ARAADNSRQK AYAVADIMAR YRFNPRAELS LNVDNLFNKH |
|
| 701 | YRTQPDRHSY GALRTVNAAF TYRFK* |
[1410]ORF23a and ORF23-1 show 99.2% identity in 725 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1411]ORF23 shows 93.4% identity over a 211aa overlap with a predicted ORF (ORF23.ng) from N. gonorrhoeae:
[0000]
[1412]The ORF23ng nucleotide sequence <SEQ ID 669> is predicted to encode a protein comprising amino acid sequence <SEQ ID 670>:
[0000]| 1 | SAVDACRIPG YNYLFARGSR IANYQINGIP VADALADTGN ANTAAYERVE | |
|
| 51 | VVRGVAGLPD GTGEPSATVN LVRKHPTRKP LFEVRAEAGN RKHFGLGADV |
|
| 101 | SGSLNAEGTL RGRLVSTFGR GDSWRQLERS RDAELYGILE YDIAPQTRVH |
|
| 151 | AGMDYQQAKE TADAPLSYAV YDSQGYATAF GPKDNPATNW SNSRNRALNL |
|
| 201 | FAGIEHRFNQ DWKLKAEYDY TRSRFRQPYG VAGVLSIDHS TAATDLIPGY |
|
| 251 | WHADPRTHSA SMSLTGKYRL FGREHDLIAG INGYKYASNK YGERSIIPNA |
|
| 301 | IPNAYEFSRT GAYPQPSSFA QTIPQYDTRR QIGGYLATRF RAADNLSLIL |
|
| 351 | GGRYSRYRAG SYNSRTQGMT YVSANRFTPY TGIVFDLTGN LSLYGSYSSL |
|
| 401 | FVPQLQKDEH GSYLKPVTGN NLEADIKGEW LEGRLNASAA VYRARKNNLA |
|
| 451 | TAAGRDQSGN TYYRAANQAK THGWEIEVGG RITPEWQIQA GYSQSKPRDQ |
|
| 501 | DGSRLNPDSV PERSFKLFTA YHLAPEAPSG RTIGAGVRRQ GETHTDPAAL |
|
| 551 | RIPNPAAKAR AVANSRQKAY AVADIMARYR FNPRTELSLN VDNLFNKHYR |
|
| 601 | TQPDRHSYGA LRTVNAAFTY RFK* |
[1413]Further work revealed the complete nucleotide sequence <SEQ ID 671>:
[0000]| 1 | ATGACACGCT TCAAATACTC CCTGCTTTTT GCCGCCCTGC TACCCGTGTA | |
|
| 51 | CGCGCAGGCC GATGTTTCTG TTTCAGACGA CCCCAAACCG CAGGAAAGCA |
|
| 101 | CCGAATTGCC GACCATCACC GTTACCGCCG ACCGCACCGC GAGTTCCAAC |
|
| 151 | GACGGCTACA CCGTTTCCGG CACGCACACC CCGTTCGGGC TGCCCATGAC |
|
| 201 | CCTGCGCGAA ATCCCGCAGA GCGTCAGCGT CATCACATCG CAACAAATGC |
|
| 251 | GCGACCAAAA CATCAAAACG CTCGACCGCG CCCTGTTGCA GGCGACCGGC |
|
| 301 | ACCAGCCGCC AGATTTACGG CTCCGACCGC GCGGGCTACA ACTACCTGTT |
|
| 351 | CGCGCGCGGC AGCCGCATCG CCAACTACCA AATCAACGGC ATCCCCGTTG |
|
| 401 | CCGACGCGCT GGCCGATACG GGCAATGCCA ACACCGCCGC CTATGAGCGC |
|
| 451 | GTAGAAGTCG TGCGCGGCGT GGCGGGGCTG CCGGACGGCA CGGGCGAGCC |
|
| 501 | TTCTGCCACC GTCAATCTGG TACGCAAACA CCCGACCCGC AAGCCATTGT |
|
| 551 | TTGAAGTCCG CGCCGAAGCC GGCAACCGCA AACATTTCGG GCTGGGCGCG |
|
| 601 | GACGTATCGG GCAGCCTGAA CGCCGAAGGC ACGCTGCGCG GCCGCCTGGT |
|
| 651 | TTCCACCTTC GGACGCGGCG ACTCGTGGCG GCAGCTCGAA CGCAGCCGCG |
|
| 701 | ATGCCGAACT CTACGGCATT TTGGAATACG ACATCGCACC GCAAACCCGC |
|
| 751 | GTCCACGCAG GCATGGACTA CCAGCAGGCG AAAGAAACCG CAGACGCGCC |
|
| 801 | GCTCAGCTAC GCCGTGTACG ACAGCCAAGG TTATGCCACC GCCTTCGGCC |
|
| 851 | CAAAAGACAA CCCCGCCACA AATTGGTCGA ACAGCCGCAA CCGTGCGCTC |
|
| 901 | AACCTGTTCG CCGGCATAGA ACACCGCTTC AACCAAGACT GGAAACTCAA |
|
| 951 | AGCCGAATAC GACTACACCC GTAGCCGCTT CCGCCAGCCC TACGGTGTGG |
|
| 1001 | CAGGCGTACT TTCCATCGAC CACAGCACTG CCGCCACCGA CCTGATTCCC |
|
| 1051 | GGTTATTGGC ACGCcgatcc GCGCACCCAC AGCGCCAGCA TGTCATTGAC |
|
| 1101 | CGGCAAATAC CgcctGTTCG GCCGCGAGCA CGATTTAATC GCGGGTATCA |
|
| 1151 | ACGGCTACAA ATACGCCAGC AACAAATACG GCGAACGCAG CATCATTCCC |
|
| 1201 | AACGCCATTC CCAACGCCTA CGAATTTTCC CGCACGGGCG CCTATCCGCA |
|
| 1251 | GCCATCATCG TTTGCCCAAA CCATCCCGCA ATACGACACC AGGCGGCAAA |
|
| 1301 | TCGGCGGCTA TCTCGCCACC CGTTTCCGCG CCGCCGACAA CCTTTCGCTG |
|
| 1351 | ATACTCGGCG GCAGATACAG CCGCTACCGC GCAGGCAGCT ACAACAGCCG |
|
| 1401 | CACACAAGGC ATGACCTATG TGTCCGCCAA CCGTTTCACC CCCTACACAG |
|
| 1451 | GCATCGTGTT CGATCTGACC GGCAACCTGT CGCTTTACGG CTCGTACAGC |
|
| 1501 | AGCCTGTTCG TCCCGCAATT GCAAAAAGAC GAACACGGCA GCTACCTGAA |
|
| 1551 | ACCCGTAACC GGCAACAATC TGGAAGCCGA CATCAAAGGC GAATGGCTTG |
|
| 1601 | AAGGGCGTCT GAACGCATCC GCCGCCGTGT ACCGCGCCCG TAAAAACAAC |
|
| 1651 | CTCGCCACCG CAGCAGGACG CGACCAGAGC GGCAACACCT ACTATCGCGC |
|
| 1701 | CGCCAACCAA GCCAAAACCC ACGGCTGGGA AATCGAAGTC GGCGGCCGCA |
|
| 1751 | TCACGCCCGA ATGGCAGATA CAGGCAGGCT ACAGCCAAAG CAAACCCCGC |
|
| 1801 | GACCAAGACG GCAGCCGCCT GAACCCCGAC AGCGTAcCCG AACGCAGCTT |
|
| 1851 | CAAACTCTTC ACCGCCTACC ACTTAGCCCC CGAAGCCCCC AGCGGCCGGA |
|
| 1901 | CCATcggTGC GGGTGTGCGC CGGCAGGGCG AAACCCACAC CGACCCAGCC |
|
| 1951 | GCGCTCCGCA TCCCCAACCC CGCCGCCAAA GCCCGCGCCG TCGCCAACAG |
|
| 2001 | CCGCCAGAAA GCCTACGCCG TCGCCGACAT CATGGCGCGT TACCGCTTCA |
|
| 2051 | ATCCGCGCAC CGAACTGTCG CTGAACGTGG ACAACCTGTT CAACAAACAC |
|
| 2101 | TACCGCACCC AGCCCGACCG CCACAGCTAC GGCGCACTGC GGACAGTGAA |
|
| 2151 | CGCGGCGTTT ACCTATCGGT TTAAATAA |
[1414]This corresponds to the amino acid sequence <SEQ ID 672; ORF23ng-1>:
[0000]| 1 | MTRFKYSLLF AALLPVYAQA DVSVSDDPKP QESTELPTIT VTADRTASSN | |
|
| 51 | DGYTVSGTHT PFGLPMTLRE IPQSVSVITS QQMRDQNIKT LDRALLQATG |
|
| 101 | TSRQIYGSDR AGYNYLFARG SRIANYQING IPVADALADT GNANTAAYER |
|
| 151 | VEVVRGVAGL PDGTGEPSAT VNLVRKHPTR KPLFEVRAEA GNRKHFGLGA |
|
| 201 | DVSGSLNAEG TLRGRLVSTF GRGDSWRQLE RSRDAELYGI LEYDIAPQTR |
|
| 251 | VHAGMDYQQA KETADAPLSY AVYDSQGYAT AFGPKDNPAT NWSNSRNRAL |
|
| 301 | NLFAGIEHRF NQDWKLKAEY DYTRSRFRQP YGVAGVLSID HSTAATDLIP |
|
| 351 | GYWHADPRTH SASMSLTGKY RLFGREHDLI AGINGYKYAS NKYGERSIIP |
|
| 401 | NAIPNAYEFS RTGAYPQPSS FAQTIPQYDT RRQIGGYLAT RFRAADNLSL |
|
| 451 | ILGGRYSRYR AGSYNSRTQG MTYVSANRFT PYTGIVFDLT GNLSLYGSYS |
|
| 501 | SLFVPQLQKD EHGSYLKPVT GNNLEADIKG EWLEGRLNAS AAVYRARKNN |
|
| 551 | LATAAGRDQS GNTYYRAANQ AKTHGWEIEV GGRITPEWQI QAGYSQSKPR |
|
| 601 | DQDGSRLNPD SVPERSFKLF TAYHLAPEAP SGRTIGAGVR RQGETHTDPA |
|
| 651 | ALRIPNPAAK ARAVANSRQK AYAVADIMAR YRFNPRTELS LNVDNLFNKH |
|
| 701 | YRTQPDRHSY GALRTVNAAF TYRFK* |
[1415]ORF23ng-1 and ORF23-1 show 95.9% identity in 725 aa overlap:
[0000]
[1416]In addition, ORF1ing-1 shows significant homology with an OMP from E. coli:
[0000]| sp|P16869|FHUE_ECOLI OUTER-MEMBRANE RECEPTOR FOR FE(III)-COPROGEN, FE(III)- | |
| FERRIOXAMINE B AND FE(III)-RHODOTRULIC ACID PRECURSOR |
| >gi|1651542|gnl|PID|d1015403 |
| (D90745) Outer membrane protein FhuE precursor [Escherichia coli] |
| >gi|1651545|gnl|PID|d1015405 (D90746) Outer membrane protein FhuE precursor |
| [Escherichia coli] >gi|1787344 (AE000210) outer-membrane receptor for |
| Fe(III)-coprogen, Fe(III)-ferrioxamine B and Fe(III)-rhodotrulic acid precursor |
| [Escherichia coli] Length = 729 |
| Score = 332 bits (843), Expect = 3e−90 |
| Identities = 228/717 (31%), Positives = 350/717 (48%), Gaps = 60/717 (8%) |
|
| Query: | 38 | TITVTADRTASSN--DGYTVSGTHTPFGLPMTLREIPQSVSVITSQQMRDQNIKTLDRAL | 95 | |
| | T+ V TA + + Y+V+ T + MT R+IPQSV++++ Q+M DQ ++TL + |
| Sbjct: | 43 | TVIVEGSATAPDDGENDYSVTSTSAGTKMQMTQRDIPQSVTIVSQQRMEDQQLQTLGEVM | 102 |
|
| Query: | 96 | LQATGTSRQIYGSDRAGYNYLFARGSRIANYQINGIP--------VADALADTGNANTAA | 147 |
| | G S+ SDRA Y ++RG +I NY ++GIP + DAL+D A |
| Sbjct: | 103 | ENTLGISKSQADSDRALY---YSRGFQIDNYMVDGIPTYFESRWNLGDALSDM-----AL | 154 |
|
| Query: | 148 | YERVEVVRGVAGLPDGTGEPSATVNLVRKHPTRKPLF-EVRAEAGNRKHFGLGADVSGSL | 206 |
| | +ERVEVVRG GL GTG PSA +N+VRKH T + +V AE G+ AD+ L |
| Sbjct: | 155 | FERVEVVRGATGLMTGTGNPSAAINMVRKHATSREFKGDVSAEYGSWNKERYVADLQSPL | 214 |
|
| Query: | 207 | NAEGTLRGRLVSTFGRGDSWRQLERSRDAELYGILEYDIAPQTRVHAGMDYQQAKETADA | 266 |
| | +G +R R+V + DSW S GI++ D+ T + AG +YQ+ + |
| Sbjct: | 215 | TEDGKIRARIVGGYQNNDSWLDRYNSEKTFFSGIVDADLGDLTTLSAGYEYQRIDVNSPT | 274 |
|
| Query: | 267 | PLSYAVYDSQGYATAFGPKDNPATNWSNSRNRALNLFAGIEHRFNQDWKLKAEYDYTRSR | 326 |
| | +++ G + ++ + A +W+ + +F ++ +F w+ ++ |
| Sbjct: | 275 | WGGLPRWNTDGSSNSYDRARSTAPDWAYNDKEINKVFMTLKQQFADTWQATLNATHSEVE | 334 |
|
| Query: | 327 | F--RQPYGVAGVLSIDHSTAA--TDLIPGY-------WHADPRTHSA-SMSLTGKYRLFG | 374 |
| | F + Y A V D ++ PG+ W++ R A + G Y LFG |
| Sbjct: | 335 | FDSKMMYVDAYVNKADGMLVGPYSNYGPGFDYVGGTGWNSGKRKVDALDLFADGSYELFG | 394 |
|
| Query: | 375 | REHDLIAGINGYKYASNKYGER--SIIPNAIPNAYEFSRTGAYPQPSSFAQTIPQYDTRR | 432 |
| | R+H+L+ G Y +N+Y +I P+ I + Y F+ G +PQ Q++ Q DT |
| Sbjct: | 395 | RQHNLMFG-GSYSKQNNRYFSSWANIFPDEIGSFYNFN--GNFPQTDWSPQSLAQDDTTH | 451 |
|
| Query: | 433 | QIGGYLATRFRAADNLSLILGGRYSRYRAGSYNSRTQGMTY-VSANRFTPYTGIVFDXXX | 491 |
| | Y ATR AD L LILG RY+ +R + +TY + N TPY G+VFD |
| Sbjct: | 452 | MKSLYAATRVTLADPLHLILGARYTNWRVDT-------LTYSMEKNHTTPYAGLVFDIND | 504 |
|
| Query: | 492 | XXXXXXXXXXXFVPQLQKDEHGSYLKPVTGNNLEADIKGEWLEGRLNASAAVYRARKNNL | 551 |
| | F PQ +D G YL P+TGNN E +K +W+ RL + A++R ++N+ |
| Sbjct: | 505 | NWSTYASYTSIFQPQNDRDSSGKYLAPITGNNYELGLKSDWMNSRLTTTLAIFRIEQDNV | 564 |
|
| Query: | 552 | ATAAGR---DQSGNTYYRAANQAKTHGWEIEVGGRITPEWQIQAGYSQSKPRDQDGSRLN | 608 |
| | A + G +G T Y+A + + G E E+ G IT WQ+ G ++ D +G+ +N |
| Sbjct: | 565 | AQSTGTPIPGSNGETAYKAVDGTVSKGVEFELNGAITDNWQLTFGATRYIAEDNEGNAVN | 624 |
|
| Query: | 609 | PDSVPERSFKLFTAYHLAPEAPSGRTIGAGVRRQGETHTDPAALRIPNPAAKARAVANSR | 668 |
| | P ++P + K+FT+Y L P P T+G GV Q +TD P RA |
| Sbjct: | 625 | P-NLPRTTVKMFTSYRL-PVMPE-LTVGGGVNWQNRVYTDTV-----TPYGTFRA----E | 672 |
|
| Query: | 669 | QKAYAVADIMARYRFNPRTELSLNVDNLFNKHYRTQPDRH-SYGALRTVNAAFTYRF | 724 |
| | Q +YA+ D+ RY+ L NV+NLF+K Y T + YG R + TY+F |
| Sbjct: | 673 | QGSYALVDLFTRYQVTKNFSLQGNVNNLFDKTYDTNVEGSIVYGTPRNFSITGTYQF | 729 |
[1417]Based on this analysis, it was predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1418]ORF23-1 (77.5 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 15A shows the results of affinity purification of the His-fusion protein, and FIG. 15B shows the results of expression of the GST-fusion in E. coli. Purified His-fusion protein was used to immunise mice, whose sera were used for Western blot (FIG. 15C) and for ELISA (positive result). These experiments confirm that ORF23-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 80
[1419]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 673>:
[0000]| 1 | ATGCGCACGG CAGTGGTTTT GCTGTTGATC ATGCCGATGG CGGCTTCGTC | |
|
| 51 | GGCAATGATG CCGGAAATGG TGTGCGCGGG CGTGTCGCCG GGAACGGCAA |
|
| 101 | TCATATCCAA GCCGACCGAA CAAACGGCGG TCATGGCTTC GAGTTTGTCC |
|
| 151 | AGCGTCAgcA CGCCTGCTTC GGCGgcGgCa ATCATACCTT CGTCTTCGGA |
|
| 201 | AACGGGGATA AACGcGCCAC TCAAACCCCC GACCGCGCTG GAAGCCATCA |
|
| 251 | TGCCGCCTTT TTTCACGGCA TCGTTCAGCA ATGCCAAAGC TGCTGTTGTG |
|
| 301 | CCGTGCGTAC CGCAGACGCT CAAGCCCATT TnTTCAAGAA TGCGTGCCAC |
|
| 351 | TnAGTCGCCG ACGGGG.. |
[1420]This corresponds to the amino acid sequence <SEQ ID 674; ORF24>:
[0000]| 1 | MRTAVVLLLI MPMAASSAMM PEMVCAGVSP GTAIISKPTE QTAVMASSLS | |
|
| 51 | SVSTPASAAA IIPSSSETGI NAPLKPPTAL EAIMPPFFTA SFSNAKAAVV |
|
| 101 | PCVPQTLKPI XSRMRATXSP TG.. |
[1421]Further work revealed the complete nucleotide sequence <SEQ ID 675>:
[0000]| 1 | ATGCGCACGG CAGTGGTTTT GCTGTTGATC ATGCCGATGG CGGCTTCGTC | |
|
| 51 | GGCAATGATG CCGGAAATGG TGTGCGCGGG CGTGTCGCCG GGAACGGCAA |
|
| 101 | TCATATCCAA GCCGACCGAA CAAACGGCGG TCATGGCTTC GAGTTTGTCC |
|
| 151 | AGCGTCAGCA CGCCTGCTTC GGCGGCGGCA ATCATACCTT CGTCTTCGGA |
|
| 201 | AACGGGGATA AACGCGCCAC TCAAACCCCC GACCGCGCTG GAAGCCATCA |
|
| 251 | TGCCGCCTTT TTTCACGGCA TCGTTCAGCA ATGCCAAAGC TGCTGTTGTG |
|
| 301 | CCGTGCGTAC CGCAGACGCT CAAGCCCATT TCTTCAAGAA TGCGTGCCAC |
|
| 351 | TGAGTCGCCG ACGGCGGGGG TCGGCGCCAG CGACAAGTCG AGAATACCAA |
|
| 401 | ACGGGATATT CAGCATTTTT GAGGCTTCGC GGCCGATGAG TTCGCCCACG |
|
| 451 | CGGGTAATTT TGAAAGCAGT TTTCTTCACT ACTTCCGCAA CTTCGGTCAA |
|
| 501 | TGTCGTTGCA TCTGAATTTT CCAACGCGGC TTTTACGACA CCTGGGCCGG |
|
| 551 | ATACGCCGAC ATTGATAACG GCATCCGCTT CGCCCGAACC ATGAAACGCG |
|
| 601 | CCCGCCATAA ACGGGTTGTC TTCCACCGCG TTGCAGAACA CGACAATTTT |
|
| 651 | AGCGCAGCCG AAACCTTCGG GCGTGATTTC CGCCGTGCGT TTGACGGTTT |
|
| 701 | CGCCCGCCAG CTTGACCGCA TCCATATTGA TACCGGCACG CGTACTGCCG |
|
| 751 | ATATTGATGG AGCTGCACAC AATATCGGTA GTCTTCATCG CTTCGGGAAT |
|
| 801 | GGAGCGGATT AACACCTCAT CCGAAGGCGA CATCCCTTTT TGCACCAACG |
|
| 851 | CGGAAAAACC GCCGATAAAA GACACACCGA TGGCTTTGGC AGCTTTATCC |
|
| 901 | AAAGTTTGCG CCACGCTGAC GTAA |
[1422]This corresponds to the amino acid sequence <SEQ ID 676; ORF24-1>:
[0000]| 1 | MRTAVVLLLI MPMAASSAMM PEMVCAGVSP GTAIISKPTE QTAVMASSLS | |
|
| 51 | SVSTPASAAA IIPSSSETGI NAPLKPPTAL EAIMPPFFTA SFSNAKAAVV |
|
| 101 | PCVPQTLKPI SSRMRATESP TAGVGASDKS RIPNGIFSIF EASRPMSSPT |
|
| 151 | RVILKAVFFT TSATSVNVVA SEFSNAAFTT PGPDTPTLIT ASASPEP*NA |
|
| 201 | PAINGLSSTA LQNTTILAQP KPSGVISAVR LTVSPASLTA SILIPARVLP |
|
| 251 | ILMELHTISV VFIASGMERI NTSSEGDIPF CTNAEKPPIK DTPMALAALS |
|
| 301 | KVCATLT* |
[1423]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1424]ORF24 shows 96.4% identity over a 307 aa overlap with an ORF (ORF24a) from strain A of N. meningitidis.
[0000]
[1425]The complete length ORF24a nucleotide sequence <SEQ ID 677> is:
[0000]| 1 | ATGCGCACGG CAGTGGTTTT GCTGTTGATC ATGCCGATGG CGGCTTCGTC | |
|
| 51 | GGCAATGATG CCGGAAATGG TGTGCGCGGG TGTGTCGCCG GGAACGGCAA |
|
| 101 | TCATATCCAA NCCGACCGAA CAAACGGCGG TCATCGCTTC GAGTTTATCC |
|
| 151 | AACGTCAGCA CGCCTGCTTC GGCGGCGGCA ATCATACCTT CGTCTTCGGA |
|
| 201 | NACGGGGATA AACGCGCCAC TCAAACCGCC AACCGCGCTC GAAGCCATCA |
|
| 251 | TGCCGCCCTT TTTCACGGCA TCGTTCAGCA ATGCCAAAGC TGCTGTTGTG |
|
| 301 | CCGTGCGTAC CGCAGACGCT CAAACCCATT TCTTCAAGAA TGCGCGCCAC |
|
| 351 | CGAGTCGCCG ACGGCAGGGG TCGGTGCCAG CGACAAGTCG AGAATACCAA |
|
| 401 | ACGGGATATT CAGCATTTTT GAGGCTTCGC GGCCGATGAG TTCGCCCACG |
|
| 451 | CGGGTAATTT TGAAGGCGGT TTTCTTCACA ACTTCGGCAA CTTCGGTCAA |
|
| 501 | TGTCGTTGCA TCCGAATTTT CCAACGCGGC TTTTACGACA CCCGGGCCGG |
|
| 551 | ATACGCCGAC ATTAATCACA GCATCCGCTT CGCCTGAGCC GTGAAACGCG |
|
| 601 | CCCGCCATAN ACGGGTTGTC TTCCNCCGCG TTGCAGAACA CGACGATTTT |
|
| 651 | GGCGCAGCCG AAACCTTCTA GTGTGATTTC ANCCGTGCGT TTGATGGTTT |
|
| 701 | CGCCCGCCAG TCTGACCGCG TCCATATTGA TACCGGCGCG CGTACTGCCG |
|
| 751 | ATATTGATGG AGCTGCACAC GATATCAGTA GTCTTCATCG CTTCGGGAAT |
|
| 801 | GGAACGGATN AACACCTCGT CAGAAGGCGA CATACCTTTT TGCACCAGCG |
|
| 851 | CGGAAAAGCC GCCAATAAAA GACACGCCGA TGGCTTTGGC AGCCTTATCC |
|
| 901 | AAAGTTTGCG CCACGCTGAC GTAA |
[1426]This encodes a protein having amino acid sequence <SEQ ID 678>:
[0000]| 1 | MRTAVVLLLI MPMAASSAMM PEMVCAGVSP GTAIISXPTE QTAVIASSLS | |
|
| 51 | NVSTPASAAA IIPSSSXTGI NAPLKPPTAL EAIMPPFFTA SFSNAKAAVV |
|
| 101 | PCVPQTLKPI SSRMRATESP TAGVGASDKS RIPNGIFSIF EASRPMSSPT |
|
| 151 | RVILKAVFFT TSATSVNVVA SEFSNAAFTT PGPDTPTLIT ASASPEP*NA |
|
| 201 | PAIXGLSSXA LQNTTILAQP KPSSVISXVR LMVSPASLTA SILIPARVLP |
|
| 251 | ILMELHTISV VFIASGMERX NTSSEGDIPF CTSAEKPPIK DTPMALAALS |
|
| 301 | KVCATLT* |
[1427]It should be noted that this protein includes a stop codon at position 198.
[1428]ORF24a and ORF24-1 show 96.4% identity in 307 aa overlap:
[0000]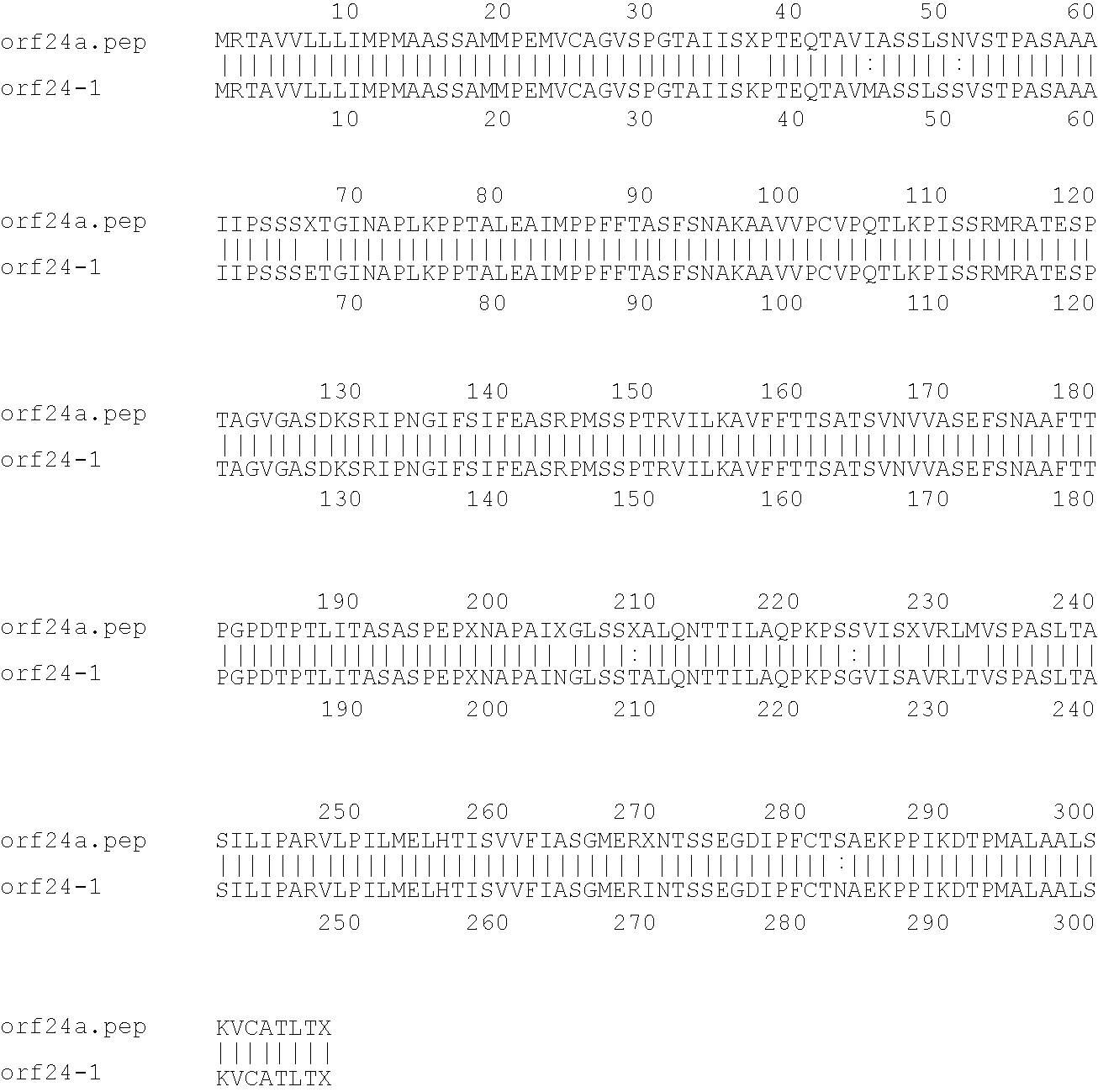
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1429]ORF24 shows 96.7% identity over a 121 aa overlap with a predicted ORF (ORF24ng) from N. gonorrhoeae:
[0000]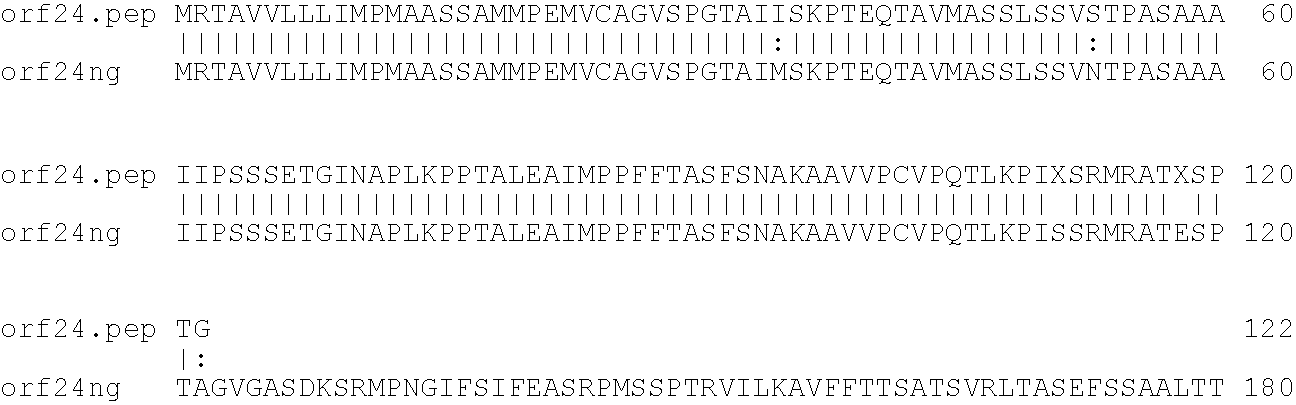
[1430]The complete length ORF24ng nucleotide sequence <SEQ ID 679> is:
[0000]| 1 | ATGCGCACGG CGGTGGTTTT GCTGTTGATC ATGCCGATGG CGGCTTCGTC | |
|
| 51 | GGCGATGATG CCGGAAATGG TGTGCGCGGG CGTGTCGCCG GGAACGGCAA |
|
| 101 | TCATGTCCAA ACCAACGGAG CAGACGGCGG TCATGGCTTC GAGTTTGTCC |
|
| 151 | AGCGTCAACA CGCCTGCCTC GGCGGCGGCA ATCATACCTT CGTCTTCGGA |
|
| 201 | AACGGGGATA AACGCGCCGC TCAAACCGCC GACCGCGCTG GAAGCCATCA |
|
| 251 | TGCCGCCCTT TTTCACGGCA TCGTTCAGCA ATGCCAAAGC TGCTGTTGTG |
|
| 301 | CCGTGCGTAC CGCAGACGCT CAAGCCCATT TCTTCAAGAA TGCGCGCCAC |
|
| 351 | CGAGTCGCCG ACGGCGGGGG TCGGTGCCAG CGACAAATCG AGAATGCCGA |
|
| 401 | ACGGGATATT CAGCATTTTT GAGGCTTCGC GACCGATGAG TTCGCCCACG |
|
| 451 | CGGGTGATTT TGAAAGCGGT TTTCTTCACG ACTTCGGCGA CCTCGGTCAG |
|
| 501 | GCTGACCGCG TCCGAATTTT CCAGCGCGGC TTTGACCACG CCTGGACCGG |
|
| 551 | ATACGCCGAC ATTAATCACA GCATCCGCTT CGCCCGAGCC GTGGAACGCA |
|
| 601 | CCCGCCATAA ACGGATTGTC TTCCACCGCG TTGCAGAACA CGACGATTTT |
|
| 651 | GGCGCAGCCG AAACCTTCGG GTGTGATTTC AGCCGTGCGT TTGATGGTTT |
|
| 701 | CGCCTGCCAG CTTGACCGCA TCCATATTGA TACCGGCACG CGTGCTGCCG |
|
| 751 | ATATTGATGG AGCTGCACAC GATATCGGTA GTTTTCATCG CTTCGGGAAC |
|
| 801 | GGAACGGATC AACACCTCAT CCGAAGGCGA CATACCTTTT TGCACCAGCG |
|
| 851 | CGGAAAAGCC GCCGATAAAG GACACGCCGA TGGCTTTGGC TGCCTTGTCC |
|
| 901 | AAAGTCTGCG CCACGCTGAC ATAA |
[1431]This encodes a protein having amino acid sequence <SEQ ID 680>:
[0000]| 1 | MRTAVVLLLI MPMAASSAMM PEMVCAGVSP GTAIMSKPTE QTAVMASSLS | |
|
| 51 | SVNTPASAAA IIPSSSETGI NAPLKPPTAL EAIMPPFFTA SFSNAKAAVV |
|
| 101 | PCVPQTLKPI SSRMRATESP TAGVGASDKS RMPNGIFSIF EASRPMSSPT |
|
| 151 | RVILKAVFFT TSATSVRLTA SEFSSAALTT PGPDTPTLIT ASASPEPWNA |
|
| 201 | PAINGLSSTA LQNTTILAQP KPSGVISAVR LMVSPASLTA SILIPARVLP |
|
| 251 | ILMELHTISV VFIASGTERI NTSSEGDIPF CTSAEKPPIK DTPMALAALS |
|
| 301 | KVCATLT* |
[1432]ORF24ng and ORF24-1 show 96.1% identity in 307 aa overlap:
[0000]
[1433]Based on this analysis, including the presence of a putative leader sequence (first 18 aa—double-underlined) and putative transmembrane domains (single-underlined) in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 81
[1434]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 681>:
[0000]| 1 | ..ACCGACGTGC AAAAAGAGTT GGTCGGCGAA CAACGCAAGT GGGCGCAGGA | |
|
| 51 | AAAAATCAGC AACTGCCGAC AAGCCGCCGC GCAGGCAGAC CGGCAGGAAT |
|
| 101 | ACGCCGAATA CCTCAAGCTG CAATGCGACA CGCGGATGAC GCGCGAACGG |
|
| 151 | ATACAGTATC TTCGCGGCTA TTCCATCGAT TAG |
[1435]This corresponds to the amino acid sequence <SEQ ID 682; ORF25>:
[0000]| 1 | ..TDVQKELVGE QRKWAQEKIS NCRQAAAQAD RQEYAEYLKL QCDTRMTRER | |
|
| 51 | IQYLRGYSID * |
[1436]Further work revealed the complete nucleotide sequence <SEQ ID 683>:
[0000]| 1 | ATGTATCGGA AACTCATTGC GCTGCCGTTT GCCCTGCTGC TTGCCGCTTG | |
|
| 51 | CGGCAGGGAA GAACCGCCCA AGGCATTGGA ATGCGCCAAC CCCGCCGTGT |
|
| 101 | TGCAAGGCAT ACGCGGCAAT ATTCAGGAAA CGCTCACGCA GGAAGCGCGT |
|
| 151 | TCTTTCGCGC GCGAAGACGG CAGGCAGTTT GTCGATGCCG ACAAAATTAT |
|
| 201 | CGCCGCCGCC TACGGTTTGG CGTTTTCTTT GGAACACGCT TCGGAAACGC |
|
| 251 | AGGAAGGCGG GCGCACGTTC TGTATCGCCG ATTTGAACAT TACCGTGCCG |
|
| 301 | TCTGAAACGC TTGCCGATGC CAAGGCAAAC AGCCCCCTGT TGTACGGGGA |
|
| 351 | AACTGCTTTG TCGGATATTG TGCGGCAGAA GACGGGCGGC AATGTCGAGT |
|
| 401 | TTAAAGACGG CGTATTGACG GCAGCCGTCC GCTTCCTGCC CGTCAAAGAC |
|
| 451 | GGTCAGACGG CATTTGTCGA CAACACGGTC GGTATGGCGG CGCAAACGCT |
|
| 501 | GTCTGCCGCG CTGCTGCCTT ACGGCGTGAA GAGCATCGTG ATGATAGACG |
|
| 551 | GCAAGGCGGT GAAAAAAGAA GACGCGGTCA GGATTTTGAG CGGAAAAGCC |
|
| 601 | CGTGAAGAAG AACCGTCCAA ACCCACGCCC GAAGACATTT TGGAACACAA |
|
| 651 | TGCCGCCGGC GGCGATGCGG GCGTACCCCA AGCCGCAGAA GGCGCGCCCG |
|
| 701 | AACCGGAAAT CCTGCATCCT GACGACGGCG AGCGTGCCGA TACCGTTACC |
|
| 751 | GTATCACGGG GCGAAGTGGA AGAGGCGCGC GTACAAAACC AGCGTGCGGA |
|
| 801 | ATCCGAAATT ACCAAACTTT GGGGAGGACT CGATACCGAC GTGCAAAAAG |
|
| 851 | AGTTGGTCGG CGAACAACGC AAGTGGGCGC AGGAAAAAAT CAGCAACTGC |
|
| 901 | CGACAAGCCG CCGCGCAGGC AGACCGGCAG GAATACGCCG AATACCTCAA |
|
| 951 | GCTGCAATGC GACACGCGGA TGACGCGCGA ACGGATACAG TATCTTCGCG |
|
| 1001 | GCTATTCCAT CGATTAG |
[1437]This corresponds to the amino acid sequence <SEQ ID 684; ORF25-1>:
[0000]| 1 | MYRKLIALPF ALLLAACGRE EPPKALECAN PAVLQGIRGN IQETLTQEAR | |
|
| 51 | SFAREDGRQF VDADKIIAAA YGLAFSLEHA SETQEGGRTF CIADLNITVP |
|
| 101 | SETLADAKAN SPLLYGETAL SDIVRQKTGG NVEFKDGVLT AAVRFLPVKD |
|
| 151 | GQTAFVDNTV GMAAQTLSAA LLPYGVKSIV MIDGKAVKKE DAVRILSGKA |
|
| 201 | REEEPSKPTP EDILEHNAAG GDAGVPQAAE GAPEPEILHP DDGERADTVT |
|
| 251 | VSRGEVEEAR VQNQRAESEI TKLWGGLDTD VQKELVGEQR KWAQEKISNC |
|
| 301 | RQAAAQADRQ EYAEYLKLQC DTRMTRERIQ YLRGYSID* |
[1438]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1439]ORF25 shows 98.3% identity over a 60aa overlap with an ORF (ORF25a) from strain A of N. meningitidis:
[0000]
[1440]The complete length ORF25a nucleotide sequence <SEQ ID 685> is:
[0000]| 1 | ATGTATCGGA AACTCATTGC GCTGCCGTTT GCCCTGCTGC TTGCCGCTTG | |
|
| 51 | CGGCAGGGAA GAACCGCCCA AGGCATTGGA ATGCGCCAAC CCCGCCGTGT |
|
| 101 | TGCAANGCAT ACGCNGCAAT ATTCAGGAAA CGCTCACGCA GGAAGCGCGT |
|
| 151 | TCTTTCGCGC GCGAAGACNG CANGCAGTTT GTCGATGCCG ACNAAATTAT |
|
| 201 | CGCCGCCGCC TANGNTNNGN NGNTNTCTTT GGAACACGCT TCGGAAACGC |
|
| 251 | AGGAAGGCGG GCGCACGTTC TGTNTCGCCG ATTTGAACAT TACCGTGCCG |
|
| 301 | TCTGAAACGC TTGCCGATGC CAAGGCAAAC AGCCCCCTGC TGTACGGGGA |
|
| 351 | AACCGCTTTG TCGGATATTG TGCGGCAGAA GACGGGCGGC AATGTCGAGT |
|
| 401 | TTAAAGACGG CGTATTGACG GCAGCCGTCC GCTTCCTACC CGTCAAAGAC |
|
| 451 | GGTCAGANGG CATTTGTCGA CAACACGGTC GGTATGGCGG CGCAAACGCT |
|
| 501 | GTCTGCCGCG TTGCTGCCTT ACGGCGTGAA GAGCATCGTG ATGATAGACG |
|
| 551 | GCAAGGCGGT AAAAAAAGAA GACGCGGTCA GGATTNTGAG CNGANAAGCC |
|
| 601 | CGTGAANAAG AACCGTCCAA ANCCNNGCCC GAAGACATTT TGGAACATAA |
|
| 651 | TGCCGCCGGA GGGGATGCAG ACGTACCCCA AGCCGGAGAA GACGCGCCCG |
|
| 701 | AACCGGAAAT CCTGCATCCT GACGACGGCG AGCGTGCCGA TACCGTTACC |
|
| 751 | GTATCACGGG GCGAAGTGGA AGAGGCGCGN GTACAAAACC AGCGTGCGGA |
|
| 801 | ATCCGAAATT ACCAAACTTT GGGGAGGACT CGATACCGAC GTGCAAAAAG |
|
| 851 | AGTTGGTCGG CGAANAACGC AAGTGGGCGC AGGAAAAAAT CAGCAACTGC |
|
| 901 | CGACAAGCCG CCGCGCAGGC AGACCGGCAG GAATACGCCG AATACCTCAA |
|
| 951 | GCTGCAATGC GACACGCGGA TGACGCGCGA ACGGATACAG TATCTTCGCG |
|
| 1001 | GCTATTCCAT CGATTAG |
[1441]This encodes a protein having amino acid sequence <SEQ ID 686>:
[0000]| 1 | MYRKLIALPF ALLLAACGRE EPPKALECAN PAVLQXIRXN IQETLTQEAR | |
|
| 51 | SFAREDXXQF VDADXIIAAA XXXXXSLEHA SETQEGGRTF CXADLNITVP |
|
| 101 | SETLADAKAN SPLLYGETAL SDIVRQKTGG NVEFKDGVLT AAVRFLPVKD |
|
| 151 | GQXAFVDNTV GMAAQTLSAA LLPYGVKSIV MIDGKAVKKE DAVRIXSXXA |
|
| 201 | REXEPSKXXP EDILEHNAAG GDADVPQAGE DAPEPEILHP DDGERADTVT |
|
| 251 | VSRGEVEEAR VQNQRAESEI TKLWGGLDTD VQKELVGEXR KWAQEKISNC |
|
| 301 | RQAAAQADRQ EYAEYLKLQC DTRMTRERIQ YLRGYSID* |
[1442]ORF25a and ORF25-1 show 93.5% identity in 338 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1443]ORF25 shows 100% identity over a 60aa overlap with a predicted ORF (ORF25ng) from N. gonorrhoeae:
[0000]
[1444]The complete length ORF25ng nucleotide sequence <SEQ ID 687> is:
[0000]| 1 | ATGTATCGGA AACTCATTGC GCTGCCGTTT GCCCTGCTGC TTGCAGCGTG | |
|
| 51 | CGGCAGGGAA GAACCGCCCA AGGCGTTGGA ATGCGCCAAC CCCGCCGTGT |
|
| 101 | TGCAGGACAT ACGCGGCAGT ATTCAGGAAA CGCTCACGCA GGAAGCGCGT |
|
| 151 | TCTTTCGCGC GCGAAGACGG CAGGCAGTTT GTCGATGCCG ACAAAATTAT |
|
| 201 | CGCCGCCGCC TACGGTTTGG CGTTTTCTTT GGAACACGCT TCGGAAACGC |
|
| 251 | AGGAAGGCGG GCGCACGTTC TGTATCGCCG ATTTGAACAT TACCGTGCCG |
|
| 301 | TCTGAAACGC TTGCCGATGC CGAGGCAAAC AGCCCCCTGC TGTATGGGGA |
|
| 351 | AACGTCTTTG GCAGACATCG TGCAGCAGAA GACGGGCGGC AATGTCGAGT |
|
| 401 | TTAAAGACGG CGTATTGACG GCAGCCGTCC GCTTCCTGCC CGCCAAAGAC |
|
| 451 | GCTCGGACGG CATTTATCGA CAACACGGTC GGTATGGCGA CGCAAACGCT |
|
| 501 | GTCTGCCGCG TTGCTGCCTT ACGGCGTGAA GAGCATCGTG ATGATAGACG |
|
| 551 | GCAAGGCGGT GACAAAAGAA GACGCGGTCA GGGTTTTGAG CGGCAAAGCC |
|
| 601 | CGTGAAGAAG AACCGTCCAA ACCCACCCCC GAAGACATTT TGGAACACAA |
|
| 651 | TGCCGCCGGC GGCGATGCGG GCGTACCCCA AGCCGCAGAA GGCGCACCCG |
|
| 701 | AACCCGAAAT CCTGCATCCC GACGACGTCG AGCGTGCCGA TACCGTTACC |
|
| 751 | GTATCACGGG GCGAAGTGGA AGAGGCGCGC GTACAAAACC AACGTGCGGA |
|
| 801 | ATCCGAAATT ACCAAACTTT GGGGAGGACT CGATACCGAC GTGCAAAAAG |
|
| 851 | AGTTGGTCGG CGAACAGCGC AAGTGGGCGC AGGAAAAAAT CAGcaactgc |
|
| 901 | cgACAAGCCG CCGCGCAGGC AGACCGGCAG GAATACGCCG AATACCTCAA |
|
| 951 | GCTCCAATGC GACACGCGGA TGACGCGCGA ACggaTACAG TATCTTCGCG |
|
| 1001 | GCTATTCCAT CGATTAG |
[1445]This encodes a protein having amino acid sequence <SEQ ID 688>:
[0000]| 1 | MYRKLIALPF ALLLAACGRE EPPKALECAN PAVLQDIRGS IQETLTQEAR | |
|
| 51 | SFAREDGRQF VDADKIIAAA YGLAFSLEHA SETQEGGRTF CIADLNITVP |
|
| 101 | SETLADAEAN SPLLYGETSL ADIVQQKTGG NVEFKDGVLT AAVRFLPAKD |
|
| 151 | ARTAFIDNTV GMATQTLSAA LLPYGVKSIV MIDGKAVTKE DAVRVLSGKA |
|
| 201 | REEEPSKPTP EDILEHNAAG GDAGVPQAAE GAPEPEILHP DDVERADTVT |
|
| 251 | VSRGEVEEAR VQNQRAESEI TKLWGGLDTD VQKELVGEQR KWAQEKISNC |
|
| 301 | RQAAAQADRQ EYAEYLKLQC DTRMTRERIQ YLRGYSID* |
[1446]ORF25ng and ORF25-1 show 95.9% identity in 338 aa overlap:
[0000]
[1447]Based on this analysis, including the presence of a predicted prokaryotic membrane lipoprotein lipid attachment site (underlined) in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1448]ORF25-1 (37 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 16A shows the results of affinity purification of the GST-fusion protein, and FIG. 16B shows the results of expression of the His-fusion in E. coli. Purified His-fusion protein was used to immunise mice, whose sera were used for Western blot (FIG. 16C), ELISA (positive result), and FACS analysis (FIG. 16D). These experiments confirm that ORF25-1 is a surface-exposed protein, and that it is a useful immunogen. FIG. 16E shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF25-1.
Example 82
[1449]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 689>
[0000]
[1450]This corresponds to the amino acid sequence <SEQ ID 690; ORF26>:
[0000]
[1451]Further work revealed the complete nucleotide sequence <SEQ ID 691>:
[0000]| 1 | ATGCAGCTGA TCGACTATTC ACATTCATTT TTCTCGGTTG TGCCACCCTT | |
|
| 51 | TTTGGCACTG GCACTTGCCG TCATTACCCG CCGCGTACTG CTGTCTTTAG |
|
| 101 | GCATCGGTAT TCTGGTCGGC GTTGCCTTTT TGGTCGGCGG CAACCCCGTC |
|
| 151 | GACGGTCTGA CACACCTGAA AGACATGGTC GTCGGCTTGG CTTGGTCAGA |
|
| 201 | CGGCGATTGG TCGCTGGGCA AACCAAAAAT CTTGGTTTTC CTGATACTTT |
|
| 251 | TGGGTATTTT TACTTCCCTG CTGACCTACT CCGGCAGCAA TCAGGCGTTT |
|
| 301 | GCCGACTGGG CAAAACGGCA CATTAAAAAC CGGCGCGGCG CGAAAATGCT |
|
| 351 | GACCGCCTGC CTCGTGTTCG TAACCTTTAT CGACGACTAT TTCCACAGTC |
|
| 401 | TCGCCGTCGG TGCGATTGCC CGCCCCGTTA CCGACAAGTT TAAAGTTTCC |
|
| 451 | CGCACCAAAC TCGCCTACAT CCTCGACTCC ACTGCCGCTC CTATGTGCGT |
|
| 501 | GCTGATGCCC GTTTCAAGCT GGGGCGCGTC GATTATCGCC ACGCTTGCCG |
|
| 551 | GACTGCTCGT TACCTACAAA ATCACCGAAT ACACGCCGAT GGGGACGTTT |
|
| 601 | GTCGCCATGA GCCTGATGAA CTATTACGCA CTGTTTGCCC TGATTATGGT |
|
| 651 | GTTCGTCGTC GCATGGTTTT CCTTCGACAT CGGCTCGATG GCACGTTTCG |
|
| 701 | AACAAGCCGC GTTGAACGAA GCCCACGATG AAACTGCCGT TTCAGACGCT |
|
| 751 | ACCAAAGGTC GTGTTTACGC ACTGATTATT CCCGTTTTGG CCTTAATCGC |
|
| 801 | CTCAACGGTT TCCGCCATGA TCTACACCGG CGCGCAGGCA AGCGAAACCT |
|
| 851 | TCAGCATTTT GGGGGCATTT GAAAACACGG ACGTAAACAC TTCGCTGGTA |
|
| 901 | TTCGGCGGCA CTTGCGGCGT CCTTGCCGTC GTTCTCTGCA CGCTCGGCAC |
|
| 951 | GATTAAAACC GCCGACTATC CCAAAGCCGT TTGGCAGGGT GCGAAATCTA |
|
| 1001 | TGTTCGGCGC AATCGCCATT TTAATCCTCG CTTGGCTCAT CAGTACGGTT |
|
| 1051 | GTCGGCGAAA TGCACACCGG CGATTACCTC TCCACACTGG TTGCGGGCAA |
|
| 1101 | CATCCATCCC GGCTTCCTGC CCGTCATCCT CTTCCTGCTC GCCAGCGTGA |
|
| 1151 | TGGCGTTTGC CACAGGCACA AGCTGGGGGA CGTTCGGCAT TATGCTGCCG |
|
| 1201 | ATTGCCGCCG CCATGGCGGT CAAAGTCGAA CCCGCGCTGA TTATCCCGTG |
|
| 1251 | TATGTCCGCA GTAATGGCGG GGGCGGTATG CGGCGACCAC TGCTCGCCCA |
|
| 1301 | TTTCCGACAC GACCATCCTG TCGTCCACCG GCGCGCGCTG CAACCACATC |
|
| 1351 | GACCACGTTA CCTCGCAACT GCCTTACGCC TTAACCGTTG CCGCCGCCGC |
|
| 1401 | CGCATCGGGC TACCTCGCAT TGGGTCTGAC AAAATCCGCG CTGTTGGGCT |
|
| 1451 | TTGGCACGAC AGGCATTGTA TTGGCGGTGC TGATTTTTCT GTTGAAAGAT |
|
| 1501 | AAAAAACGCG CCAACGCCTG A |
[1452]This corresponds to the amino acid sequence <SEQ ID 692; ORF26-1>:
[0000]| 1 | MQLIDYSHSF FSVVPPFLAL ALAVITRRVL LSLGIGILVG VAFLVGGNPV | |
|
| 51 | DGLTHLKDMV VGLAWSDGDW SLGKPKILVF LILLGIFTSL LTYSGSNQAF |
|
| 101 | ADWAKRHIKN RRGAKMLTAC LVFVTFIDDY FHSLAVGAIA RPVTDKFKVS |
|
| 151 | RTKLAYILDS TAAPMCVLMP VSSWGASIIA TLAGLLVTYK ITEYTPMGTF |
|
| 201 | VAMSLMNYYA LFALIMVFVV AWFSFDIGSM ARFEQAALNE AHDETAVSDA |
|
| 251 | TKGRVYALII PVLALIASTV SAMIYTGAQA SETFSILGAF ENTDVNTSLV |
|
| 301 | FGGTCGVLAV VLCTLGTIKT ADYPKAVWQG AKSMFGAIAI LILAWLISTV |
|
| 351 | VGEMHTGDYL STLVAGNIHP GFLPVILFLL ASVMAFATGT SWGTFGIMLP |
|
| 401 | IAAAMAVKVE PALIIPCMSA VMAGAVCGDH CSPISDTTIL SSTGARCNHI |
|
| 451 | DHVTSQLPYA LTVAAAAASG YLALGLTKSA LLGFGTTGIV LAVLIFLLKD |
|
| 501 | KKRANA* |
[1453]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the Hypothetical Transmembrane Protein HI1586 of H. influenzae (Accession Number P44263)
[1454]ORF26 and HI1586 show 53% and 49% amino acid identity in 97 and 221 aa overlap at the N-terminus and C-terminus, respectively:
[0000]
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1455]ORF26 shows 58.2% identity over a 502aa overlap with an ORF (ORF26a) from strain A of N. meningitidis.
[0000]
[1456]The complete length ORF26a nucleotide sequence <SEQ ID 693> is:
[0000]| 1 | ATGCAGCTGA TCGACTATTC ACATTCATTT TTCTCGGTTG TGCCACCCTT | |
|
| 51 | TTTGGCACTG GCACTTGCCG TCATTACCCG CCGCGTACTG CTGTCTTTAG |
|
| 101 | GCATCGGTAT TCTGGTCGGC GTTGCCTTTT TGGTCGGCGG CAACCCCGTC |
|
| 151 | GACGGTCTGA CACACCTGAA AGACATGGTC GTCGGCTTGG CTTGGTCAGA |
|
| 201 | CGGCGATTGG TCGCTGGGCA AACCAAAANT CTTGGTTTTC CTGATACTTT |
|
| 251 | TGGGTATTTT TACTTCCCTG CTGACCTACT CCGGCAGCAA TCAGGCGTTT |
|
| 301 | GCCGACTGGG CAAAACGGCA CATTAAAAAC CGGCGCGGCG CGAAAATGCT |
|
| 351 | GACCGCCTGC CTCGTGTTCG TAACCTTTAT CGACGACTAT TTCCACAGTC |
|
| 401 | TCGCCGTCGG TGCGNTTGCC CGCCCCGTTA CCGACAAGTT TAAAGTTTCC |
|
| 451 | CGCGCCAAAC TCGCCTACAT CCTCGACTCC ACTGCCGCGC CTATGTGCGT |
|
| 501 | GCTGATGCCC GTTTCAAGCT GGGGCGCGTC GATTATCGCC ACGCTTGCCG |
|
| 551 | GACTGCTCGT TACCTACAAA ATCACCGAAT ACACGCCGAT GGGGACGTTT |
|
| 601 | GTCGCCATGA GCCTGATGAA CTATTACGCA CTGTTTGCCC TGATTATGGT |
|
| 651 | GTTCGTCGTC GCATGGTTCT CCTTCGACAT CGGCTCGATG GCACGTTTCG |
|
| 701 | AACAAGCCGC GTTGAACGAA GCCCACGATG AAACTGCCGT TTCAGACGGC |
|
| 751 | AGCTGGGGCA GGGTTTACGC ATTGATTATT CCCGTTTTGG CCTTAATCGC |
|
| 801 | CTCAACGGTT TCCGCCATGA TCTACACCGG TGCACAGGCA AGCGAAACCT |
|
| 851 | TCAGCATTTT GGGTGCATTT GAAAATACGG ACGTGAACAC TTCGCTGGTA |
|
| 901 | TTCGGCGGCA CTTGCGGCGT GCTTGCCGTC GTCCTCTGCA CGCTCGGCAC |
|
| 951 | GATTAAAATC GCCGATTATC CCAAAGCCGT TTGGCAGGGT GCGAAATCCA |
|
| 1001 | TGTTCGGCGC AATCGCCATT TTAATCCTTG CCTGGCTCAT CAGTACGGTT |
|
| 1051 | GTCGGCGAAA TGCACACAGG CGACTACCTC TCCACGCTGG TTGCGGGCAA |
|
| 1101 | CATCCATCCC GGCTTCCTGN CCGTCATCCT TTTCCTGCTC GCCAGCGTGA |
|
| 1151 | TGGCGTTTGC CACAGGCACA AGCTGGGGGA CGTTCGGCAT CATGCTGCCG |
|
| 1201 | ATTGCCGCCG CCATGGCGGT CAAAGTCGAT CCCTCACTGA TTATCCCGTG |
|
| 1251 | TATGTCCGCC GTGATGGCGG GGGCGGTATG CGGCGACCAC TGCTCGCCCA |
|
| 1301 | TTTCCGACAC GACCATCCTG TCGTCCACCG GCGCGCGCTG CAACCACATC |
|
| 1351 | GACCACGTTA CNTCGCAACT GCCTTACGCC TTAACCGTTG CCGCCGCCGC |
|
| 1401 | CGCATCGGGN TACCTCGCAT TGGGTCTGAC AAAATCCGCG CTGTTGGGTT |
|
| 1451 | TTGGCANGAC AGGCATTGTA TTGGCGGTGC TGATTTTTCT GTTGAAAGAT |
|
| 1501 | AAAAAACGCG CCAACGCCTG A |
[1457]This encodes a protein having amino acid sequence <SEQ ID 694>:
[0000]| 1 | MQLIDYSHSF FSVVPPFLAL ALAVITRRVL LSLGIGILVG VAFLVGGNPV | |
|
| 51 | DGLTHLKDMV VGLAWSDGDW SLGKPKXLVF LILLGIFTSL LTYSGSNQAF |
|
| 101 | ADWAKRHIKN RRGAKMLTAC LVFVTFIDDY FHSLAVGAXA RPVTDKFKVS |
|
| 151 | RAKLAYILDS TAAPMCVLMP VSSWGASIIA TLAGLLVTYK ITEYTPMGTF |
|
| 201 | VAMSLMNYYA LFALIMVFVV AWFSFDIGSM ARFEQAALNE AHDETAVSDG |
|
| 251 | SWGRVYALII PVLALIASTV SAMIYTGAQA SETFSILGAF ENTDVNTSLV |
|
| 301 | FGGTCGVLAV VLCTLGTIKI ADYPKAVWQG AKSMFGAIAI LILAWLISTV |
|
| 351 | VGEMHTGDYL STLVAGNIHP GFLXVILFLL ASVMAFATGT SWGTFGIMLP |
|
| 401 | IAAAMAVKVD PSLIIPCMSA VMAGAVCGDH CSPISDTTIL SSTGARCNHI |
|
| 451 | DHVTSQLPYA LTVAAAAASG YLALGLTKSA LLGFGXTGIV LAVLIFLLKD |
|
| 501 | KKRANA* |
[1458]ORF26a and ORF26-1 show 97.8% identity in 506 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1459]ORF26 shows 94.8% and 99% identity in 97 and 206 aa overlap at the N-terminus and C-terminus, respectively, with a predicted ORF (ORF26ng) from N. gonorrhoeae:
[0000]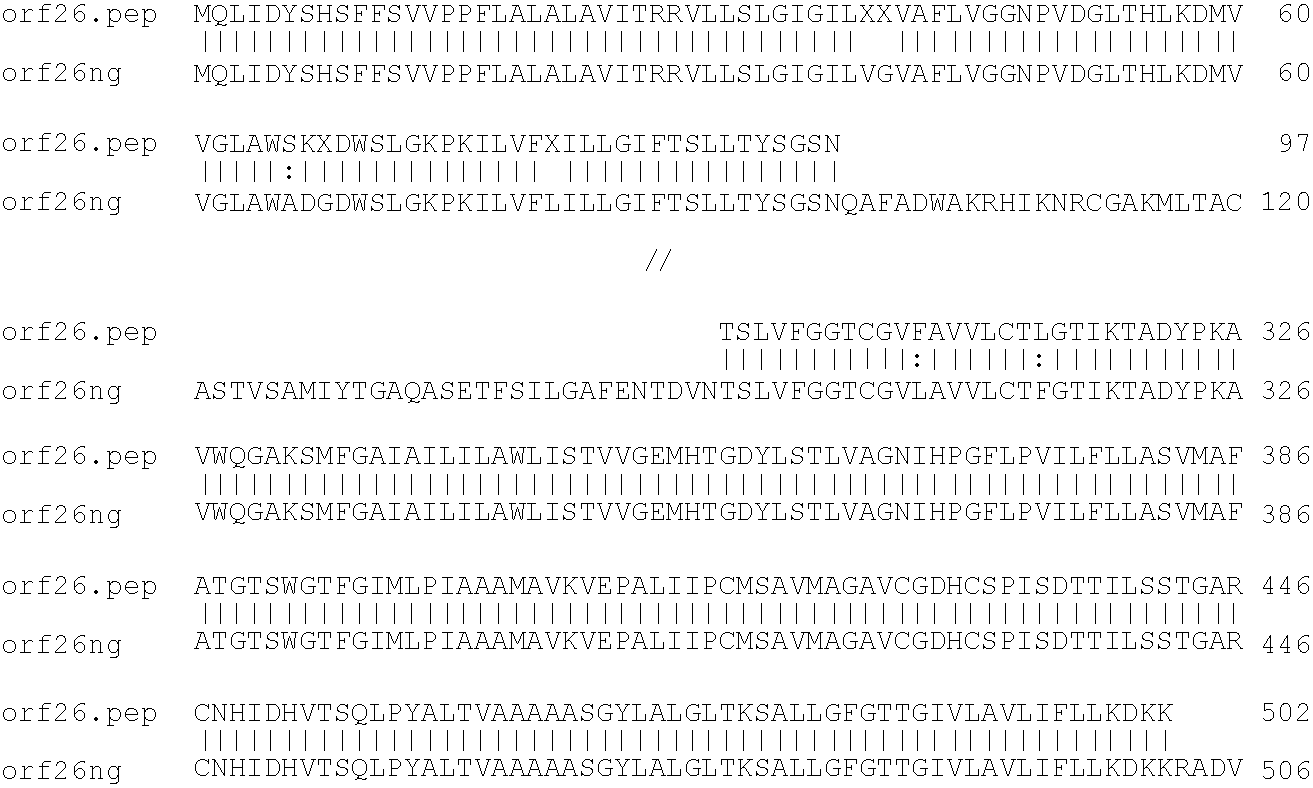
[1460]The complete length ORF26ng nucleotide sequence <SEQ ID 695> is:
[0000]| 1 | ATGCAGCTGA TTGACTATTC ACATTCATTT TTCTCGGTTG TGCCACCCTT | |
|
| 51 | TTTGGCACTG GCACTTGCCG TCATTACCCG CCGCGTACTG CTGTCTTTAG |
|
| 101 | GCATCGGTAT TTTGGTCGGC GTTGCCTTTT TGGTCGGCGG CAACCCCGTC |
|
| 151 | GACGGTCTGA CACACCTGAA AGACATGGTC GTCGGCTTGG CTTGGGCAGA |
|
| 201 | CGGCGATTGG TCGCTGGGCA AACCAAAAAT CTTGGTTTTC CTGATACTTT |
|
| 251 | TGGGCATTTT CACTTCACTG CTGACCTACT CCGGCAGCAA TCAGGCGTTT |
|
| 301 | GCCGACTGGG CAAAACGGCA CATTAAAAAC CGGTGCGGCG CGAAAATGCT |
|
| 351 | GACCGCCTGC CTCGTGTTCG TAACCTTTAT CGACGACTAT TTCCACAGCC |
|
| 401 | TCGCCGTCGG TGCGATTGCC CGCCCCGTTA CCGACAAGTT TAAAGTTTCC |
|
| 451 | CGCGCCAAAC TCGCCTACAT CCTCGACTCC ACTGCCTCGC CCATGTGCGT |
|
| 501 | GCTGATGCCC GTTTCAAGCT GGGGCGCGTC GATTATCGCC ACGCTTGCCG |
|
| 551 | GATTGCTCGT TACCTACAAA ATTACCGAAT ACACGCCGAT GGGGACGTTT |
|
| 601 | GTCGCCATGA GCCTGATGAA CTATTACGCG CTGTTTGCCC TGATTATGGT |
|
| 651 | ATTCGTCGTC GCATGGTTCT CCTTCGACAT CGGCTCGAtg gCGCGTTTCG |
|
| 701 | AACAGGCTGC GTTGAACGAA gcccaggacg aaaccgccgc tTCAGACgCT |
|
| 751 | ACCAAAGGTC GTGTTTACGC ATTGATTATT CCCGTTTTGG CCTTAATCGC |
|
| 801 | CTCAACGGTT TCCGCCATGA TCTACACCGG CGCGCAGGCA AGCGAAACCT |
|
| 851 | TCAGCATTTT GGGGGCATTT GAAAATACCG ACGTAAACAC TTCGCTGGTA |
|
| 901 | TTCGGCGGCA CTTGCGGCGT GCTTGCCGTC GTCCTCTGCA CGTTCGGCAC |
|
| 951 | GATTAAAACC GCCGATTATC CCAAAGCCGT GTGGCAGGGT GCGAAATCCA |
|
| 1001 | TGTTCGGCGC AATCGCCATT TTAATCCTCG CCTGGCTCAT CAGTACGGTT |
|
| 1051 | GTCGGCGAAA TGCACACGGG CGACTACCTC TCCACGCTGG TTGCGGGCAA |
|
| 1101 | CATCCATCCC GGCTTCCTGC CCGTCATCCT CTTCCTGCTC GCCAGCGTGA |
|
| 1151 | TGGCGTTTGC CACAGGCACA AGCTGGGGGA CGTTCGGCAT TATGCTGCCG |
|
| 1201 | ATTGCCGCCG CCATGGCGGT CAAAGTCGAA CCCGCGCTGA TTAtcccGTG |
|
| 1251 | TATGTCCGCA GTAATGGCGG GGGCGGTATG CGGCGACCAC TGTTCGCCCA |
|
| 1301 | TCTCCGACAC GACCATCCTG TCGTCCACCG GCGCGCGCTG CAACCACATC |
|
| 1351 | GACCACGTTA CCTCGCAACT GCCTTATGCC CTGACGGTTG CCGCCGCCGC |
|
| 1401 | CGCATCGGGC TACCTCGCAT TGGGTCTGAC AAAATCCGCG CTGTTGGGCT |
|
| 1451 | TTGGCACGAC CGGTATTGTA TTGGCGGTGC TGATTTTTCT GTTGAAAGAT |
|
| 1501 | AAAAAACGCG CCGACGTTTG A |
[1461]This encodes a protein having amino acid sequence <SEQ ID 696>:
[0000]| 1 | MQLIDYSHSF FSVVPPFLAL ALAVITRRVL LSLGIGILVG VAFLVGGNPV | |
|
| 51 | DGLTHLKDMV VGLAWADGDW SLGKPKILVF LILLGIFTSL LTYSGSNQAF |
|
| 101 | ADWAKRHIKN RCGAKMLTAC LVFVTFIDDY FHSLAVGAIA RPVTDKFKVS |
|
| 151 | RAKLAYILDS TASPMCVLMP VSSWGASIIA TLAGLLVTYK ITEYTPMGTF |
|
| 201 | VAMSLMNYYA LFALIMVFVV AWFSFDIGSM ARFEQAALNE AQDETAASDA |
|
| 251 | TKGRVYALII PVLALIASTV SAMIYTGAQA SETFSILGAF ENTDVNTSLV |
|
| 301 | FGGTCGVLAV VLCTFGTIKT ADYPKAVWQG AKSMFGAIAI LILAWLISTV |
|
| 351 | VGEMHTGDYL STLVAGNIHP GFLPVILFLL ASVMAFAGTFGIMLP |
|
| 401 | IAAAMAVKVE PALIIPCMSA VMAGAVCGDH CSPISDTTIL SSTGARCNHI |
|
| 451 | DHVTSQLPYA LTVAAAAASG YLALGLTKSA LLGFGTTGIV LAVLIFLLKD |
|
| 501 | KKRADV* |
[1462]ORF26ng and ORF26-1 show 98.4% identity in 505 aa overlap:
[0000]
[1463]In addition, ORF26ng shows significant homology to a hypothetical H. influenzae protein:
[0000]
[1464]Based on this analysis, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 83
[1465]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 697>:
[0000]| 1 | ..AAGCAATGGT ATGCCGACGN .AGTATCAAG ACGGAAATGG TTATGGTCAA | |
|
| 51 | CGATGAGCCT GCCAAAATTC TGACTTGGGA TGAAAGCGGC CGATTACTCT |
|
| 101 | CGGAACTGTC TATCCGCCAC CATCAACGCA ACGGGGTGGT TTTGGAGTGG |
|
| 151 | TATGAAGATG GTTCTAAAAA GAGCGAAGT. GTTTATCAGG ATGACAAGTT |
|
| 201 | GGTCAGGAAA ACCCAGTGGG ATAAGGATGG TTATTTAATC GAACCCTGA |
[1466]This corresponds to the amino acid sequence <SEQ ID 698; ORF27>:
[0000]| 1 | ..KQWYADXSIK TEMVMVNDEP AKILTWDESG RLLSELSIRH HQRNGVVLEW | |
|
| 51 | YEDGSKKSEX VYQDDKLVRK TQWDKDGYLI EP* |
[1467]Further work revealed the complete nucleotide sequence <SEQ ID 699>:
[0000]| 1 | ATGAAAAAAT TATCTCGGAT TGTATTTTCA ACTGTCCTGT TGGGTTTTTC | |
|
| 51 | GGCCGCTTTG CCGGCGCAGA CCTATTCTGT TTATTTTAAT CAGAACGGAA |
|
| 101 | AGCTGACGGC GACGATGTCT TCTGCCGCTT ATATCAGGCA ATATAGTGTG |
|
| 151 | GTGGCGGGTA TTGCGCACGC GCAGGATTTT TATTATCCGT CGATGAAGAA |
|
| 201 | ATATTCTGAA CCTTATATCG TTGCTTCAAC GCAAATCAAA TCTTTTGTGC |
|
| 251 | CTACCCTGCA AAACGGTATG TTGATTTTGT GGCATTTTAA TGGTCAGAAA |
|
| 301 | AAAATGGCGG GGGGCTTCAG CAAGGGTAAG CCGGACGGGG AGTGGGTCAA |
|
| 351 | CTGGTATCCG AACGGTAAAA AATCTGCCGT TATGCCTTAT AAAAATGGCT |
|
| 401 | TGAGTGAGGG TACGGGATAC CGCTATTACC GTAACGGCGG CAAGGAAAGC |
|
| 451 | GAAATCCAGT TTAAGCAAAA TAAGGCAAAC GGCGTATGGA AGCAATGGTA |
|
| 501 | TGCCGACGGC AGTATCAAGA CGGAAATGGT TATGGTCAAC GATGAGCCTG |
|
| 551 | CCAAAATTCT GACTTGGGAT GAAAGCGGCC GATTACTCTC GGAACTGTCT |
|
| 601 | ATCCGCCACC ATCAACGCAA CGGGGTGGTT TTGGAGTGGT ATGAAGATGG |
|
| 651 | TTCTAAAAAG AGCGAAGCTG TTTATCAGGA TGACAAGTTG GTCAGGAAAA |
|
| 701 | CCCAGTGGGA TAAGGATGGT TATTTAATCG AACCCTGA |
[1468]This corresponds to the amino acid sequence <SEQ ID 700; ORF27-1>:
[0000]| 1 | MKKLSRIVFS TVLLGFSAAL PAQTYSVYFN QNGKLTATMS SAAYIRQYSV | |
|
| 51 | VAGIAHAQDF YYPSMKKYSE PYIVASTQIK SFVPTLQNGM LILWHFNGQK |
|
| 101 | KMAGGFSKGK PDGEWVNWYP NGKKSAVMPY KNGLSEGTGY RYYRNGGKES |
|
| 151 | EIQFKQNKAN GVWEQWYADG SIKTEMVMVN DEPAKILTWD ESGRLLSELS |
|
| 201 | IRHHQRNGVV LEWYEDGSKK SEAVYQDDKL VRKTQWDKDG YLIEP* |
[1469]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1470]ORF27 shows 91.5% identity over a 82aa overlap with an ORF (ORF27a) from strain A of N. meningitidis:
[0000]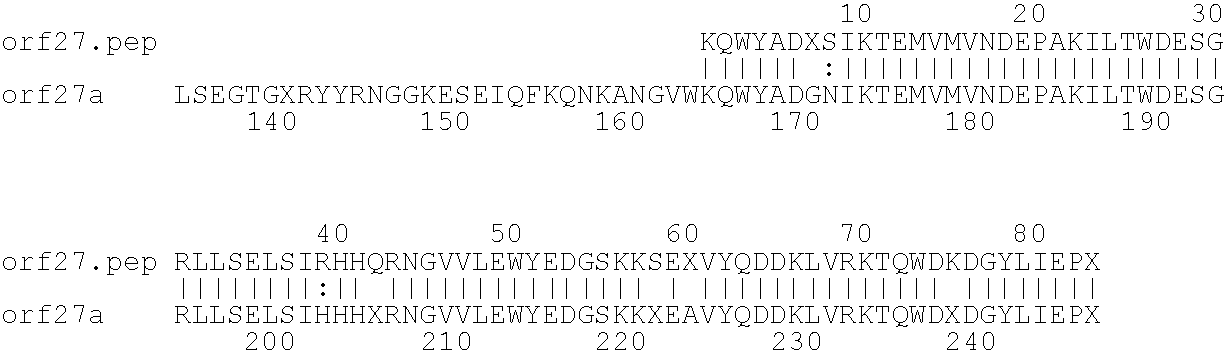
[1471]The complete length ORF27a nucleotide sequence <SEQ ID 701> is:
[0000]| 1 | ATGAAAAAAT TATCTCGGAT TGTATTTTCA ACTGTCCTGT TGGGTTTTTC | |
|
| 51 | GGCCGCTTTG CCGGCGCAGA NCTATTCTGT TTATTTTAAT CAGAACGGGA |
|
| 101 | AACTGACGGC GACGNTGTCT TCTGCCGCNT ATATCAGGCA ATATAGTGTG |
|
| 151 | GCGGAGGGTA TTGCGCACGC GCAGGANTTT TANTATCCGT CGATGAAGAA |
|
| 201 | ATATTCCGAA CCTTATATCG TTGCTTCAAC GCAAATCAAA TCTTTTGTGC |
|
| 251 | CTACCCTGCA AAACGGTATG TTGATTTTGT GGCATTTTAA NGGTCAGAAA |
|
| 301 | AAAATGGCNG GGGGCTTCAG CAAGGGTAAG CCGGACGGGG AGTGGGTCAA |
|
| 351 | CTGGTATCCG AACGGTAAAA AATCTGCCGT TATGCCTTAT AAAAATGGTT |
|
| 401 | TGAGTGAAGG TACGGGGTNN CGCTATTACC GTAACGGCGG CAAGGAAAGC |
|
| 451 | GAAATCCAGT TTAAACAGAA TAAGGCAAAC GGCGTATGGA AGCAATGGTA |
|
| 501 | TGCCGACGGC AATATCAAAA CGGAAATGGT TATGGTCAAT GATGAGCCTG |
|
| 551 | CCAAAATTCT GACATGGGAT GAAAGCGGTC GATTACTCTC GGAACTGTCT |
|
| 601 | ATCCATCATC ATNAACGTAA TGGAGTAGTC TTAGAGTGGT ATGAAGATGG |
|
| 651 | TTCTAAAAAG ANTGAAGCTG TTTATCAGGA TGATAAGTTG GTCAGGAAAA |
|
| 701 | CCCAGTGGGA TAANGATGGT TATTTAATCG AACCCTGA |
[1472]This encodes a protein having amino acid sequence <SEQ ID 702>:
[0000]| 1 | MKKLSRIVFS TVLLGFSAAL PAQXYSVYFN QNGKLTATXS SAAYIRQYSV | |
|
| 51 | AEGIAHAQXF XYPSMKKYSE PYIVASTQIK SFVPTLQNGM LILWHFXGQK |
|
| 101 | KMAGGFSKGK PDGEWVNWYP NGKKSAVMPY KNGLSEGTGX RYYRNGGKES |
|
| 151 | EIQFKQNKAN GVWKQWYADG NIKTEMVMVN DEPAKILTWD ESGRLLSELS |
|
| 201 | IHHHXRNGVV LEWYEDGSKK XEAVYQDDKL VRKTQWDXDG YLIEP* |
[1473]ORF27a and ORF27-1 show 94.7% identity in 245 aa overlap:
[0000]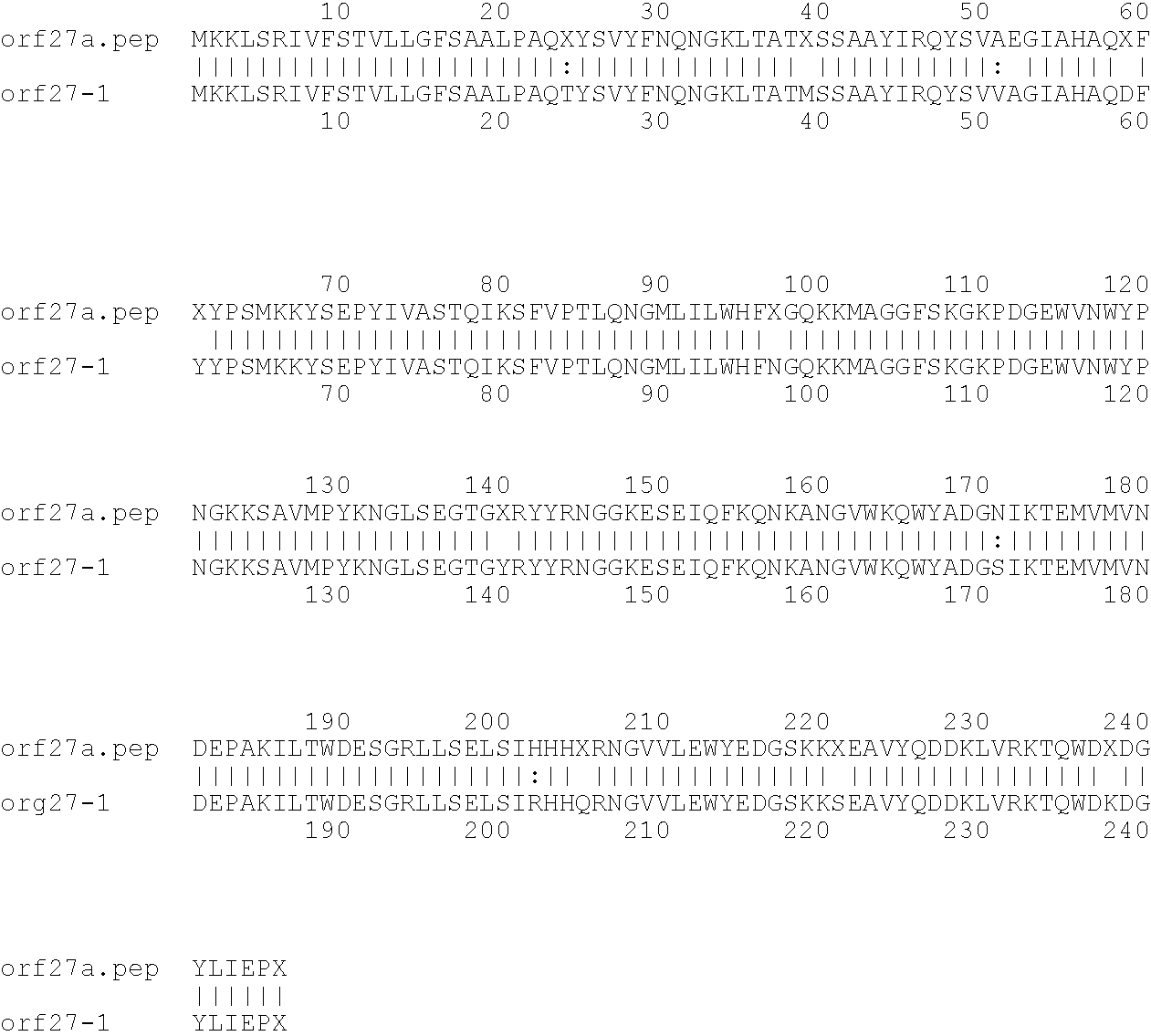
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1474]ORF27 shows 96.3% identity over 82 aa overlap with a predicted ORF (ORF27ng) from N. gonorrhoeae:
[0000]
[1475]The complete length ORF27ng nucleotide sequence <SEQ ID 703> is:
[0000]| 1 | ATGAAGAAAT TATCTCGGAT TGTATTTTCA ATCGTACTGT TGGGTTTTTC | |
|
| 51 | GGCCGCTTTG CCGGCGCAGA CCTATTCTGT TTATTTTAAT CAGAACGGGA |
|
| 101 | AACTGACGGC GACGATGTCT TCTGCCGCTT ATATCAGGCA ATATAGTGTG |
|
| 151 | GCGGCGGGTA TCGCACACGC GCAGGATTTT TATTATCCGT CGATGAAGAA |
|
| 201 | ATATTCCGAA CCTTATATCG TTGCTTCAAC GCAAATCAAA TCTTTTGTGC |
|
| 251 | CTACCCTGCA AAACGGTATG TTGATTTTGT GGCATTTTAA TGGTCAGAAA |
|
| 301 | AAAATGGCGG GGGGCTTCAG CAAGGGTAAG CCGGACGGGG AATGGGTCAA |
|
| 351 | CTGGTATCCG AACGGTAAAA AATCTGCGGT TATGCCTTAT AAAAATGGCT |
|
| 401 | TGAGTGAGGG TACGGGATAC CGTTATTACC GTAACGGCGG CAAGGAAAGC |
|
| 451 | GAAATCCAGT TTAAGCAAAA TAAGGCGAAC GGCGTATGGA AGCAATGGTA |
|
| 501 | TGCCGATGGA AGTATCAAGA CGGAAATGGT TATGGTCAAC GATGAGCCTG |
|
| 551 | CCAAAATTCT GACTTGGGAT GAAAGCGGCC GATTACTTTC GGAACTGTCT |
|
| 601 | ATCCGCCACC ATAAACGCAA CGGGGTGGTT TTGGAGTGGT ATGAAGATGG |
|
| 651 | TTCTAAAAAG AGCGAGGCTG TTTATCAGGA TGACAAGTTG GTCAGGAAAA |
|
| 701 | CCCAATGGGA TAAGGATGGT TATTTAATCG AACCCTGA |
[1476]This encodes a protein having amino acid sequence <SEQ ID 704>:
[0000]| 1 | MKKLSRIVFS IVLLGFSAAL PAQTYSVYFN QNGKLTATMS SAAYIRQYSV | |
|
| 51 | AAGIAHAQDF YYPSMKKYSE PYIVASTQIK SFVPTLQNGM LILWHFNGQK |
|
| 101 | KMAGGFSKGK PDGEWVNWYP NGKKSAVMPY KNGLSEGTGY RYYRNGGKES |
|
| 151 | EIQFKQNKAN GVWKQWYADG SIKTEMVMVN DEPAKILTWD ESGRLLSELS |
|
| 201 | IRHHKRNGVV LEWYEDGSKK SEAVYQDDKL VRKTQWDKDG YLIEP* |
[1477]ORF27ng and ORF27-1 show 98.8% identity in 245 aa overlap:
[0000]
[1478]Based on this analysis, including the putative leader sequence in the gonococcal protein, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1479]ORF27-1 (24.5 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 17A shows the results of affinity purification of the GST-fusion protein, and FIG. 17B shows the results of expression of the His-fusion in E. coli. Purified GST-fusion protein was used to immunise mice, whose sera were used for ELISA, which gave a positive result, confirming that ORF27-1 is a surface-exposed protein and a useful immunogen.
Example 84
[1480]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 705>:
[0000]| 1 | ATGAAATTTA CCAAGCACCC CGTCTGGGCA ATGGCGTTCC GCCCATTTTA | |
|
| 51 | TTCGCTGGCG GCTCTGTACG GCGCATTGTC CGTATTGCTG TGGGGTTTCG |
|
| 101 | GCTACACGGG AACGCACkAG CTGTCCGGTT TCTATTGGCA CGCGCATGAg |
|
| 151 | ATGATTTGGG GTTATGCCGG ACTGGTCGTC ATCGCCTTCC TGCTGACCGC |
|
| 201 | CGTCGCCACT TGGACGGGGC AGCCGCCCAC GCGGGGCGGC GTaTCTGGTC |
|
| 251 | GGCTTGACTA TCTTTTGGCT GGCTGCGCGG ATTGCCGCCT TTATCCCGGG |
|
| 301 | TTGGGGTGCG TCGGCAAGCG GCATACTCGG TACGCTGTTT TTCTGGTACG |
|
| 351 | GCGCGGTGTG CATGGCTTTG CCCGTTATCC GTTCGCAGAA TCAACGCAAC |
|
| 401 | TATGTTgCCG TGTTCGCGCT GTTCGTCTTG GGCGGCACGC ATGCGGCGTT |
|
| 451 | CCACGTCCAG CTGCACAACG GCAACCTAGG CGGACTCTTG AGCGGATTGC |
|
| 501 | AGTCGGGCTT GGTGATG |
[1481]This corresponds to the amino acid sequence <SEQ ID 706; ORF47>:
[0000]| 1 | MKFTKHPVWA MAFRPFYSLA ALYGALSVLL WGFGYTGTHX LSGFYWHAHE | |
|
| 51 | MIWGYAGLVV IAFLLTAVAT WTGQPPTRGG VLVGLTIFWL AARIAAFIPG |
|
| 101 | WGASASGILG TLFFWYGAVC MALPVIRSQN QRNYVAVFAL FVLGGTHAAF |
|
| 151 | HVQLHNGNLG GLLSGLQSGL VM |
[1482]Further work revealed the complete nucleotide sequence <SEQ ID 707>:
[0000]| 1 | ATGAAATTTA CCAAGCACCC CGTCTGGGCA ATGGCGTTCC GCCCATTTTA | |
|
| 51 | TTCGCTGGCG GCTCTGTACG GCGCATTGTC CGTATTGCTG TGGGGTTTCG |
|
| 101 | GCTACACGGG AACGCACGAG CTGTCCGGTT TCTATTGGCA CGCGCATGAG |
|
| 151 | ATGATTTGGG GTTATGCCGG ACTGGTCGTC ATCGCCTTCC TGCTGACCGC |
|
| 201 | CGTCGCCACT TGGACGGGGC AGCCGCCCAC GCGGGGCGGC GTTCTGGTCG |
|
| 251 | GCTTGACTAT CTTTTGGCTG GCTGCGCGGA TTGCCGCCTT TATCCCGGGT |
|
| 301 | TGGGGTGCGT CGGCAAGCGG CATACTCGGT ACGCTGTTTT TCTGGTACGG |
|
| 351 | CGCGGTGTGC ATGGCTTTGC CCGTTATCCG TTCGCAGAAT CAACGCAACT |
|
| 401 | ATGTTGCCGT GTTCGCGCTG TTCGTCTTGG GCGGCACGCA TGCGGCGTTC |
|
| 451 | CACGTCCAGC TGCACAACGG CAACCTAGGC GGACTCTTGA GCGGATTGCA |
|
| 501 | GTCGGGCTTG GTGATGGTGT CGGGTTTTAT CGGTCTGATT GGTACGCGGA |
|
| 551 | TTATTTCGTT TTTTACGTCC AAACGCTTGA ATGTGCCGCA GATTCCCAGT |
|
| 601 | CCGAAATGGG TGGCGCAGGC TTCGCTGTGG CTGCCCATGC TGACTGCCAT |
|
| 651 | GCTGATGGCG CACGGTGTGT TGGCTTGGCT GTCTGCCGTT TTTGCCTTTG |
|
| 701 | CGGCAGGTGT GATTTTTACC GTGCAGGTGT ACCGCTGGTG GTATAAACCC |
|
| 751 | GTGTTGAAAG AGCCGATGCT GTGGATTCTG TTTGCCGGCT ATCTGTTTAC |
|
| 801 | CGGATTGGGG CTGATTGCGG TCGGCGCGTC TTATTTCAAA CCCGCTTTCC |
|
| 851 | TCAATCTGGG TGTGCATCTG ATCGGGGTCG GCGGTATCGG CGTGCTGACT |
|
| 901 | TTGGGCATGA TGGCGCGTAC CGCGCTTGGT CATACGGGCA ATCCGATTTA |
|
| 951 | TCCGCCGCCC AAAGCCGTTC CCGTTGCGTT TTGGCTGATG ATGGCGGCAA |
|
| 1001 | CCGCCGTCCG TATGGTTGCC GTATTTTCTT CCGGCACTGC CTACACGCAC |
|
| 1051 | AGCATCCGCA CCTCTTCGGT TTTGTTTGCA CTCGCGCTTT TGGTGTATGC |
|
| 1101 | GTGGAAGTAT ATTCCTTGGC TGATTCGTCC GCGTTCGGAC GGCAGGCCCG |
|
| 1151 | GTTGA |
[1483]This corresponds to the amino acid sequence <SEQ ID 708; ORF47-1>:
[0000]| 1 | MKFTKHPVWA MAFRPFYSLA ALYGALSVLL WGFGYTGTHE LSGFYWHAHE | |
|
| 51 | MIWGYAGLVV IAFLLTAVAT WTGQPPTRGG VLVGLTIFWL AARIAAFIPG |
|
| 101 | WGASASGILG TLFFWYGAVC MALPVIRSQN QRNYVAVFAL FVLGGTHAAF |
|
| 151 | HVQLHNGNLG GLLSGLQSGL VMVSGFIGLI GTRIISFFTS KRLNVPQIPS |
|
| 201 | PKWVAQASLW LPMLTAMLMA HGVLAWLSAV FAFAAGVIFT VQVYRWWYKP |
|
| 251 | VLKEPMLWIL FAGYLFTGLG LIAVGASYFK PAFLNLGVHL IGVGGIGVLT |
|
| 301 | LGMMARTALG HTGNPIYPPP KAVPVAFWLM MAATAVRMVA VFSSGTAYTH |
|
| 351 | SIRTSSVLFA LALLVYAWKY IPWLIRPRSD GRPG* |
[1484]Computer analysis of this amino acid sequence predicts a leader peptide and also gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1485]ORF47 shows 99.4% identity over a 172aa overlap with an ORF (ORF47a) from strain A of N. meningitidis.
[0000]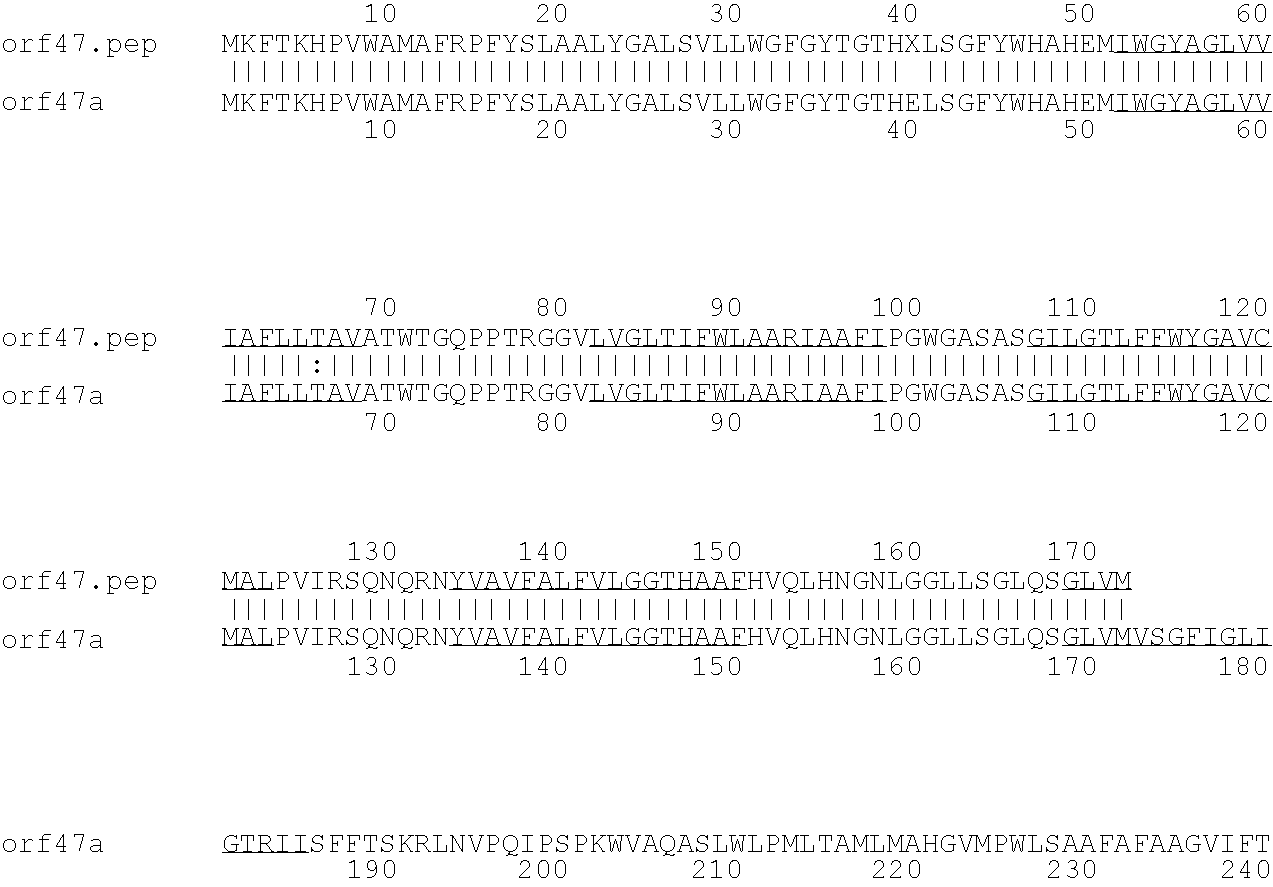
[1486]The complete length ORF47a nucleotide sequence <SEQ ID 709> is:
[0000]| 1 | ATGAAATTTA CCAAGCACCC CGTTTGGGCA ATGGCGTTCC GCCCGTTTTA | |
|
| 51 | TTCACTGGCG GCTCTGTACG GCGCATTGTC CGTATTGCTG TGGGGTTTCG |
|
| 101 | GCTACACGGG AACGCACGAG CTGTCCGGTT TCTATTGGCA CGCGCATGAG |
|
| 151 | ATGATTTGGG GTTATGCCGG ACTGGTCGTC ATCGCCTTCC TGCTGACCGC |
|
| 201 | CGTCGCCACT TGGACGGGGC AGCCGCCCAC GCGGGGCGGC GTTCTGGTCG |
|
| 251 | GCTTGACTAT CTTTTGGCTG GCTGCGCGGA TTGCCGCCTT TATCCCGGGT |
|
| 301 | TGGGGTGCGT CGGCAAGCGG CATACTCGGT ACGCTGTTTT TCTGGTACGG |
|
| 351 | CGCGGTGTGC ATGGCTTTGC CCGTTATCCG TTCGCAGAAT CAACGCAATT |
|
| 401 | ATGTTGCCGT GTTCGCGCTG TTCGTCTTGG GCGGTACGCA CGCGGCGTTC |
|
| 451 | CACGTCCAGC TGCACAACGG CAACCTAGGC GGACTCTTGA GCGGATTGCA |
|
| 501 | GTCGGGCTTG GTGATGGTGT CGGGTTTTAT CGGTCTGATT GGTACGCGGA |
|
| 551 | TTATTTCGTT TTTTACGTCC AAACGGTTGA ATGTGCCGCA GATTCCCAGT |
|
| 601 | CCGAAATGGG TGGCGCAGGC TTCGCTGTGG CTGCCCATGC TGACCGCCAT |
|
| 651 | GCTGATGGCG CACGGCGTGA TGCCTTGGCT GTCGGCGGCT TTCGCGTTTG |
|
| 701 | CGGCAGGTGT GATTTTTACC GTGCAGGTGT ACCGCTGGTG GTATAAGCCT |
|
| 751 | GTGTTGAAAG AGCCGATGCT GTGGATTCTG TTTGCCGGCT ATCTGTTTAC |
|
| 801 | CGGATTGGGG CTGATTGCGG TCGGCGCGTC TTATTTCAAA CCCGCTTTCC |
|
| 851 | TCAATCTGGG TGTGCATCTG ATCGGGGTCG GCGGTATCGG CGTGCTGACT |
|
| 901 | TTGGGCATGA TGGCGCGTAC CGCGCTCGGT CATACGGGCA ATCCGATTTA |
|
| 951 | TCCGCCGCCC AAAGCCGTTC CCGTTGCGTT TTGGCTGATG ATGGCGGCAA |
|
| 1001 | CCGCCGTCCG TATGGTTGCC GTATTTTCTT CCGGCACTGC CTACACGCAC |
|
| 1051 | AGCATACGCA CCTCTTCGGT TTTGTTTGCA CTCGCGCTTT TGGTGTATGC |
|
| 1101 | GTGGAAGTAT ATTCCTTGGC TGATTCGTCC GCGTTCGGAC GGCAGGCCCG |
|
| 1151 | GTTGA |
[1487]This encodes a protein having amino acid sequence <SEQ ID 710>:
[0000]| 1 | MKFTKHPVWA MAFRPFYSLA ALYGALSVLL WGFGYTGTHE LSGFYWHAHE | |
|
| 51 | MIWGYAGLVV IAFLLTAVAT WTGQPPTRGG VLVGLTIFWL AARIAAFIPG |
|
| 101 | WGASASGILG TLFFWYGAVC MALPVIRSQN QRNYVAVFAL FVLGGTHAAF |
|
| 151 | HVQLHNGNLG GLLSGLQSGL VMVSGFIGLI GTRIISFFTS KRLNVPQIPS |
|
| 201 | PKWVAQASLW LPMLTAMLMA HGVMPWLSAA FAFAAGVIFT VQVYRWWYKP |
|
| 251 | VLKEPMLWIL FAGYLFTGLG LIAVGASYFK PAFLNLGVHL IGVGGIGVLT |
|
| 301 | LGMMARTALG HTGNPIYPPP KAVPVAFWLM MAATAVRMVA VFSSGTAYTH |
|
| 351 | SIRTSSVLFA LALLVYAWKY IPWLIRPRSD GRPG* |
[1488]ORF47a and ORF47-1 show 99.2% identity in 384 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1489]ORF47 shows 97.1% identity over 172 aa overlap with a predicted ORF (ORF47ng) from N. gonorrhoeae:
[0000]
[1490]The ORF47ng nucleotide sequence <SEQ ID 711> is predicted to encode a protein comprising amino acid sequence <SEQ ID 712>:
[0000]| 1 | MKFTKHPVWA MAFRPFYSLA ALYGALSVLL WGFGYTGTHE LSGFYWHAHE | |
|
| 51 | MIWGYAGLVV IAFLLTAVAT WTGQPPTRGG VLVGLTAFWL AARIAAFIPG |
|
| 101 | WGAAASGILG TLFFWYGAVC MALPVIRSQN RRNYVAVFAI FVLGGTHAAF |
|
| 151 | HVQLHNGNLG GLLSGLQSGL VMVWGFIGLI GMKIISFFTS KRLKLPQIPS |
|
| 201 | PKWVAHASLW LPMLNAILMA HRVMPWLSAA FPFAAGVIFT VQVYAGGITP |
|
| 251 | IEETSCGSVA GICYRLGNSS G |
[1491]The predicted leader peptide and transmembrane domains are identical (except for an Ile/Ala substitution at residue 87 and an Leu/Ile substitution at position 140) to sequences in the meningococcal protein (see also Pseudomonas stutzeri orf396, accession number e246540):
[0000] |
| INTEGRAL | Likelihood = −5.63 | Transmembrane | 52-68 |
| INTEGRAL | Likelihood = −3.88 | Transmembrane | 169-185 |
| INTEGRAL | Likelihood = −3.08 | Transmembrane | 82-98 |
| INTEGRAL | Likelihood = −1.91 | Transmembrane | 134-150 |
| INTEGRAL | Likelihood = −1.44 | Transmembrane | 107-123 |
| INTEGRAL | Likelihood = −1.38 | Transmembrane | 227-243 |
|
[1492]Further work revealed the complete gonococcal DNA sequence <SEQ ID 713>:
[0000]| 1 | ATGAAATTTA CCAAACATCC CGTCTGGGCA ATGGCGTTCC GCCCGTTTTA | |
|
| 51 | TTCACTGGCG GCACTGTACG GCGCATTGTC CGTATTGCTG TGGGGTTTCG |
|
| 101 | GCTACACGGG AACGCACGAG CTGTCCGGTT TCTATTGGCA CGCGCATGAG |
|
| 151 | ATGATTTGGG GTTATGCCGG TCTCGTCGTC ATCGCCTTCC TGCTGACCGC |
|
| 201 | CGTCGCCACT TGGACGGGAC AGCCGCCCAC GAGGGGCGGC GTTCTGGTCG |
|
| 251 | GCTTGACCGC CTTTTGGCTG GCTGCGCGGA TTGCCGCCTT TATCCCGGGT |
|
| 301 | TGGGGTGCGG CGGCAAGCGG CATACTCGGT ACGCTGTTTT TCTGGTACGG |
|
| 351 | CGCGGTGTGC ATGGCTTTGC CCGTTATCCG TtcgCAAAAC CGGCGCAACT |
|
| 401 | ATGtcgCCGT ATTCGCAATA TTTGTGCTGG GCGGTACGCA TGCGgcgTTC |
|
| 451 | CACGtccAgc tGCACAACGG CAACCTAGGC GGACTCTTGA GCGGATTGCA |
|
| 501 | GTCGGGCCTG GTTATGGTGT CGGGCTTTAT CGGCCTGATT GGGATGAGGA |
|
| 551 | TTATTTCGTT TTTTACGTCC AAACGGTTGA ACGTGCCGCA GATTCCCAGT |
|
| 601 | CCGAAATGGG TGGCGCAGGC TTCGCTGTGG CTACCCATGC TGACCGCCAT |
|
| 651 | ACTGATGGCG CACGGCGTGA TGCCTTGGCT GTCGGCGGCT TTCGCGTTTG |
|
| 701 | CGGCGGGCGT GATTTTTACC GTACAGGTGT ACCGCTGGTG GTATAAACCC |
|
| 751 | GTATTGAAAG AACCGATGCT GTGGATTCTG TTTGCCGGCT ATCTGTTTAC |
|
| 801 | CGGATTGGGG CTGATTGCGG TCGGCGCGTC TTATTTCAAA CCTGCCTTCC |
|
| 851 | TCAATCTGGG CGTACATCTG ATCGGGGTCG GCGGTATCGG CGTGCTGACT |
|
| 901 | TTGGGCATGA TGGCGCGTAC CGCGCTCGGT CATACGGGCA ATTCGATTTA |
|
| 951 | TCCGCCGCCC AAAGCCGTTC CCGTTGCGTT TTGGCTGATG ATGGCGGCAA |
|
| 1001 | CCGCCGTCCG TATGGTTGCC GTATTTTCTT CCGGCACTGC CTACACGCAC |
|
| 1051 | AGCATCCGCA CGTCTTCGGT TTTGTTTGCA CTCGCGCTGC TGGTGTATGC |
|
| 1101 | GTGGAAATAC ATTCCGTGGC TGATCCGTCC GCGTTCGGAC GGCAGGCCCG |
|
| 1151 | GTTGA |
[1493]This encodes a protein having amino acid sequence <SEQ ID 714; ORF47ng-1>:
[0000]| 1 | MKFTKHPVWA MAFRPFYSLA ALYGALSVLL WGFGYTGTHE LSGFYWHAHE | |
|
| 51 | MIWGYAGLVV IAFLLTAVAT WTGQPPTRGG VLVGLTAFWL AARIAAFIPG |
|
| 101 | WGAAASGILG TLFFWYGAVC MALPVIRSQN RRNYVAVFAI FVLGGTHAAF |
|
| 151 | HVQLHNGNLG GLLSGLQSGL VMVSGFIGLI GMRIISFFTS KRLNVPQIPS |
|
| 201 | PKWVAQASLW LPMLTAILMA HGVMPWLSAA FAFAAGVIFT VQVYRWWYKP |
|
| 251 | VLKEPMLWIL FAGYLFTGLG LIAVGASYFK PAFLNLGVHL IGVGGIGVLT |
|
| 301 | LGMMARTALG HTGNSIYPPP KAVPVAFWLM MAATAVRMVA VFSSGTAYTH |
|
| 351 | SIRTSSVLFA LALLVYAWKY IPWLIRPRSD GRPG* |
[1494]ORF47ng-1 and ORF47-1 show 97.4% identity in 384 aa overlap:
[0000]
[1495]Furthermore, ORF47ng-1 shows significant homology to an ORF from Pseudomonas stutzeri:
[0000]
[1496]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 85
[1497]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 715>:
[0000]| 1 | ..ATGCCGTCTG AAGGTTCAGA CGGCmTCGGT GyCGGGGAAy CAGAAGyGGT | |
|
| 51 | AGCGCATGCC CAATGAGACT TCGTGGGTTT TGAAGCGGGT GTTTTCCAAG |
|
| 101 | CGTCCCCAGT TGTGGTAACG GTATCCGGTG TCyAArGTCA GCTTGGGyGT |
|
| 151 | GATGTCGAAa CCGACACCGG CGATGACACC AAGACCyAmG CTGCTGATrC |
|
| 201 | TGTkGCTTTC GTGATAGGsA GGTTTGyTGG kmksAsyTTG TAyrATwkkG |
|
| 251 | CCTssCwsTG kAGmGCCkTk CkyTGGTkkA swGrwArTAG TCGTGGTTTy |
|
| 301 | TkTTyyCACC GAATGAACyT GATGTTTAAC GTGTCCGTAG GCGACGCGCG |
|
| 351 | CGCCGATATA GGGTTTGAAT TTATCGTTGA GTTTGAAATC GTAAATGGCG |
|
| 401 | GACAAGCCGA GAGAAGAAAC GGCGTGGAAG CTGCCGTTTC CCTGATGTTT |
|
| 451 | TGTTTGGGTT TCTTTGTAGT TGTTGTTTAT CTCTTCAGTA ACTTTTTTAG |
|
| 501 | TAGAAGAATT ACTTTCTTTC CATTTTCTGT AACTGGCATA ATCTGCCGCT |
|
| 551 | ATTCTCCAGC CGCCGAAATC .. |
[1498]This corresponds to the amino acid sequence <SEQ ID 716; ORF67>:
[0000]| 1 | ..MPSEGSDGXG XGEXEXVAHA QXDFVGFEAG VFQASPVVVT VSGVXXQLGX | |
|
| 51 | DVETDTGDDT KTXAADXVAF VIGRFXGXXL YXXAXXXXAX XWXXXXSRGF |
|
| 101 | XXHRMNLMFN VSVGDARADI GFEFIVEFEI VNGGQAERRN GVEAAVSLMF |
|
| 151 | CLGFFVVVVY LFSNFFSRRI TFFPFSVTGI ICRYSPAAEI .. |
[1499]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1500]ORF67 shows 51.8% identity over 199 aa overlap with a predicted ORF (ORF67ng) from N. gonorrhoeae:
[0000]
[1501]The ORF67ng nucleotide sequence <SEQ ID 717> is predicted to encode a protein comprising amino acid sequence <SEQ ID 718>:
[0000]| 1 | MPSETVGSIV NVGVDESVGF SPPFPSIQHF YRFHRIHRIR LFRPPGPMQL | |
|
| 51 | NRHSHGSGNL GRGVWATVLS DKFPCGQVRI PACAGMTNFE IAVLSGMTVR |
|
| 101 | VFYCARPAPV NGGRLKMPSE GSDGIGIGES EAVAHAQRGF VGFEAGVFQA |
|
| 151 | SPVVVAVAGV QGQAGRDVYA HARHRAEAQA AAAVAFLIGV FLRMSVRINR |
|
| 201 | NCCVSITRVG GKSTCYFFSR IDAVSDVSVG DARTDIGFEF VVEFEIVNGG |
|
| 251 | QAERRNGVEC AVFLMFRLLV FYVKLVAAKS FIILSFQLFY VHGIFIVVPF |
|
| 301 | PVTGIIRGDA PAAEVVADRH PGVDGMRTDV SEIIAYRAYF VFAWSGWFRI |
|
| 351 | IVGNAFGGVG * |
[1502]Based on the presence of a several putative transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 86
[1503]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 719>
[0000]| 1 | ATGTTTGCTT TTTTAGAAGC CTTTTTTGTC GAATACGGTT ATGCGGCTGT | |
|
| 51 | TTTTTTTGTA TTGGTCATCT GCGGTTTCGG CGTGCCGATT CCCGAGGATT |
|
| 101 | TGACCTTGGT AACAGGCGGC GTGATTTCGG GTATGGGTTA TACCAATCCG |
|
| 151 | CATATTATGT TTGCAGTCGG TATGCTCGGC GTATTGGTCG GGGACGGCAT |
|
| 201 | CATGTTCGCC GCCGGACGAA TTTGGGGGCA GArArTCCTA rGGTTCArAC |
|
| 251 | CTATTGCGsG CATCATGACG CCGrAACGTT ATGAGCAGGT TCAGGAAAAA |
|
| 301 | TTCGACAAAT ACGGTAACTG GGTCTTATTT GTCGCCCGTT TCCTGCCCGG |
|
| 351 | TTTGAGAACG GCCGTATTTG TTACAGCCGG TATCAGCCGC AAGGTTTCAT |
|
| 401 | ACTTGCGTTT TATCATTATG GATGGACTGG CCGCA... |
[1504]This corresponds to the amino acid sequence <SEQ ID 720; ORF78>:
[0000]| 1 | MFAFLEAFFV EYGYAAVFFV LVICGFGVPI PEDLTLVTGG VISGMGYTNP | |
|
| 51 | HIMFAVGMLG VLVGDGIMFA AGRIWGQXXL XFXPIAXIMT PXRYEQVQEK |
|
| 101 | FDKYGNWVLF VARFLPGLRT AVFVTAGISR KVSYLRFIIM DGLAA... |
[1505]Further work revealed the complete nucleotide sequence <SEQ ID 721>:
[0000]| 1 | ATGTTTGCTT TTTTAGAAGC CTTTTTTGTC GAATACGGTT ATGCGGCTGT | |
|
| 51 | TTTTTTTGTA TTGGTCATCT GCGGTTTCGG CGTGCCGATT CCCGAGGATT |
|
| 101 | TGACCTTGGT AACAGGCGGC GTGATTTCGG GTATGGGTTA TACCAATCCG |
|
| 151 | CATATTATGT TTGCAGTCGG TATGCTCGGC GTATTGGTCG GGGACGGCAT |
|
| 201 | CATGTTCGCC GCCGGACGAA TTTGGGGGCA GAAAATCCTA AGGTTCAAAC |
|
| 251 | CTATTGCGCG CATCATGACG CCGAAACGTT ATGAGCAGGT TCAGGAAAAA |
|
| 301 | TTCGACAAAT ACGGTAACTG GGTCTTATTT GTCGCCCGTT TCCTGCCCGG |
|
| 351 | TTTGAGAACG GCCGTATTTG TTACAGCCGG TATCAGCCGC AAGGTTTCAT |
|
| 401 | ACTTGCGTTT TATCATTATG GATGGACTGG CCGCACTGAT TTCCGTCCCT |
|
| 451 | ATTTGGATTT ATCTGGGCGA ATACGGTGCG CACAACATCG ATTGGCTGAT |
|
| 501 | GGCGAAAATG CACAGCCTGC AATCGGGTAT TTTTGTTATC TTGGGTATAG |
|
| 551 | GTGCGACCGT TGTCGCTTGG ATTTGGTGGA AAAAACGCCA ACGTATCCAG |
|
| 601 | TTTTACCGCA GCAAATTGAA AGAAAAGCGG GCGCAACGCA AAGCCGCCAA |
|
| 651 | GGCAGCCAAA AAAGCCGCGC AAAGCAAACA ATAA |
[1506]This corresponds to the amino acid sequence <SEQ ID 722; ORF78-1>:
[0000]| 1 | MFAFLEAFFV EYGYAAVFFV LVICGFGVPI PEDLTLVTGG VISGMGYTNP | |
|
| 51 | HIMFAVGMLG VLVGDGIMFA AGRIWGQKIL RFKPIARIMT PKRYEQVQEK |
|
| 101 | FDKYGNWVLF VARFLPGLRT AVFVTAGISR KVSYLRFIIM DGLAALISVP |
|
| 151 | IWIYLGEYGA HNIDWLMAKM HSLQSGIFVI LGIGATVVAW IWWKKRQRIQ |
|
| 201 | FYRSKLKEKR AQRKAAKAAK KAAQSKQ* |
[1507]Computer analysis of this amino acid sequence predicts several transmembrane domains, and also gave the following results:
[0000]Homology with the dedA Homologue of H. influenzae (Accession Number P45280)
[1508]ORF78 and the dedA homologue show 58% aa identity in 144aa overlap:
[0000]| Orf78: | 4 | FLEAFFVEYGYAAVFFVLVICGFGVPIPEDLTLVTGGVISGM--GYTNPHIMFAVGMLGV | 61 | |
| | FL FF EYGY AV FVL+ICGFGVPIPED+TLV+GGVI+G+ N H+M V M+GV |
| DedA: | 20 | FLIGFFTEYGYWAVLFVLIICGFGVPIPEDITLVSGGVIAGLYPENVNSHLMLLVSMIGV | 79 |
|
| Orf78: | 62 | LVGDGIMFAAGRIWGQXXLXFXPIAXIMTPXRYEQVQEKFDKYGNWVLFVARFLPGLRTA | 121 |
| | L GD M+ GRI+G L F PI I+T R V+EKF +YGN VLFVARFLPGLR |
| DedA: | 80 | LAGDSCMYWLGRIYGTKILRFRPIRRIVTLQRLRMVREKFSQYGNRVLFVARFLPGLRAP | 139 |
|
| Orf78: | 122 | VFVTAGISRKVSYLRFIIMDGLAA | 145 |
| | +++ +GI+R+VSY+RF+++D AA |
| DedA: | 140 | IYMVSGITRRVSYVRFVLIDFCAA | 163 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1509]ORF78 shows 93.8% identity over a 145aa overlap with an ORF (ORF78a) from strain A of N. meningitidis.
[0000]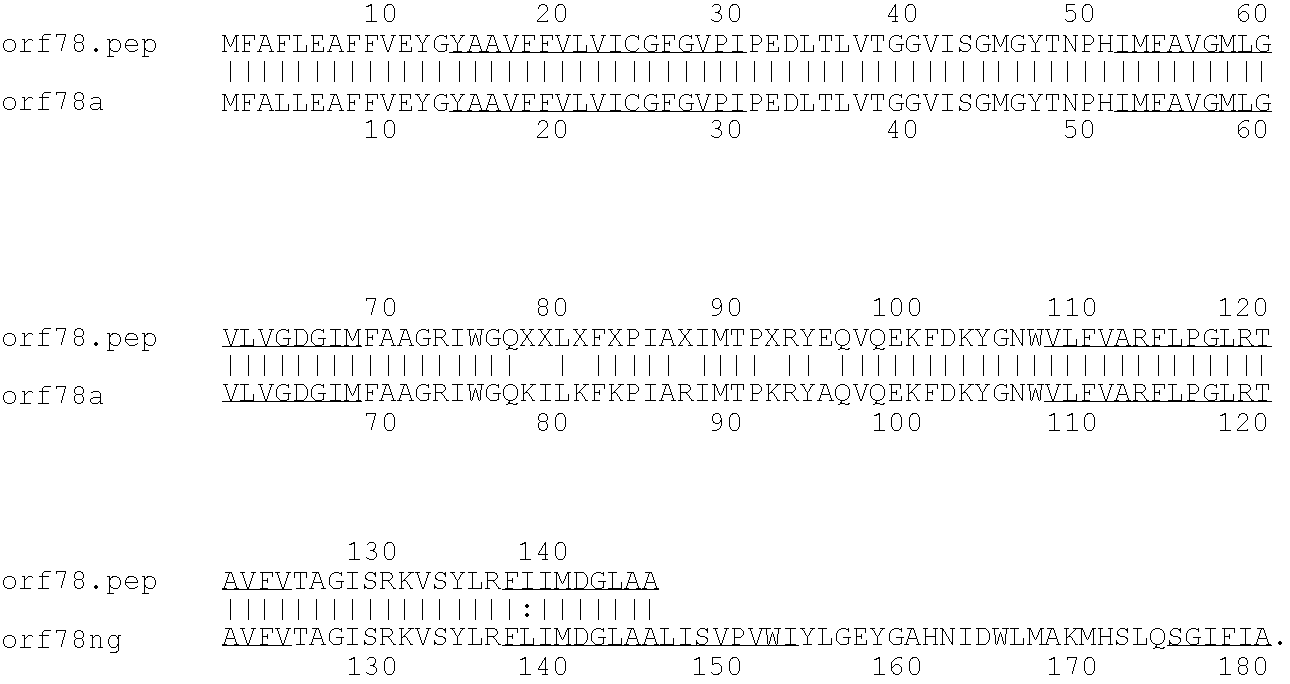
[1510]The complete length ORF78a nucleotide sequence <SEQ ID 723> is:
[0000]| 1 | ATGTTTGCCC TTTTGGAAGC CTTTTTTGTC GAATACGGCT ATGCGGCCGT | |
|
| 51 | GTTTTTCGTT TTGGTCATCT GCGGTTTCGG CGTGCCGATT CCCGAGGATT |
|
| 101 | TGACCTTGGT AACAGGCGGC GTGATTTCGG GTATGGGTTA TACCAATCCG |
|
| 151 | CATATTATGT TTGCAGTCGG TATGCTCGGC GTATTGGTCG GGGACGGCAT |
|
| 201 | CATGTTCGCC GCCGGACGCA TCTGGGGGCA GAAAATCCTC AAGTTCAAAC |
|
| 251 | CGATTGCGCG CATCATGACG CCGAAACGTT ACGCACAGGT TCAGGAAAAA |
|
| 301 | TTCGACAAAT ACGGCAACTG GGTGTTATTT GTCGCTCGTT TCCTGCCCGG |
|
| 351 | TTTGCGGACT GCCGTTTTCG TTACCGCCGG CATCAGCCGC AAAGTATCGT |
|
| 401 | ATCTGCGCTT TCTGATTATG GACGGGCTTG CCGCGCTGAT TTCCGTGCCC |
|
| 451 | GTTTGGATTT ACTTGGGCGA GTACGGCGCG CACAACATCG ATTGGCTGAT |
|
| 501 | GGCGAAAATG CACAGCCTGC AATCCGGCAT CTTCATCGCA TTGGGCGTGC |
|
| 551 | TGGCGGCGGC GCTGGCGTGG TTCTGGTGGC GCAAACGCCG ACATTATCAG |
|
| 601 | CTTTACCGCG CACAATTGAG CGAAAAACGC GCCAAACGCA AGGCGGAAAA |
|
| 651 | GGCAGCGAAA AAAGCGGCAC AGAAGCAGCA GTAA |
[1511]This encodes a protein having amino acid sequence <SEQ ID 724>:
[0000]| 1 | MFALLEAFFV EYGYAAVFFV LVICGFGVPI PEDLTLVTGG VISGMGYTNP | |
|
| 51 | HIMFAVGMLG VLVGDGIMFA AGRIWGQKIL KFKPIARIMT PKRYAQVQEK |
|
| 101 | FDKYGNWVLF VARFLPGLRT AVFVTAGISR KVSYLRFLIM DGLAALISVP |
|
| 151 | VWIYLGEYGA HNIDWLMAKM HSLQSGIFIA LGVLAAALAW FWWRKRRHYQ |
|
| 201 | LYRAQLSEKR AKRKAEKAAK KAAQKQQ* |
[1512]ORF78a and ORF78-1 show 89.0% identity in 227 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1513]ORF78 shows 97.4% identity over 38 aa overlap with a predicted ORF (ORF78ng) from N. gonorrhoeae:
[0000]
[1514]The ORF78ng nucleotide sequence <SEQ ID 725> is predicted to encode a protein comprising amino acid sequence <SEQ ID 726>:
[0000]| 1 | ..YPVLFVARFL PGLRTAVFVT AGISRKVSYL RFLIMDGLAA LISVPVWIYL | |
|
| 51 | GEYGAHNIDW LMAKMHSLQS GIFIALGVLA AALAWFWWRK RRHYQLYRAQ |
|
| 101 | LSEKRAKRKA EKAAKKAAQKN QQ* |
[1515]Further work revealed the complete gonococcal nucleotide sequence <SEQ ID 727>:
[0000]| 1 | atgtttgccc tttTggaagc CTTTTTTGTC GAAtacggCt atgcGGCCGT | |
|
| 51 | GTTTTTCGTT TTGGTCATCT GCGGTTTCGG CGTGCCGATT CCCGAAGATT |
|
| 101 | TGACCTTGGT AACGGGCGGC GTGATTTCGG GTATGGGTTA TACCAATCCG |
|
| 151 | CATATTATGT TTGCGGTCGG TATGCTCGGC GTGTTGGCGG GCGACGGCGT |
|
| 201 | GATGTTTGCC GCCGGACGCA TCTGGGGGCA GAAAATCCTC AAGTTCAAAC |
|
| 251 | CGATTGCGCG CATCATGACG CCGAAACGTT ACGCGCAGGT TCAGGAAAAA |
|
| 301 | TTCGACAAAT ACGGCAACTG GGTTCTGTTT GTCGCCCGTT TCCTGCCGGG |
|
| 351 | TTTGCGGACT GCCGTTTTCG TTACCGCCGG CATCAGCCGC AAAGTATCGT |
|
| 401 | ATCTGCGCTT TCTGATTATG GACGGGCTGG CCGCGCTGAT TTCCGTGCCC |
|
| 451 | GTTTGGATTT ACTTGGGCGA GTACGGCGCG CACAACATCG ATTGGCTGAT |
|
| 501 | GGCGAAAATG CACAGCCTGC AATCGGGCAT CTTCATCGCA TTGGGCGTGC |
|
| 551 | TGGCGGCGGC GCTGGCGTGG TTCTGGTGGC GCAAACGCCG ACATTATCAG |
|
| 601 | CTTTACCGCG CACAATTGAG CGAAAAACGC GCCAAACGCA AGGCGGAAAA |
|
| 651 | GGCAGCGAAA AAAGCGGCAC AGAAGCAGCA GTAa |
[1516]This corresponds to the amino acid sequence <SEQ ID 728; ORF78ng-1>:
[0000]| 1 | MFALLEAFFV EYGYAAVFFV LVICGFGVPI PEDLTLVTGG VISGMGYTNP | |
|
| 51 | HIMFAVGMLG VLAGDGVMFA AGRIWGQKIL KFKPIARIMT PKRYAQVQEK |
|
| 101 | FDKYGNWVLF VARFLPGLRT AVFVTAGISR KVSYLRFLIM DGLAALISVP |
|
| 151 | VWIYLGEYGA HNIDWLMAKM HSLQSGIFIA LGVLAAALAW FWWRKRRHYQ |
|
| 201 | LYRAQLSEKR AKRKAEKAAK KAAQKQQ* |
[1517]ORF78ng-1 and ORF78-1 show 88.1% identity in 227 aa overlap:
[0000]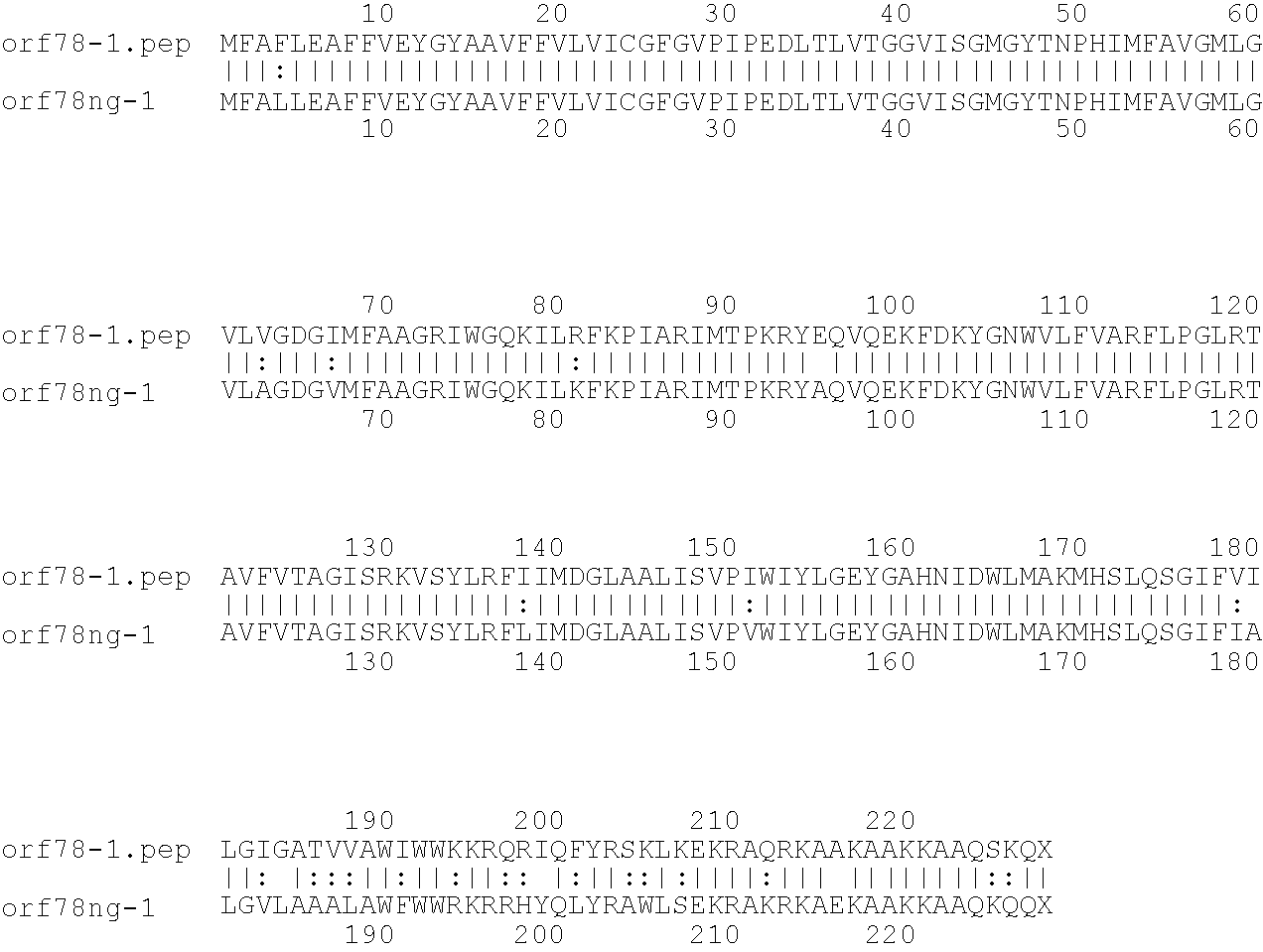
[1518]Furthermore, orf78ng-1 shows homology to the dedA protein from H. influenzae:
[0000]| sp|P45280|YG29_HAEIN HYPOTHETICAL PROTEIN HI1629 >gi|1073983|pir||D64133 | |
| dedA protein (dedA) homolog - Haemophilus influenzae (strain Rd KW20) |
| >gi|1574476 (U32836) dedA protein (dedA) [Haemophilus |
| influenzae] Length = 212 |
| Score = 223 bits (563), Expect = 7e−58 |
| Identities = 108/182 (59%), Positives = 140/182 (76%), Gaps = 2/182 (1%) |
|
| Query: | 5 | LEAFFVEYGYAAVFFVLVICGFGVPIPEDLTLVTGGVISGM--GYTNPHIMFAVGMLGVL | 62 | |
| | L FF EYGY AV FVL+ICGFGVPIPED+TLV+GGVI+G+ N H+M V M+GVL |
| Sbjct: | 21 | LIGFFTEYGYWAVLFVLIICGFGVPIPEDITLVSGGVIAGLYPENVNSHLMLLVSMIGVL | 80 |
|
| Query: | 63 | AGDGVMFAAGRIWGQKILKFKPIARIMTPKRYAQVQEKFDKYGNWVLFVARFLPGLRTAV | 122 |
| | AGD M+ GRI+G KIL+F+PI RI+T +R V+EKF +YGN VLFVARFLPGLR + |
| Sbjct: | 81 | AGDSCMYWLGRIYGTKILRFRPIRRIVTLQRLRMVREKFSQYGNRVLFVARFLPGLRAPI | 140 |
|
| Query: | 123 | FVTAGISRKVSYLRFLIMDGLAALISVPVWIYLGEYGAHNIDWLMAKMHSLQSGIFIALG | 182 |
| | ++ +GI+R+VSY+RF+++D AA+ISVP+WIYLGE GA N+DWL ++ Q I+I +G |
| Sbjct: | 141 | YMVSGITRRVSYVRFVLIDFCAAIISVPIWIYLGELGAKNLDWLHTQIQKGQIVIYIFIG | 200 |
|
| Query: | 183 | VL | 184 |
| | L |
| Sbjct: | 201 | YL | 202 |
[1519]Based on this analysis, including the presence of putative transmembrane domains, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 87
[1520]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 729>:
[0000]| 1 | ATGAAAAAAT TATTGGCGGC CGTGATGATG GCAGGTTTGG CAGGCGCGGT | |
|
| 51 | TTCCGCCGCC GGAGTCCACG TTGAGGACGG CTGGGCGCGC ACCACCGTCG |
|
| 101 | AAGGTATGAA AATAGGCGGC GCGTTCATGA AAATCCACAA CGACGAAGCC |
|
| 151 | AAACAAGACT TTTTGCTCGG CGGAAGCAGC CCCGTTGCCG ACCGCGTCGA |
|
| 201 | AGTGCATACC CACATCAACG ACAACGGCGT GATGCGGATG CGCGAAGTCG |
|
| 251 | AAGGCGGCGT GCCTTTGGAA GCGAAATCCG TTACCGAACT CAAACCCGGC |
|
| 301 | AGCTATCATG TGATGTTTAT GGGTTTGAAA AAACAATTAA AAGAGGGCGA |
|
| 351 | TAAAATTCCC GTTACCCTGA AATTTAAAAA CGCCAAAGCG CAAACCGTCC |
|
| 401 | AACTGGAAGT CAAAATCGCG CCGATGCCGG CAATGAACCA C... |
[1521]This corresponds to the amino acid sequence <SEQ ID 730; ORF79>:
[0000]| 1 | MKKLLAAVMM AGLAGAVSAA GVHVEDGWAR TTVEGMKIGG AFMKIHNDEA | |
|
| 51 | KQDFLLGGSS PVADRVEVHT HINDNGVMRM REVEGGVPLE AKSVTELKPG |
|
| 101 | SYHVMFMGLK KQLKEGDKIP VTLKFKNAKA QTVQLEVKIA PMPAMNH.. |
[1522]Further work revealed the complete nucleotide sequence <SEQ ID 731>:
[0000]| 1 | ATGAAAAAAT TATTGGCGGC CGTGATGATG GCAGGTTTGG CAGGCGCGGT | |
|
| 51 | TTCCGCCGCC GGAGTCCACG TTGAGGACGG CTGGGCGCGC ACCACCGTCG |
|
| 101 | AAGGTATGAA AATAGGCGGC GCGTTCATGA AAATCCACAA CGACGAAGCC |
|
| 151 | AAACAAGACT TTTTGCTCGG CGGAAGCAGC CCCGTTGCCG ACCGCGTCGA |
|
| 201 | AGTGCATACC CACATCAACG ACAACGGCGT GATGCGGATG CGCGAAGTCG |
|
| 251 | AAGGCGGCGT GCCTTTGGAA GCGAAATCCG TTACCGAACT CAAACCCGGC |
|
| 301 | AGCTATCATG TGATGTTTAT GGGTTTGAAA AAACAATTAA AAGAGGGCGA |
|
| 351 | TAAAATTCCC GTTACCCTGA AATTTAAAAA CGCCAAAGCG CAAACCGTCC |
|
| 401 | AACTGGAAGT CAAAATCGCG CCGATGCCGG CAATGAACCA CGGTCATCAC |
|
| 451 | CACGGCGAAG CGCATCAGCA CTAA |
[1523]This corresponds to the amino acid sequence <SEQ ID 732; ORF79-1>:
[0000]| 1 | MKKLLAAVMM AGLAGAVSAA GVHVEDGWAR TTVEGMKIGG AFMKIHNDEA | |
|
| 51 | KQDFLLGGSS PVADRVEVHT HINDNGVMRM REVEGGVPLE AKSVTELKPG |
|
| 101 | SYHVMFMGLK KQLKEGDKIP VTLKFKNAKA QTVQLEVKIA PMPAMNHGHH |
|
| 151 | HGEAHQH* |
[1524]Computer analysis of this amino acid sequence revealed a putative leader peptide and also gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1525]ORF79 shows 94.6% identity over a 147aa overlap with an ORF (ORF79a) from strain A of N. meningitidis.
[0000]
[1526]The complete length ORF79a nucleotide sequence <SEQ ID 733> is:
[0000]| 1 | ATGAAANAAC TATTGGCAGC CGTGATGATG GCAGGTTTGG CAGGCGCGGT | |
|
| 51 | TTCCGCCGCC GGAATCCACG TTGAGGACGG CTGGGCGCGC ACCACCGTCG |
|
| 101 | AAGGTATGAA AATGGGCGGC GCGTTCATGA AAATCCACAA CGACGAAGCC |
|
| 151 | AAACAAGACT TTTTGCTCGG CGGAAGCAGC CCTGTTGCCG ACCGCGTCGA |
|
| 201 | AGTGCATACC CATATCAATG ATAACGGTGT GATGCGGATG CGCGAAGTCG |
|
| 251 | AAGGCGGCGT GCCTTTGGAG GCGAAATCCG TTACCGAACT CAAACCCGGC |
|
| 301 | AGCTATCATG TCATGTTTAT GGGTNTGAAA AAACAATTAA AAGANGGCGA |
|
| 351 | CAAGATTCCC GTTACCCTGA AATTTAAAAA CGCCAAAGCA CAAACCGTCC |
|
| 401 | AACTGGAAGT CAAAACCGCG CCGATGTCGG CAATGGACCA CGGTCATCAC |
|
| 451 | CACGGCGAAG CGCATCAGCA CTAA |
[1527]This encodes a protein having amino acid sequence <SEQ ID 734>:
[0000]| 1 | MKXLLAAVMM AGLAGAVSAA GIHVEDGWAR TTVEGMKMGG AFMKIHNDEA | |
|
| 51 | KQDFLLGGSS PVADRVEVHT HINDNGVMRM REVEGGVPLE AKSVTELKPG |
|
| 101 | SYHVMFMGXK KQLKXGDKIP VTLKFKNAKA QTVQLEVKTA PMSAMDHGHH |
|
| 151 | HGEAHQH* |
[1528]ORF79a and ORF79-1 show 94.9% identity in 157 aa overlap:
[0000]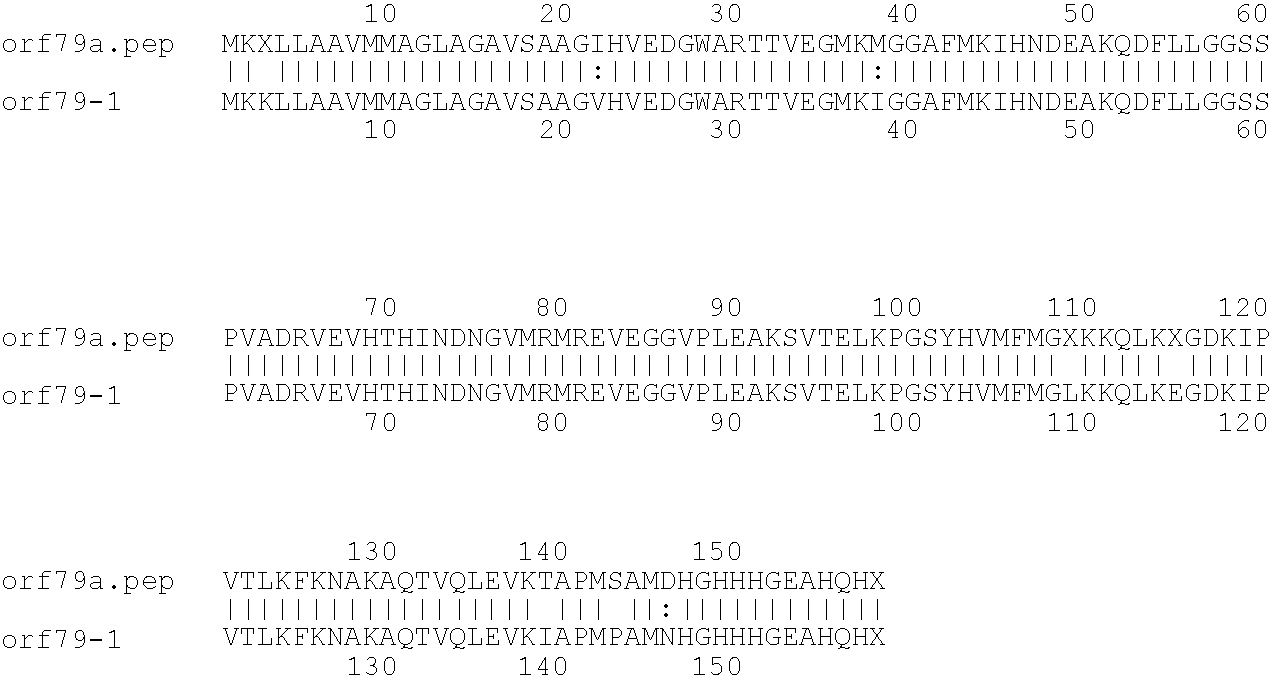
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1529]ORF79 shows 96.1% identity over 76 aa overlap with a predicted ORF (ORF79ng) from N. gonorrhoeae:
[0000]
[1530]An ORF79ng nucleotide sequence <SEQ ID 735> was predicted to encode a protein comprising amino acid sequence <SEQ ID 736>:
[0000]| 1 | ..INDNGVMRMR EVKGGVPLEA KSVTELKPGS YHVMFMGLKK QLKEGDKIPV | |
|
| 51 | TLKFKNAKAQ TVQLEVKTAP MSAMNHGHHH GEAHQH* |
[1531]Further work revealed the complete gonococcal DNA sequence <SEQ ID 737>:
[0000]| 1 | ATGAAAAAAT TATTGGCAGC CGTGATGATG GCAGGTTTGG CAGGCGCGGT | |
|
| 51 | TTccgccgCc GGagTccAtG TCGAggACGG CTGGGCGCGc accaCTGtcg |
|
| 101 | aaggtATgaa aatggGCGGC GCgttCATga aaATCCACAA CGACGaaGcc |
|
| 151 | atacaaGACt ttgtgcTCgg CGGaagcatg cccgttgccg accgcGTCGA |
|
| 201 | AGTGCAtaca cacATCAACG ACAACGGCGT GATGCGTATG CGCGAAGTCA |
|
| 251 | AAGGCGGCGT GCCTTTGGAG GCGAAATCCG TTACCGAACT CAAACCCGGC |
|
| 301 | AGCTATCACG TGATGTTTAT GGGTTTGAAA AAACAACTGA AAGAGGGCGA |
|
| 351 | CAAGATTCCC GTTACCCTGA AATTTAAAAA CGCCAAAGCG CAAACCGTCC |
|
| 401 | AACTGGAAGT CAAAACCGCG CCGATGTCGG CAATGAACCA CGGTCATCAC |
|
| 451 | CACGGCGAAG CGCATCAGCA CTAA |
[1532]This corresponds to the amino acid sequence <SEQ ID 738; ORF79ng-1>:
[0000]| 1 | MKKLLAAVMM AGLAGAVSAA GVHVEDGWAR TTVEGMKMGG AFMKIHNDEA | |
|
| 51 | IQDFVLGGSM PVADRVEVHT HINDNGVMRM REVKGGVPLE AKSVTELKPG |
|
| 101 | SYHVMFMGLK KQLKEGDKIP VTLKFKNAKA QTVQLEVKTA PMSAMNHGHH |
|
| 151 | HGEAHQH* |
[1533]ORF79ng-1 and ORF79-1 show 95.5% identity in 157 aa overlap:
[0000]
[1534]Furthermore, ORF79ng-1 shows significant homology to a protein from Aquifex aeolicus:
[0000]| gi|2983695 (AE000731) putative protein [Aquifex aeolicus] Length = 151 | |
| Score = 63.6 bits (152), Expect = 6e−10 |
| Identities = 38/114 (33%), Positives = 58/114 (50%), Gaps = 1/114 (0%) |
|
| Query: | 24 | VEDGWARTTVEGMKMGGAFMKIHNDEAIQDFVLGGSMPVADRVEVHTHINDNGVMRMREV | 83 | |
| | V+ W G M I N+ D+++G +A RVE+H + +N V +M |
| Sbjct: | 27 | VKHPWVMEPPPGPNTTMMGMIIVNEGDEPDYLIGAKTDIAQRVELHKTVIENDVAKMVPQ | 86 |
|
| Query: | 84 | KGGVPLEAKSVTELKPGSYHVMFMGLKKQLKEGDKIPVTLKFKNAKAQTVQLEV | 137 |
| | + + + K E K YHVM +GLKK++KEGDK+ V L F+ + TV+ V |
| Sbjct: | 87 | ER-IEIPPKGKVEFKHHGYHVMIIGLKKRIKEGDKVKVELIFEKSGKITVEAPV | 139 |
[1535]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1536]ORF79-1 (15.6 kDa) was cloned in the pET vector and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 18A shows the results of affinity purification of the His-fusion protein. Purified His-fusion protein was used to immunise mice, whose sera were used for ELISA (positive result) and FACS analysis (FIG. 18B) These experiments confirm that ORF79-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 88
[1537]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 739>:
[0000]| 1 | ATGACGGTAA CTGCGGCCGA AGGCGGCAAA GCTGCCAAGG CGTTAAAAAA | |
|
| 51 | ATATCTGATT ACGGGCATTT TGGTCTGGCT GCCGATTGCG GTAACGGTTT |
|
| 101 | GGGTGGTTTC CTATATCGTT TCCGCGTCCG ATCAGCTCGT CAACCTGCTG |
|
| 151 | CCGAAGCAAT GGCGGCCGCA ATATGTTTTG GGGTTTAATA TCCCGGGGCT |
|
| 201 | GGGCGTTATC GTTGCCATTG CCGTATTGTT TGTAACCGGA TTGTTTGCCG |
|
| 251 | CCAACGTATT GGGTCGGCAG ATCCTCGCCG CGTGGGACAG CCTGTTGGGG |
|
| 301 | CGGATTCCGG TTGTGAAAtC CATCTATTCG AGTGTGAAAA AAGTATCCGA |
|
| 351 | ATacgTGCTG TCCGACAGCA GCCGTTCGTT TAAAACGCCG GTACTCGTGC |
|
| 401 | CGTTTCCCCA GCCCGGTATT TGGACGATyG CTTTCGTGTC AGGGCAGGTG |
|
| 451 | TCGAATGCGG TTAAGGCCGC ATTGCCGAAs GACGGCGATT ATCTTTCCGT |
|
| 501 | GTATGTTCCG ACCACGCCGA ATCCGACCGG CGGTTACTAT ATTATGGTAA |
|
| 551 | AGAAAAGCGA TGTGCGCGAA CTCGATATGA GCGTGGACGA AsCATTGAAA |
|
| 601 | TATGTGATTT CGCTGGGTAT GGTCATCCCT GACGACCTGC CCGTCAAAAC |
|
| 651 | ATTGGCAsGA CCTATGCCGT CTGAAAAGGC GGATTTGCCC GAACAACAAT |
|
| 701 | AA |
[1538]This corresponds to the amino acid sequence <SEQ ID 740; ORF98>:
[0000]| 1 | MTVTAAEGGK AAKALKKYLI TGILVWLPIA VTVWVVSYIV SASDQLVNLL | |
|
| 51 | PKQWRPQYVL GFNIPGLGVI VAIAVLFVTG LFAANVLGRQ ILAAWDSLLG |
|
| 101 | RIPVVKSIYS SVKKVSEYVL SDSSRSFKTP VLVPFPQPGI WTIAFVSGQV |
|
| 151 | SNAVKAALPX DGDYLSVYVP TTPNPTGGYY IMVKKSDVRE LDMSVDEXLK |
|
| 201 | YVISLGMVIP DDLPVKTLAX PMPSEKADLP EQQ* |
[1539]Further work revealed the complete nucleotide sequence <SEQ ID 741>:
[0000]| 1 | ATGACGGAAC nTGCGGCCGA AGGCGGCAAA GCTGCCAArG CGTTAAAAAA | |
|
| 51 | ATATCTGATT ACGGGCATTT TGGTCTGGCT GCCGATTGCG GTAACGGTTT |
|
| 101 | GGGTGGTTTC CTATATCGTT TCCGCGTCCG ATCAGCTCGT CAACCTGCTG |
|
| 151 | CCGAAGCAAT GGCGGCCGCA ATATGTTTTG GGGTTTAATA TCCCGGGGCT |
|
| 201 | GGGCGTTATC GTTGCCATTG CCGTATTGTT TGTAACCGGA TTGTTTGCCG |
|
| 251 | CCAACGTATT GGGTCGGCAG ATCCTCGCCG CGTGGGACAG CCTGTTGGGG |
|
| 301 | CGGATTCCGG TTGTGAAATC CATCTATTCG AGTGTGAAAA AAGTATCCGA |
|
| 351 | ATCGCTGCTG TCCGACAGCA GCCGTTCGTT TAAAACGCCG GTACTCGTGC |
|
| 401 | CGTTTCCCCA GCCCGGTATT TGGACGATTG CTTTCGTGTC AGGGCAGGTG |
|
| 451 | TCGAATGCGG TTAAGGCCGC ATTGCCGAAG GACGGCGATT ATCTTTCCGT |
|
| 501 | GTATGTTCCG ACCACGCCGA ATCCGACCGG CGGTTACTAT ATTATGGTAA |
|
| 551 | AGAAAAGCGA TGTGCGCGAA CTCGATATGA GCGTGGACGA AGCATTGAAA |
|
| 601 | TATGTGATTT CGCTGGGTAT GGTCATCCCT GACGACCTGC CCGTCAAAAC |
|
| 651 | ATTGGCAGGA CCTATGCCGT CTGAAAAGGC GGATTTGCCC GAACAACAAT |
|
| 701 | AA |
[1540]This corresponds to the amino acid sequence <SEQ ID 742; ORF98-1>:
[0000]| 1 | MTEXAAEGGK AAKALKKYLI TGILVWLPIA VTVWVVSYIV SASDQLVNLL | |
|
| 51 | PKQWRPQYVL GFNIPGLGVI VAIAVLFVTG LFAANVLGRQ ILAAWDSLLG |
|
| 101 | RIPVVKSIYS SVKKVSESLL SDSSRSFKTP VLVPFPQPGI WTIAFVSGQV |
|
| 151 | SNAVKAALPK DGDYLSVYVP TTPNPTGGYY IMVKKSDVRE LDMSVDEALK |
|
| 201 | YVISLGMVIP DDLPVKTLAG PMPSEKADLP EQQ* |
[1541]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1542]ORF98 shows 96.1% identity over a 233aa overlap with an ORF (ORF98a) from strain A of N. meningitidis.
[0000]
[1543]The complete length ORF98a nucleotide sequence <SEQ ID 743> is:
[0000]| 1 | ATGACGGAAC CTGCGGCCGA AGGCGGCAAA GCTGCCAAGG CGTTAAAAAA | |
|
| 51 | ATATCTGATT ACGGGCATTT TGGTCTGGCT GCCGATTGCG GTAACGGTTT |
|
| 101 | GGGTGGTTTC CTATATCGTT TCCGCGTCCG ATCAGCTCGT CAACCTGCTG |
|
| 151 | CCGAAGCAAT GGCGGCCGCA ATATGTTTTG GGGTTTAATA TCCCGGGGCT |
|
| 201 | GGGCGTTATC GTTGCCATTG CCGTATTGTT TGTAACCGGA TTATTTGCCG |
|
| 251 | CAAACGTATT GGGCCGGCAG ATTCTTGCCG CGTGGGACAG CTTGTTGGGG |
|
| 301 | CGGATTCCGG TTGTGAAGTC CATCTATTCG AGTGTGAAAA AAGTATCCGA |
|
| 351 | NTCGTTGCTG TCCGACAGCA GCCGTTCGTT TAAAACACCA GTACTCGTGC |
|
| 401 | CGTTTCCCCA ATCGGGTATT TGGACAATCG CATTCGTGTC CGGTCAGGTG |
|
| 451 | TCGAATGCGG TTAAGGCCGC ATTGCCGAAG GACGGCGATT ATCTTTCCGT |
|
| 501 | GTATGTTCCG ACCACGCCGA ATCCGACCGG CGGTTACTAT ATTATGGTAA |
|
| 551 | AGAAAAGCGA TGTGCGCGAA CTCGATATGA GCGTGGACGA AGCGTTGAAA |
|
| 601 | TATGTGATTT CGCTGGGTAT GGTCATCCCT GACGACCTGC CCGTCAAAAC |
|
| 651 | ATTGGCAGGA CCTATGCCGT CTGAAAAGGC GGATTTGCCC GAACAACAAT |
|
| 701 | AA |
[1544]This encodes a protein having amino acid sequence <SEQ ID 744>:
[0000]| 1 | MTEPAAEGGK AAKALKKYLI TGILVWLPIA VTVWVVSYIV SASDQLVNLL | |
|
| 51 | PKQWRPQYVL GFNIPGLGVI VAIAVLFVTG LFAANVLGRQ ILAAWDSLLG |
|
| 101 | RIPVVKSIYS SVKKVSXSLL SDSSRSFKTP VLVPFPQSGI WTIAFVSGQV |
|
| 151 | SNAVKAALPK DGDYLSVYVP TTPNPTGGYY IMVKKSDVRE LDMSVDEALK |
|
| 201 | YVISLGMVIP DDLPVKTLAG PMPSEKADLP EQQ* |
[1545]ORF98a and ORF98-1 show 98.7% identity in 233 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1546]ORF98 shows 95.3% identity over a 233 aa overlap with a predicted ORF (ORF98ng) from N. gonorrhoeae:
[0000]
[1547]The complete length ORF98ng nucleotide sequence <SEQ ID 745> is predicted to encode a protein having amino acid sequence <SEQ ID 746>:
[0000]| 1 | MTEPAAEGGK AAKALKKYLI TGILVWLPIA VTVWVVSYIV SASDQLVNLL | |
|
| 51 | PKQWRPQYVL GFNIPGLGVI VAIAVLFVTG LFAANVLGRQ ILAAWDSLLX |
|
| 101 | RIPVVKSIYS SVKKVSESLL SDSSRSFKTP VLVPFPQSGI WTIAFVSGQV |
|
| 151 | SNAVKAALPQ DGDYLSVYVP TTPNPTGGYY IMVKKSDVRE LDMSVDEALK |
|
| 201 | YVISLGMVIP DDLPVKTLAG PMPPEKAELP EQQ* |
[1548]Further work revealed the complete nucleotide sequence <SEQ ID 747>:
[0000]| 1 | ATGACGGAAC CTGCGGCCGA AGGCGGCAAA GCTGCCAAGG CGTTAAAAAA | |
|
| 51 | ATATCTGATT ACAGGCATTT TGGTCTGGCT GCCGATTGCG GTAACGGTTT |
|
| 101 | GGGTGGTTTC CTATATCGTT TCCGCGTCCG ACCAGCTTGT CAACCTGCTG |
|
| 151 | CCGAAGCAAT GGCGGCCGCA ATATGTTTTG GGGTTTAATA TCCCCGGGCT |
|
| 201 | CGGCGTTATT GTTGCCATTG CCGTATTGTT TGTAACCGGA TTATTTGCCG |
|
| 251 | CAAACGTGTT GGGCCGGCAG ATTCTTGCCG CGTGGGACAG CCTGTTgggg |
|
| 301 | cggaTTCCGG TTGTCAAATC CATCTATTCG AGTGTGAAAA AAGTATCCGA |
|
| 351 | ATCGCTGCTG TCCGACAGCA GCCGTTCGTT TAAAACGCCG GTACTCGTGC |
|
| 401 | CGTTTCCCCA ATCGGGTATT TGGACAATCG CATTCGTGTC CGGTCAGGTG |
|
| 451 | TCGAATGCGG TTAAGGCCGC ATTGCCGCAG GATGGCGATT ATCTTTCCGT |
|
| 501 | GTATGTCCCG ACCACGCCCA ACCCGACCGG CGGTTACTAT ATTATGGTAA |
|
| 551 | AGAAAAGCGA TGTGCGCGAA CTCGATATGA GCGTGGACGA AGCGTTGAAA |
|
| 601 | TATGTGATTT CGCTGGGTAT GGTCATCCCT GACGACCTGC CCGTCAAAAC |
|
| 651 | ATTGGCAGGA CCTATGCCGC CTGAAAAGGC GGAGTTGCCC GAACAACAAT |
|
| 701 | AA |
[1549]This corresponds to the amino acid sequence <SEQ ID 748; ORF98ng-1>:
[0000]| 1 | MTEPAAEGGK AAKALKKYLI TGILVWLPIA VTVWVVSYIV SASDQLVNLL | |
|
| 51 | PKQWRPQYVL GFNIPGLGVI VAIAVLFVTG LFAANVLGRQ ILAAWDSLLG |
|
| 101 | RIPVVKSIYS SVKKVSESLL SDSSRSFKTP VLVPFPQSGI WTIAFVSGQV |
|
| 151 | SNAVKAALPQ DGDYLSVYVP TTPNPTGGYY IMVKKSDVRE LDMSVDEALK |
|
| 201 | YVISLGMVIP DDLPVKTLAG PMPPEKAELP EQQ* |
[1550]ORF98ng-1 and ORF98-1 show 97.9% identity in 233 aa overlap:
[0000]
[1551]Based on this analysis, including the fact that the putative transmembrane domains in the gonococcal protein are identical to the sequences in the meningococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 89
[1552]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 749>:
[0000]| 1 | ATgAAAACGG TAGTCTGGAT TGTCGTCCTG TTTGCCGCCG CCGTCGGACT | |
|
| 51 | GGCGCTGGCT TCGGGCATTT ACACCGGCGA CGTGTATATC GTACTCGGAC |
|
| 101 | AGACCATGCT CAGAATCAAC CTGCACGCCT TTGTGTTAGG TTCGCTGATT |
|
| 151 | GCCGTCGTGG TGTGGTATTT CTTGTTTAAA TTCATTATCG GsGgTACTCA |
|
| 201 | ATATCCCCGA AAAGATGCAG CGTTTCGGTT CGGCnCGTAA AGGCCkCAAG |
|
| 251 | ssCGsGCTTG CCTTGAACAA GGCGGGTTTG GCGTATTTTG AAGGGCGTTT |
|
| 301 | TGAAAAGGCG GAACTAGAAG CCTCACGCGT GTTGGTCAAC AAAGtAGGCC |
|
| 351 | GaGAGACAAC CGGACTTTGG CATTGATGCT GrGCGCGCAC GCCGCCGGAC |
|
| 401 | AGATGGAAAA CATCGAssTG CGCGACCGTT ATCTTGCGGA AATCGCCAAA |
|
| 451 | CTGCCGGAAA AACAGCAGCT TTCCCGTTAT CTTTTGTTGG CGGAATCGGC |
|
| 501 | GTTGAACCGG CGCGATTACG AAGCGGCGGA AGCCAATCTT CATGCGGCGG |
|
| 551 | CGAAGATGAA TGCCAACCTT ACGCGCCTCG TGCGTCTGCA .ATTCGTTAC |
|
| 601 | GCTTTCGACA GGGGCGACGC GTTGCAGGTT CTGGCAAAAA CCGAAAAACT |
|
| 651 | TTCCAAGGCG GGCGCGTTGG GCAAATCGGA AATGGAACGG TATCAAAATT |
|
| 701 | GGGCATATCC GTCGCCAGCT GGCGGATGCT GCCGATGCCG CCGCTTTGAA |
|
| 751 | AACCTGCCTG AAGCGGATTC CCGACAGCCT CAAAAACGGG GAATTGAGCG |
|
| 801 | TATCGGTTGC GGAAAAGTAC GAACGTTTGG GACTGTATGC CGATGCGGTC |
|
| 851 | AAATGGGTCA AACAGCATTA TCCGCAsAAC CGCCGCCCCG AGCTTTTGGA |
|
| 901 | AGCCTTTGTC GAAAGCGTGC GCTTTTTGGG CGAGCGCGAA CAGCAGAAAG |
|
| 951 | CCATCGATTT TGCCGATGCT TGGCTGAAAG AACAGCCCGA TAACGCGCTT |
|
| 1001 | CTGCTGATGT ATCTCGGTCG GCTCGCCTTC GGCCGCAAAC TTTGGGGCAA |
|
| 1051 | GGCAAAAGGC TACCTTGAAG CGAGCATTGC ATTAAAGCCG AGTATTTCCG |
|
| 1101 | CGCGTTTGGT TCTAACAAAG GTTTTCGACG AAATCGGAGA ACCGCAGAAG |
|
| 1151 | GCGGAGGCGC AC... |
[1553]This corresponds to the amino acid sequence <SEQ ID 750; ORF100>:
[0000]| 1 | MKTVVWIVVL FAAAVGLALA SGIYTGDVYI VLGQTMLRIN LHAFVLGSLI | |
|
| 51 | AVVVWYFLFK FIIGVLNIPE KMQRFGSARK GXKXXLALNK AGLAYFEGRF |
|
| 101 | EKAELEASRV LVNKVGRDNR TLALMLXAHA AGQMENIXXR DRYLAEIAKL |
|
| 151 | PEKQQLSRYL LLAESALNRR DYEAAEANLH AAAKMNANLT RLVRLXIRYA |
|
| 201 | FDRGDALQVL AKTEKLSKAG ALGKSEMERY QNWAYRRQLA DAADAAALKT |
|
| 251 | CLKRIPDSLK NGELSVSVAE KYERLGLYAD AVKWVKQHYP XNRRPELLEA |
|
| 301 | FVESVRFLGE REQQKAIDFA DAWLKEQPDN ALLLMYLGRL AFGRKLWGKA |
|
| 351 | KGYLEASIAL KPSISARLVL TKVFDEIGEP QKAEAH... |
[1554]Further work revealed the complete nucleotide sequence <SEQ ID 751>:
[0000]| 1 | ATGAAAACGG TAGTCTGGAT TGTCGTCCTG TTTGCCGCCG CCGTCGGACT | |
|
| 51 | GGCGCTGGCT TCGGGCATTT ACACCGGCGA CGTGTATATC GTACTCGGAC |
|
| 101 | AGACCATGCT CAGAATCAAC CTGCACGCCT TTGTGTTAGG TTCGCTGATT |
|
| 151 | GCCGTCGTGG TGTGGTATTT CTTGTTTAAA TTCATTATCG GCGTACTCAA |
|
| 201 | TATCCCCGAA AAGATGCAGC GTTTCGGTTC GGCGCGTAAA GGCCGCAAGG |
|
| 251 | CCGCGCTTGC CTTGAACAAG GCGGGTTTGG CGTATTTTGA AGGGCGTTTT |
|
| 301 | GAAAAGGCGG AACTAGAAGC CTCACGCGTG TTGGTCAACA AAGAGGCCGG |
|
| 351 | AGACAACCGG ACTTTGGCAT TGATGCTGGG CGCGCACGCC GCCGGACAGA |
|
| 401 | TGGAAAACAT CGAGCTGCGC GACCGTTATC TTGCGGAAAT CGCCAAACTG |
|
| 451 | CCGGAAAAAC AGCAGCTTTC CCGTTATCTT TTGTTGGCGG AATCGGCGTT |
|
| 501 | GAACCGGCGC GATTACGAAG CGGCGGAAGC CAATCTTCAT GCGGCGGCGA |
|
| 551 | AGATGAATGC CAACCTTACG CGCCTCGTGC GTCTGCAACT TCGTTACGCT |
|
| 601 | TTCGACAGGG GCGACGCGTT GCAGGTTCTG GCAAAAACCG AAAAACTTTC |
|
| 651 | CAAGGCGGGC GCGTTGGGCA AATCGGAAAT GGAACGGTAT CAAAATTGGG |
|
| 701 | CATACCGCCG CCAGCTGGCG GATGCTGCCG ATGCCGCCGC TTTGAAAACC |
|
| 751 | TGCCTGAAGC GGATTCCCGA CAGCCTCAAA AACGGGGAAT TGAGCGTATC |
|
| 801 | GGTTGCGGAA AAGTACGAAC GTTTGGGACT GTATGCCGAT GCGGTCAAAT |
|
| 851 | GGGTCAAACA GCATTATCCG CACAACCGCC GCCCCGAGCT TTTGGAAGCC |
|
| 901 | TTTGTCGAAA GCGTGCGCTT TTTGGGCGAG CGCGAACAGC AGAAAGCCAT |
|
| 951 | CGATTTTGCC GATGCTTGGC TGAAAGAACA GCCCGATAAC GCGCTTCTGC |
|
| 1001 | TGATGTATCT CGGTCGGCTC GCCTACGGCC GCAAACTTTG GGGCAAGGCA |
|
| 1051 | AAAGGCTACC TTGAAGCGAG CATTGCATTA AAGCCGAGTA TTTCCGCGCG |
|
| 1101 | TTTGGTTCTA GCAAAGGTTT TCGACGAAAT CGGAGAACCG CAGAAGGCGG |
|
| 1151 | AGGCGCAGCG CAACTTGGTT TTGGAAGCCG TCTCCGATGA CGAACGTCAC |
|
| 1201 | GCAGCGTTAG AGCAGCATAG CTGA |
[1555]This corresponds to the amino acid sequence <SEQ ID 752; ORF100-1>:
[0000]| 1 | MKTVVWIVVL FAAAVGLALA SGIYTGDVYI VLGQTMLRIN LHAFVLGSLI | |
|
| 51 | AVVVWYFLFK FIIGVLNIPE KMQRFGSARK GRKAALALNK AGLAYFEGRF |
|
| 101 | EKAELEASRV LVNKEAGDNR TLALMLGAHA AGQMENIELR DRYLAEIAKL |
|
| 151 | PEKQQLSRYL LLAESALNRR DYEAAEANLH AAAKMNANLT RLVRLQLRYA |
|
| 201 | FDRGDALQVL AKTEKLSKAG ALGKSEMERY QNWAYRRQLA DAADAAALKT |
|
| 251 | CLKRIPDSLK NGELSVSVAE KYERLGLYAD AVKWVKQHYP HNRRPELLEA |
|
| 301 | FVESVRFLGE REQQKAIDFA DAWLKEQPDN ALLLMYLGRL AYGRKLWGKA |
|
| 351 | KGYLEASIAL KPSISARLVL AKVFDEIGEP QKAEAQRNLV LEAVSDDERH |
|
| 401 | AALEQHS* |
[1556]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1557]ORF100 shows 93.5% identity over a 386aa overlap with an ORF (ORF100a) from strain A of N. meningitidis.
[0000]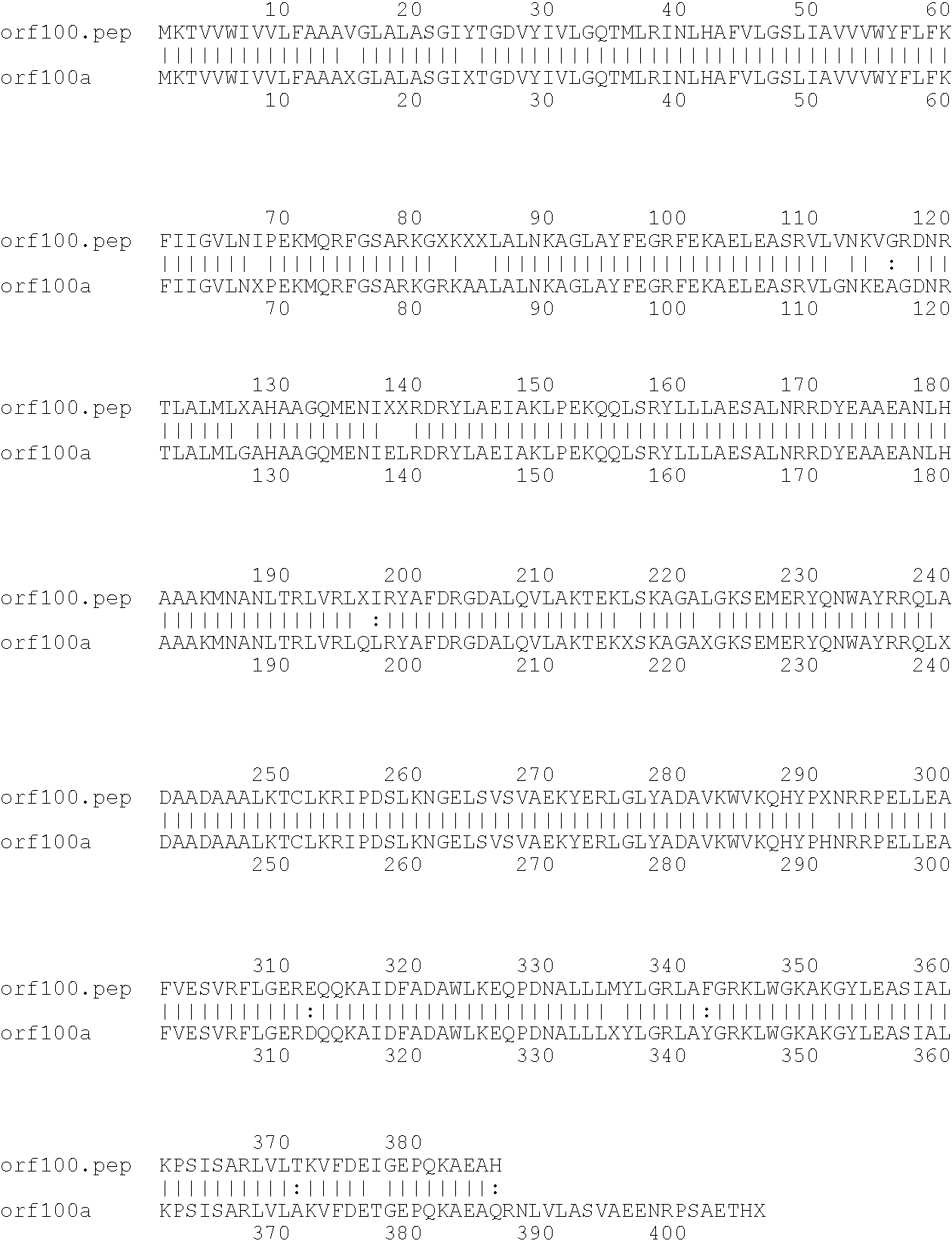
[1558]The complete length ORF100a nucleotide sequence <SEQ ID 753>
[0000]| 1 | ATGAAAACGG TAGTCTGGAT TGTCGTCCTG TTTGCCGCCG CNNTCGGGCT | |
|
| 51 | GGCATTGGCG TCGGGCATTN ACACCGGCGA CGTGTATATC GTACTCGGAC |
|
| 101 | AGACCATGCT CAGAATCAAC CTGCACGCCT TTGTGTTAGG TTCGCTGATT |
|
| 151 | GCCGTCGTGG TGTGGTATTT CCTGTTCAAA TTCATCATCG GCGTACTCAA |
|
| 201 | TANCCCCGAA AAGATGCAGC GTTTCGGTTC GGCGCGTAAA GGCCGCAAGG |
|
| 251 | CCGCGCTTGC TTTGAACAAG GCGGGTTTGG CGTATTTTGA AGGGCGTTTT |
|
| 301 | GAAAAGGCGG AACTTGAAGC CTCGCGCGTA TTGGGAAACA AAGAGGCGGG |
|
| 351 | GGATAACCGG ACTTTGGCAT TGATGTTGGG CGCACATGCC GCCGGGCAGA |
|
| 401 | TGGAAAACAT CGAGCTGCGC GACCGTTATC TTGCGGAAAT CGCCAAACTG |
|
| 451 | CCGGAAAAGC AGCAGCTTTC CCGTTATCTT TTGTTGGCGG AATCGGCGTT |
|
| 501 | GAACCGGCGC GATTACGAAG CGGCGGAAGC CAATCTTCAT GCGGCGGCGA |
|
| 551 | AGATGAATGC CAACCTTACG CGCCTCGTGC GTCTGCAACT TCGTTACGCT |
|
| 601 | TTCGACAGGG GCGACGCGTT GCAGGTTCTG GCAAAAACCG AAAAANTTTC |
|
| 651 | CAAGGCGGGC GCGTNGGGCA AATCGGAAAT GGAACGGTAT CAAAATTGGG |
|
| 701 | CATACCGCCG CCAGCTGNCG GATGCTGCCG ATGCCGCCGC TTTGAAAACC |
|
| 751 | TGCCTGAAGC GGATTCCCGA CAGCCTCAAA AACGGGGAAT TGAGCGTATC |
|
| 801 | GGTTGCGGAA AAGTACGAAC GTTTGGGACT GTATGCCGAT GCGGTCAAAT |
|
| 851 | GGGTCAAACA GCATTATCCG CACAACCGCC GACCCGAACT TTTGGAAGCN |
|
| 901 | TTTGTCGAAA GCGTGCGCTT TTTGGGCGAA CGCGATCAGC AGAAAGCCAT |
|
| 951 | CGATTTTGCC GATGCTTGGC TGAAAGAACA GCCCGATAAT GCGCTTCTGC |
|
| 1001 | TGANGTATCT CGGTCGGCTC GCCTACGGCC GCAAACTTTG GGGCAAGGCA |
|
| 1051 | AAAGGCTACC TTGAAGCGAG CATTGCATTA AAGCCGAGTA TTTCCGCGCG |
|
| 1101 | TTTGGTTCTG GCAAAGGTTT TTGACGAAAC CGGAGAACCG CAGAAGGCGG |
|
| 1151 | AGGCGCAGCG CAACTTGGTT TTGGCAAGCG TTGCCGAGGA AAACCGNCCT |
|
| 1201 | TCCGCCGAAA CCCATTGA |
[1559]This encodes a protein having amino acid sequence <SEQ ID 754>:
[0000]| 1 | MKTVVWIVVL FAAAXGLALA SGIXTGDVYI VLGQTMLRIN LHAFVLGSLI | |
|
| 51 | AVVVWYFLFK FIIGVLNXPE KMQRFGSARK GRKAALALNK AGLAYFEGRF |
|
| 101 | EKAELEASRV LGNKEAGDNR TLALMLGAHA AGQMENIELR DRYLAEIAKL |
|
| 151 | PEKQQLSRYL LLAESALNRR DYEAAEANLH AAAKMNANLT RLVRLQLRYA |
|
| 201 | FDRGDALQVL AKTEKXSKAG AXGKSEMERY QNWAYRRQLX DAADAAALKT |
|
| 251 | CLKRIPDSLK NGELSVSVAE KYERLGLYAD AVKWVKQHYP HNRRPELLEA |
|
| 301 | FVESVRFLGE RDQQKAIDFA DAWLKEQPDN ALLLXYLGRL AYGRKLWGKA |
|
| 351 | KGYLEASIAL KPSISARLVL AKVFDETGEP QKAEAQRNLV LASVAEENRP |
|
| 401 | SAETH* |
[1560]ORF100a and ORF100-1 show 95.1% identity in 406 aa overlap:
[0000]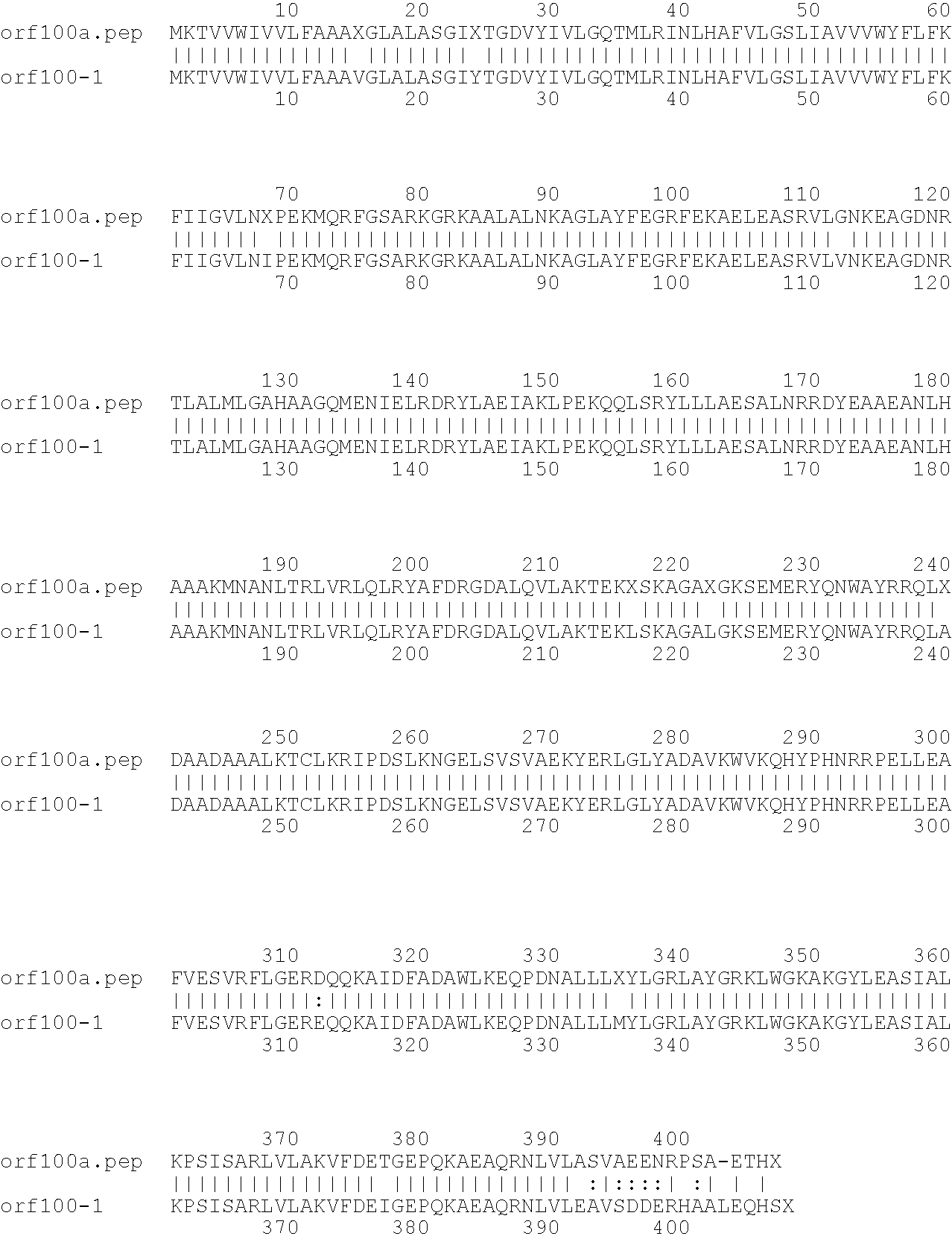
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1561]ORF100 shows 93.3% identity over a 386 aa overlap with a predicted ORF (ORF100ng) from N. gonorrhoeae:
[0000]
[1562]The complete length ORF100ng nucleotide sequence <SEQ ID 755> is:
[0000]| 1 | ATGAAAACGG TAGTCTGGAT TGTTGTCCTG TTTGCCGCCG CCGTCGGACT | |
|
| 51 | GGCGCTGGCT TCGGGCATTT ACACCGGCGA CGTGTATATC GTACTCGGAC |
|
| 101 | AGACCATGCT CAGAATCAAC CTGCACGCCT TTGTGTTAGG TTCGCTGATT |
|
| 151 | GCCGTCGTGG TGTGGTATTT CCTGTTTAAA TTCATCATCG GCGTACTCAA |
|
| 201 | TATCCCCGAA AATATGCGGC GTTCCGGTTC GGCGCGGAAA GGCCGCAAGG |
|
| 251 | CCGCGCTTGC CTTGAATAAG GCGGGTTTGG CGTATTTCGA AGGGCGTTTT |
|
| 301 | GAAAAGGCGG AACTCGAAGC CTCTCGAGTG TTGGGCAACA AAGAGGCCGG |
|
| 351 | AGACAACCGG ACTTTGGCAT TGATGCTGGG CGCGCACGCG GCAGGACAGA |
|
| 401 | TGGAAAATAT CGAGCTGCGC GACCGTTATC TTGCGGAAAT CGCCAAACTG |
|
| 451 | CCGGAAAAAC AGCAGCTTTC CCGCTATCTT CTGCTGGCGG AATCGGCGTT |
|
| 501 | AAACCGGCGC GATTACGAAG CGGCGGAAGC CAATCTTCAT GCGGCGGCGA |
|
| 551 | AGATGAATGC CAACCTTACG CGCCTCGTGC GTCTGCAACT TCGTTACGCC |
|
| 601 | TTCGATCGGG GCGATGCGTT GCAGGTTCTG GCAAAAaccG AAAAACTTTC |
|
| 651 | CAAGGCGGGC GCGTTGGGCA AATCGGAAAT GGAACGGTAT CAAAATTGGG |
|
| 701 | CATACCGCCG CCAGATGGCG GATGCTGCCG ATGCCGCCGC TTTGAAAACC |
|
| 751 | TGCCTGAAGC GGATTCCCGA CAGCCTCAAA AACGGGGAAT TGagcGTATC |
|
| 801 | GGTTGCGGAA AAGTACGAAC GTTTGGGACT GTATGCCGAT GCGGTCAAAT |
|
| 851 | GGGTCAAACA GCATTATCCG CACAACCGCC GCCCCGAGCT TTTGGAAGCC |
|
| 901 | TTTGTCGAAA GCGTGCGCTT TTTGGGCGAG CGCGAACAGC AGAAAGCCAT |
|
| 951 | CGATTTTGCC GATTCTTGGC TGAAAGAACA GCCCGATAAC GCGCTTCTGC |
|
| 1001 | TGATGTATCT CGGCCGGCTC GCCTACGGCC GCAAACTTTG GGGTAAGGCA |
|
| 1051 | AAAGGCTACC TTGAAGCGAG TATTGCACTG AAGCCGAGTA TTCCGGCGCG |
|
| 1101 | TTTGGTGTTG GCAAAGGTTT TTGACGAAAC CGCACAGTCG CAAAAAGCCG |
|
| 1151 | AAGCACAGCG CAACTTGGTT TTGGCAAGCG TTGCCGGGGA AAACCGCCCT |
|
| 1201 | TCCGCCGAAA CCCGTTGA |
[1563]This encodes a protein having amino acid sequence <SEQ ID 756>:
[0000]| 1 | MKTVVWIVVL FAAAVGLALA SGIYTGDVYI VLGQTMLRIN LHAFVLGSLI | |
|
| 51 | AVVVWYFLFK FIIGVLNIPE NMRRSGSARK GRKAALALNK AGLAYFEGRT |
|
| 101 | EKAELEASRV LGNKEAGDNR TLALMLGAHA AGQMENIELR DRYLAEIAKL |
|
| 151 | PEKQQLSRYL LLAESALNRR DYEAAEANLH AAAKMNANLT RLVRLQLRYA |
|
| 201 | FDRGDALQVL AKTEKLSKAG ALGKSEMERY QNWAYRRQMA DAADAAALKT |
|
| 251 | CLKRIPDSLK NGELSVSVAE KYERLGLYAD AVKWVKQHYP HNRRPELLEA |
|
| 301 | FVESVRFLGE REQQKAIDFA DSWLKEQPDN ALLLMYLGRL AYGRKLWGKA |
|
| 351 | KGYLEASIAL KPSIPARLVL AKVFDETAQS QKAEAQRNLV LASVAGENRP |
|
| 401 | SAETR* |
[1564]ORF100ng and ORF100-1 show 95.3% identity in 402 aa overlap:
[0000]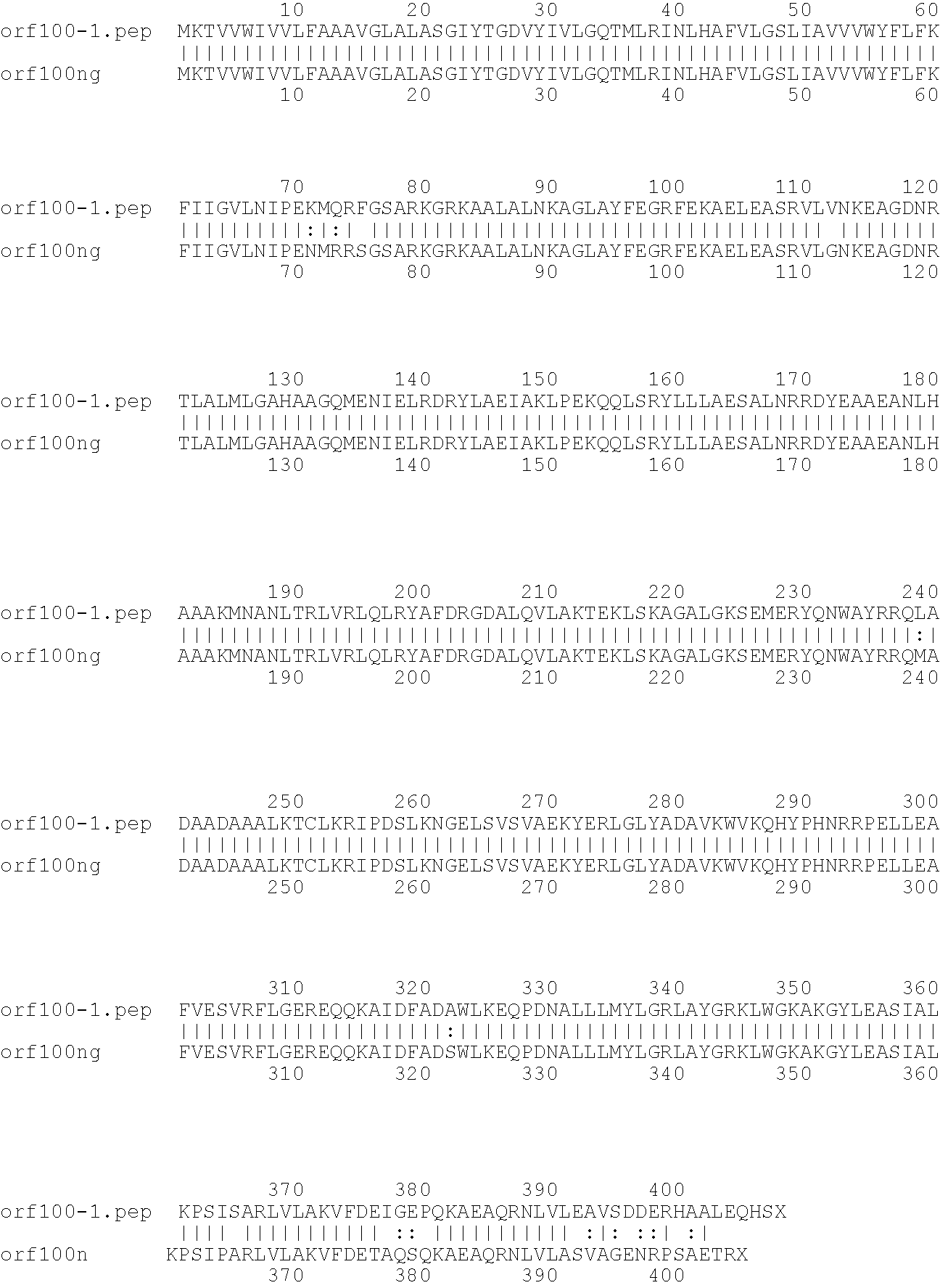
[1565]Based on this analysis, including the presence of a putative leader sequence, a putative transmembrane domain, and a RGD motif, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 90
[1566]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 757>
[0000]| 1 | ATGATGTTTT CTTGGTTCAA GCTGTTTCAC TTGTTTTTTG TCATTTCGTG | |
|
| 51 | GTTTGCAGGG CTGTTTTACC TGCCGAGGAT TTTCGTCAAT ATGGCGATGA |
|
| 101 | TTGATGTGCC GCGCGGCAAT CCCGAGTATG TGCGTCTGTC GGGCATGGCG |
|
| 151 | GTGCGGCTGT ACCGTTTTAT GTCGCCGTTG GGCTTCGGCG CGGTCGTGTT |
|
| 201 | CGGCGCGGCG ATACCGTTTG CCGCCGGCTG GTGGGGCAGC GGCTGGGTAC |
|
| 251 | ACGTCAAACT GTGTTTGGGC TTGATGCTCT TGGCTTACCA GTTGTATTGC |
|
| 301 | GGCGTGCTGC TGCGCCGTTT TCAGGATTAC AGCAATGCTT TTTCACACCG |
|
| 351 | CTGGTACCGC GTGTTCAACG AAATCCCCGT GCTGCTGATG GTTGCCGCGC |
|
| 401 | TGTATsTGGT CGTGTTCAAA CCGTTTTGA |
[1567]This corresponds to the amino acid sequence <SEQ ID 758; ORF102>:
[0000]| 1 | MMFSWFKLFH LFFVISWFAG LFYLPRIFVN MAMIDVPRGN PEYVRLSGMA | |
|
| 51 | VRLYRFMSPL GFGAVVFGAA IPFAAGWWGS GWVHVKLCLG LMLLAYQLYC |
|
| 101 | GVLLRRFQDY SNAFSHRWYR VFNEIPVLLM VAALYXVVFK PF* |
[1568]Further work revealed the complete nucleotide sequence <SEQ ID 759>:
[0000]| 1 | ATGATGTTTT CTTGGTTCAA GCTGTTTCAC TTGTTTTTTG TCATTTCGTG | |
|
| 51 | GTTTGCAGGG CTGTTTTACC TGCCGAGGAT TTTCGTCAAT ATGGCGATGA |
|
| 101 | TTGATGTGCC GCGCGGCAAT CCCGAGTATG TGCGTCTGTC GGGCATGGCG |
|
| 151 | GTGCGGCTGT ACCGTTTTAT GTCGCCGTTG GGCTTCGGCG CGGTCGTGTT |
|
| 201 | CGGCGCGGCG ATACCGTTTG CCGCCGGCTG GTGGGGCAGC GGCTGGGTAC |
|
| 251 | ACGTCAAACT GTGTTTGGGC TTGATGCTCT TGGCTTACCA GTTGTATTGC |
|
| 301 | GGCGTGCTGC TGCGCCGTTT TCAGGATTAC AGCAATGCTT TTTCACACCG |
|
| 351 | CTGGTACCGC GTGTTCAACG AAATCCCCGT GCTGCTGATG GTTGCCGCGC |
|
| 401 | TGTATCTGGT CGTGTTCAAA CCGTTTTGA |
[1569]This corresponds to the amino acid sequence <SEQ ID 760; ORF102-1>:
[0000]| 1 | MMFSWFKLFH LFFVISWFAG LFYLPRIFVN MAMIDVPRGN PEYVRLSGMA | |
|
| 51 | VRLYRFMSPL GFGAVVFGAA IPFAAGWWGS GWVHVKLCLG LMLLAYQLYC |
|
| 101 | GVLLRRFQDY SNAFSHRWYR VFNEIPVLLM VAALYLVVFK PF* |
[1570]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with HP1484 Hypothetical Integral Membrane Protein of H. pylori (Accession Number AE000647)
[1571]ORF102 and HP1484 show 33% aa identity in 143aa overlap:
[0000]| orf102 | 3 | FSWFKLFHLFFVISWFAGLFYLPRIFVNMAMIDVPRGNPEYVRLSGMAVRLYRFMSPLGF | 62 | |
| | F W K FH+ VISW A LFYLPR+FV A + V++ +LY F++ |
| HP1484 | 8 | FLWVKAFHVIAVISWMAALFYLPRLFVYHAENAHKKEFVGVVQIQEK--KLYSFIASPAM | 65 |
|
| orf102 | 63 | GAVVFGAAIPFAAG---WWGSGWVHVKLCLGLMLLAYQLYCGVLLRRFQDYSNAFSHRWY | 119 |
| | G + + + GW+H KL L ++LLAY YC +R + + R+Y |
| HP1484 | 66 | GFTLITGILMLLIEPTLFKSGGWLHAKLALVVLLLAYHFYCKKCMRELEKDPTRRNARFY | 125 |
|
| orf102 | 120 | RVFNEIPXXXXXXXXXXXXFKPF | 142 |
| | RVFNE P KPF |
| HP1484 | 126 | RVFNEAPTILMILIVILVVVKPF | 148 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1572]ORF102 shows 99.3% identity over a 142aa overlap with an ORF (ORF102a) from strain A of N. meningitidis:
[0000]
[1573]The complete length ORF102a nucleotide sequence <SEQ ID 761> is:
[0000]| 1 | ATGATGTTTT CTTGGTTCAA GCTGTTTCAC TTGTTTTTTG TCATTTCGTG | |
|
| 51 | GTTTGCAGGG CTGTTTTACC TGCCGAGGAT TTTCGTCAAT ATGGCGATGA |
|
| 101 | TTGATGTGCC GCGCGGCAAT CCCGAGTATG TGCGTCTGTC GGGCATGGCG |
|
| 151 | GTGCGGCTGT ACCGTTTTAT GTCGCCGTTG GGCTTCGGCG CGGTCGTGTT |
|
| 201 | CGGCGCGGCG ATACCGTTTG CCGCCGGCTG GTGGGGCAGC GGCTGGGTAC |
|
| 251 | ACGTCAAACT GTGTTTGGGC TTGATGCTCT TGGCTTACCA GTTGTATTGC |
|
| 301 | GGCGTGCTGC TGCGCCGTTT TCAGGATTAC AGCAATGCTT TTTCACACCG |
|
| 351 | CTGGTACCGC GTGTTCAACG AAATCCCCGT GCTGCTGATG GTTGCCGCGC |
|
| 401 | TGTATCTGGT CGTGTTCAAA CCGTTTTGA |
[1574]This encodes a protein having amino acid sequence <SEQ ID 762>:
[0000]| 1 | MMFSWFKLFH LFFVISWFAG LFYLPRIFVN MAMIDVPRGN PEYVRLSGMA | |
|
| 51 | VRLYRFMSPL GFGAVVFGAA IPFAAGWWGS GWVHVKLCLG LMLLAYQLYC |
|
| 101 | GVLLRRFQDY SNAFSHRWYR VFNEIPVLLM VAALYLVVFK PF* |
[1575]ORF102a and ORF102-1 show complete identity in 142 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1576]ORF102 shows 97.9% identity over a 142 aa overlap with a predicted ORF (ORF102ng) from N. gonorrhoeae:
[0000]
[1577]The complete length ORF102ng nucleotide sequence <SEQ ID 763> is:
[0000]| 1 | ATGATGTTTT CTTGGTTCAA GCTGTTTCAC TTGTTTTTTG TCATTTCGTG | |
|
| 51 | GTTTGCAGGG CTGTTTTACC TGCCGAGGAT TTTCGTCAAT ATGGCGATGA |
|
| 101 | TTGATGCGCC GCGCGGCAAT CCCGAGTATG TGCGCCTGTC GGGGATGGCG |
|
| 151 | GTGCGGTTGT ACCGTTTTAT GTCGCCTTTG GGTTTCGGCG CGGTCGTGTT |
|
| 201 | CGGCGCGGCG ATACCGTTTG CCGCcggccg GTGGGGCagc ggctggGTTC |
|
| 251 | ACGTCAAACT GTGTTTGGGC TTGATGCTCT TGGCTTATCA GTTGTATTGC |
|
| 301 | GGCGTGCTGC TGCGCCGTTT TCAGGATTAC AGCAATGCTT TTTCACACCG |
|
| 351 | CTGGTACCGC GTGTTCAAcg aAATCCCCGT GCTGCTGATG GTTGCCGCGC |
|
| 401 | TGTATCTGGT CGTGTTCAAA CCGTTTTGA |
[1578]This encodes a protein having amino acid sequence <SEQ ID 764>:
[0000]| 1 | MMFSWFKLFH LFFVISWFAG LFYLPRIFVN MAMIDAPRGN PEYVRLSGMA | |
|
| 51 | VRLYRFMSPL GFGAVVFGAA IPFAAGRWGS GWVHVKLCLG LMLLAYQLYC |
|
| 101 | GVLLRRFQDY SNAFSHRWYR VFNIPVLLM VAALYLVVFK PF* |
[1579]ORF102ng and ORF102-1 show 98.6% identity in 142 aa overlap:
[0000]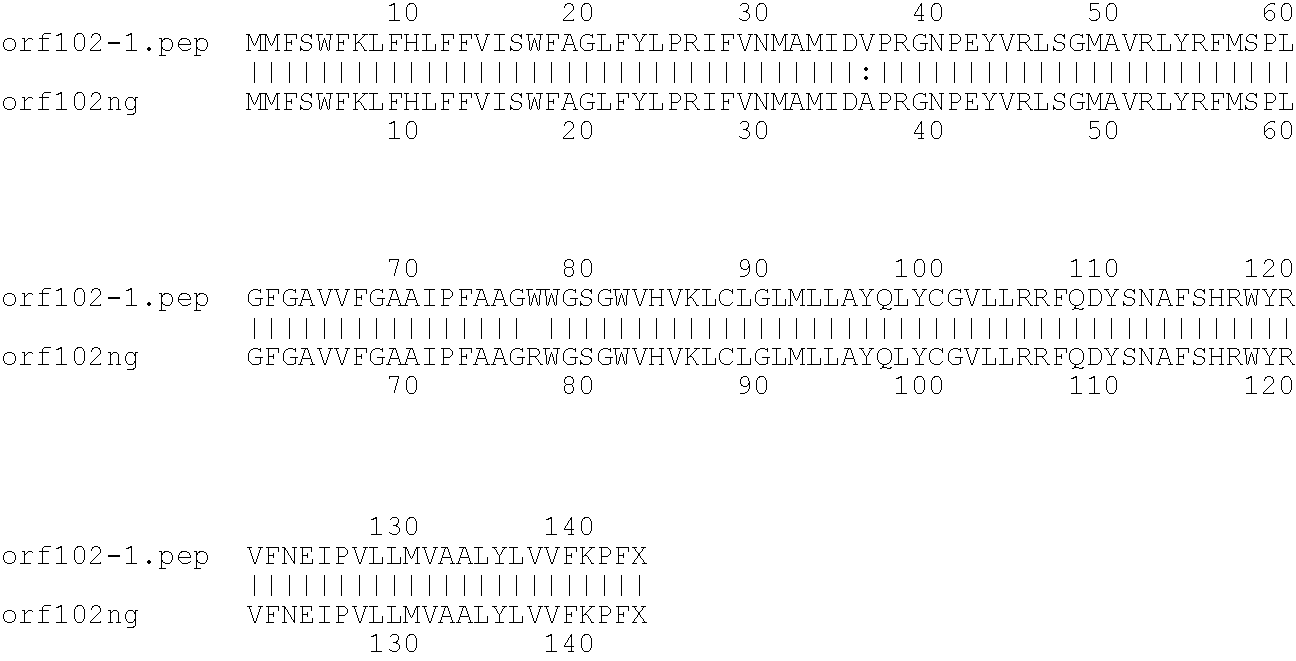
[1580]In addition, ORF102ng shows significant homology to a membrane protein from H. pylori:
[0000]| gi|2314656 (AE000647) conserved hypothetical integral membrane protein | |
| [Helicobacter pylori] Length = 148 |
| Score = 79.2 bits (192), Expect = 1e−14 |
| Identities = 50/147 (34%), Positives = 68/147 (46%), Gaps = 13/147 (8%) |
|
| Query: | 3 | FSWFKLFHLFFVISWFAGLFYLPRIFVNMAMIDAPRGNPEYVRLSGMAVRLYRFMSPLGF | 62 | |
| | F W K FH+ VISW A LFYLPR+FV A + V++ +LY F++ |
| Sbjct: | 8 | FLWVKAFHVIAVISWMAALFYLPRLFVYHAENAHKKEFVGVVQIQEK--KLYSFIASPAM | 65 |
|
| Query: | 63 | GAVVFGAAIP-------FAAGRWGSGWVHVKLCLGLMLLAYQLYCGVLLRRFQDYSNAFS | 115 |
| | G + + F +G GW+H KL L ++LLAY YC +R + + |
| Sbjct: | 66 | GFTLITGILMLLIEPTLFKSG----GWLHAKLALVVLLLAYHFYCKKCMRELEKDPTRRN | 121 |
|
| Query: | 116 | HRWYRVFNEIPXXXXXXXXXXXXFKPF | 142 |
| | R+YRVFNE P KPF |
| Sbjct: | 122 | ARFYRVFNEAPTILMILIVILVVVKPF | 148 |
[1581]Based on this analysis, it is predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 91
[1582]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 765>:
[0000]
[1583]This corresponds to the amino acid sequence <SEQ ID 766; ORF85>:
[0000]| 1 | MAKMMKWAAV AAVAAAAVWG GWS.LKPEPH VLDITETVRR G......... | |
|
| 51 | ........... ........... ........... ........... ........... |
|
| 101 | ........... ........... ........... ........... ........... |
|
| 151 | ........... ........... ........... ........... ........... |
|
| 201 | ........... ........... ........... ..........I SFTILSEPDT |
|
| 251 | PIKAKLDSVD PGLTTMSSGG YNSSTDTASN AVYYYARSFV PNPDGKLATG |
|
| 301 | MTTQNTVEID GVKNVLIIPS LTVKNRGGKA FVRVLGADGK AAEREIRTGM |
|
| 351 | RDSMNTEVKS GLKEGDKVVI SEITAAEQQE SGERALGGPP RR* |
[1584]Further work revealed the further partial nucleotide sequence <SEQ ID 767>:
[0000]| 1 | ..GTATCGGTCG GCGCGCAGGC ATCGGGGCAG ATTAAGATAC TTTATGTCAA | |
|
| 51 | ACTCGGGCAA CAGGTTAAAA AGGGCGATTT GATTGCGGAA ATCAATTCGA |
|
| 101 | CCTCGCAGAC CAATACGCTC AATACGGAAA AATCCAAGTT GGAAACGTAT |
|
| 151 | CAGGCGAAGC TGGTGTCGGC ACAGATTGCA TTGGGCAGCG CGGAGAAGAA |
|
| 201 | ATATAAGCGT CAGGCGGCGT TATGGAAGGA AAACGCGACT TCCAAAGAGG |
|
| 251 | ATTTGGAAAG CGCGCAGGAT GCGTTTGCCG CCGCCAAAGC CAATGTTGCC |
|
| 301 | GAGCTGAAGG CTTTAATCAG ACAGAGCAAA ATTTCCATCA ATACCGCCGA |
|
| 351 | GTCGGAATTG GGCTACACGC GCATTACCGC AACGATGGAC GGCACGGTGG |
|
| 401 | TGGCGATTCT CGTGGAAGAG GGGCAGACTG TGAACGCGGC GCAGTCTACG |
|
| 451 | CCGACGATTG TCCAATTGGC GAATCTGGAT ATGATGTTGA ACAAAATGCA |
|
| 501 | GATTGCCGAG GGCGATATTA CCAAGGTGAA GGCGGGGCAG GATATTTCGT |
|
| 551 | TTACGATTTT GTCCGAACCG GATACGCCGA TTAAGGCGAA GCTCGACAGC |
|
| 601 | GTCGACCCCG GGCTGACCAC GATGTCGTCG GGCGGTTACA ACAGCAGTAC |
|
| 651 | GGATACGGCT TCCAATGCGG TCTACTATTA TGCCCGTTCG TTTGTGCCGA |
|
| 701 | ATCCGGACGG CAAACTCGCC ACGGGGATGA CGACGCAGAA TACGGTTGAA |
|
| 751 | ATCGACGGCG TGAAAAATGT GCTGATTATT CCGTCGCTGA CCGTGAAAAA |
|
| 801 | TCGCGGCGGC AAGGCGTTTG TGCGCGTGTT GGGTGCGGAC GGCAAGGCGG |
|
| 851 | CGGAACGCGA AATCCGGACC GGTATGAGAG ACAGTATGAA TACCGAAGTA |
|
| 901 | AAAAGCGGGT TGAAAGAGGG GGACAAAGTG GTCATCTCCG AAATAACCGC |
|
| 951 | CGCCGAGCAA CAGGAAAGCG GCGAACGCGC CCTAGGCGGC CCGCCGCGCC |
|
| 1001 | GATAA |
[1585]This corresponds to the amino acid sequence <SEQ ID 768; ORF85-1>:
[0000]| 1 | ..VSVGAQASGQ IKILYVKLGQ QVKKGDLIAE INSTSQTNTL NTEKSKLETY | |
|
| 51 | QAKLVSAQIA LGSAEKKYKR QAALWKENAT SKEDLESAQD AFAAAKANVA |
|
| 101 | ELKALIRQSK ISINTAESEL GYTRITATMD GTVVAILVEE GQTVNAAQST |
|
| 151 | PTIVQLANLD MMLNKMQIAE GDITKVKAGQ DISFTILSEP DTPIKAKLDS |
|
| 201 | VDPGLTTMSS GGYNSSTDTA SNAVYYYARS FVPNPDGKLA TGMTTQNTVE |
|
| 251 | IDGVKNVLII PSLTVKNRGG KAFVRVLGAD GKAAEREIRT GMRDSMNTEV |
|
| 301 | KSGLKEGDKV VISEITAAEQ QESGERALGG PPRR* |
[1586]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1587]ORF85 shows 87.8% identity over a41aa overlap and 99.3% identity over a 153aa overlap with an ORF (ORF85a) from strain A of N. meningitidis:
[0000]
[1588]The complete length ORF85a nucleotide sequence <SEQ ID 769> is:
[0000]| 1 | ATGGCAAAAA TGATGAAATG GGCGGCTGTT GCGGCGGTCG CGGCGGCAGC | |
|
| 51 | GGTTTGGGGC GGATGGTCTT ATCTGAAGCC CGAGCCGCAG GCTGCTTATA |
|
| 101 | TTACGGAAAC GGTCAGGCGC GGCGACATCA GCCGGACGGT TTCTGCAACA |
|
| 151 | GGGGAGATTT CGCCGTCCAA CCTGGTATCG GTCGGCGCGC AGGCATCGGG |
|
| 201 | GCAGATTAAG AAACTTTATG TCAAACTCGG GCAACAGGTT AAAAAGGGCG |
|
| 251 | ATTTGATTGC GGAAATCAAT TCGACCTCGC AGACCAATAC GCTCAATACG |
|
| 301 | GAAAAATCCA AATTGGAAAC GTATCAGGCG AAGCTGGTGT CGGCACAGAT |
|
| 351 | TGCATTGGGC AGCGCGGAGA AGAAATATAA GCGTCAGGCG GCGTTGTGGA |
|
| 401 | AGGATGATGC GACCGCTAAA GAAGATTTGG AAAGCGCACA GGATGCGCTT |
|
| 451 | GCCGCCGCCA AAGCCAATGT TGCCGAGCTG AAGGCTCTAA TCAGACAGAG |
|
| 501 | CAAAATTTCC ATCAATACCG CCGAGTCGGA ATTGGGCTAC ACGCGCATTA |
|
| 551 | CCGCAACGAT GGACGGCACG GTGGTGGCGA TTCTCGTGGA AGAGGGGCAG |
|
| 601 | ACTGTGAACG CGGCGCAGTC TACGCCGACG ATTGTCCAAT TGGCGAATCT |
|
| 651 | GGATATGATG TTGAACAAAA TGCAGATTGC CGAGGGCGAT ATTACCAAGG |
|
| 701 | TGAAGGCGGG GCAGGATATT TCGTTTACGA TTTTGTCCGA ACCGGATACG |
|
| 751 | CCGATTAAGG CGAAGCTCGA CAGCGTCGAC CCCGGGCTGA CCACGATGTC |
|
| 801 | GTCGGGCGGC TACAACAGCA GTACGGATAC GGCTTCCAAT GCGGTCTACT |
|
| 851 | ATTATGCCCG TTCGTTTGTG CCGAATCCGG ACGGCAAACT CGCCACGGGG |
|
| 901 | ATGACGACGC AGAATACGGT TGAAATCGAC GGTGTGAAAA ATGTGCTGAT |
|
| 951 | TATTCCGTCG CTGACCGTGA AAAATCGCGG CGGCAGGGCG TTTGTGCGCG |
|
| 1001 | TGTTGGGTGC AGACGGCAAG GCGGCGGAAC GCGAAATCCG GACCGGTATG |
|
| 1051 | AGAGACAGTA TGAATACCGA AGTAAAAAGC GGGTTGAAAG AGGGGGACAA |
|
| 1101 | AGTGGTCATC TCCGAAATAA CCGCCGCCGA GCAGCAGGAA AGCGGCGAAC |
|
| 1151 | GCGCCCTAGG CGGCCCGCCG CGCCGATAA |
[1589]This encodes a protein having amino acid sequence <SEQ ID 770>:
[0000]| 1 | MAKMMKWAAV AAVAAAAVWG GWSYLKPEPQ AAYITETVRR GDISRTVSAT | |
|
| 51 | GEISPSNLVS VGAQASGQIK KLYVKLGQQV KKGDLIAEIN STSQTNTLNT |
|
| 101 | EKSKLETYQA KLVSAQIALG SAEKKYKRQA ALWKDDATAK EDLESAQDAL |
|
| 151 | AAAKANVAEL KALIRQSKIS INTAESELGY TRITATMDGT VVAILVEEGQ |
|
| 201 | TVNAAQSTPT IVQLANLDMM LNKMQIAEGD ITKVKAGQDI SFTILSEPDT |
|
| 251 | PIKAKLDSVD PGLTTMSSGG YNSSTDTASN AVYYYARSFV PNPDGKLATG |
|
| 301 | MTTQNTVEID GVKNVLIIPS LTVKNRGGRA FVRVLGADGK AAEREIRTGM |
|
| 351 | RDSMNTEVKS GLKEGDKVVI SEITAAEQQE SGERALGGPP RR* |
[1590]ORF85a and ORF85-1 show 98.2% identity in 334 aa overlap:
[0000]
[1591]FIG. 19D shows plots of hydrophilicity, antigenic index, and AMPHI regions for ORF85a.
[0000]Homology with a predicted ORF from N. gonorrhoeae
[1592]ORF85 shows a high degree of identity with a predicted ORF (ORF85ng) from N. gonorrhoeae:
[0000]
[1593]The complete length ORF85ng nucleotide sequence <SEQ ID 771> is:
[0000]| 1 | ATGGCAAAAA TGATGAAATG GGCGGCTGTT GCGGCGGTCG CGGCGGCaac | |
|
| 51 | GGTTTGGGGC GGATGGTCTT ATCTGAAGCC CGAACCGCAG GCTGCTTATA |
|
| 101 | TTACGGAaac ggTCAGGCGC GGCGATATCA GCCGGACGGT TTCCGCGACG |
|
| 151 | GgcgAGATTT CGCCGTCCAA CCTGGTATCG GTCGGCGCGC AGGCTTCGGG |
|
| 201 | GCAGATTAAA AAGCTTTATG TCAAACTCGG GCAACAGGTC AAAAAGGGCG |
|
| 251 | ATTTGATTGC GGAAATCAAT TCGACCACGC AGACCAACAC GATCGATATG |
|
| 301 | GAAAAATCCA AATTGGAAAC GTATCAGGCG AAGCTGGTGT CGGCACAGAT |
|
| 351 | TGCATTGGGC AGCGCGGAGA AGAAATATAA GCGTCAGGCG GCGTTGTGGA |
|
| 401 | AGGATGATGC GACCTCTAAA GAAGATTTGG AAAGCGCGCA GGATGCGCTT |
|
| 451 | GCCGCCGCCA AAGCCAATGT TGCCGAGTTG AAGGCTTTAA TCAGACAGAG |
|
| 501 | CAAAATTTCC ATCAATACCG CCGAGTCGGA TTTGGGCTAC ACGCGCATTA |
|
| 551 | CCGCGACGAT GGACGGCACG GTGGTGGCGA TTCCCGTGGA AGAGGGGCAG |
|
| 601 | ACTGTGAACG CGGCGCAGTC TACGCCGACG ATTGTCCAAT TGGCGAATCT |
|
| 651 | GGATATGATG TTGAACAAAA TGCAGATTGC CGAGGGCGAT ATTACCAAGG |
|
| 701 | TGAAGGCGGG GCAGGATATT TCGTTTACGA TTTTGTCCGA ACCGGATACG |
|
| 751 | CCGATTAAGG CGAAGCTCGA CAGCGTCGAC CCCGGGCTGA CCACGATGTC |
|
| 801 | GTCGGGCGGC TACAACAGCA GTACGGATAC GGCTTCCAAT GCGGTCTATT |
|
| 851 | ATTATGCCCG TTCGTTTGTG CCGAATCCGG ACGGCAAACT CGCCACGGGG |
|
| 901 | ATGACGACGC AGAATACGGT TGAAATCGAC GGTGTGAAAA ATGTGTTGCT |
|
| 951 | TATTCCGTCG CTGACCGTGA AAAATCGCGG CGGCAAGGCG TTCGTACGCG |
|
| 1001 | TGTTGGGTGC GGACGGCAAG GCAGTGGAAC GCGAAATCCG GACCGGTATG |
|
| 1051 | AAAGACAGTA TGAATACCGA AGTGAAAAGC GGGTTGAAAG AGGGGGACAA |
|
| 1101 | AGTGGTCATC TCCGAAATAA CCGCCGCCGA GCAGCAGGAA AGCGGCGAAC |
|
| 1151 | GCGCCCTAGG CGGCCCGCCG CGCCGATAA |
[1594]This encodes a protein having amino acid sequence <SEQ ID 772>:
[0000]| 1 | MAKMMKWAAV AAVAAAAVWG GWSYLKPEPQ AAYITEAVRR GDISRTVSAT | |
|
| 51 | GEISPSNLVS VGAQASGQIK KLYVKLGQQV KKGDLIAEIN STTQTNTIDM |
|
| 101 | EKSKLETYQA KLVSAQIALG SAEKKYKRQA ALWKDDATSK EDLESAQDAL |
|
| 151 | AAAKANVAEL KALIRQSKIS INTAESDLGY TRITATMDGT VVAIPVEEGQ |
|
| 201 | TVNAAQSTPT IVQLANLDMM LNKMQIAEGD ITKVKAGQDI SFTILSEPDT |
|
| 251 | PIKAKLDSVD PGLTTMSSGG YNSSTDTASN AVYYYARSFV PNPDGKLATG |
|
| 301 | MTTQNTVEID GVKNVLLIPS LTVKNRGGKA FVRVLGADGK AVEREIRTGM |
|
| 351 | KDSMNTEVKS GLKEGDKVVI SEITAAEQQE SGERALGGPP RR* |
[1595]ORF85ng and ORF85-1 show 96.1% identity in 334 aa overlap:
[0000]
[1596]In addition, ORF85ng shows significant homology to an E. coli membrane fusion protein:
[0000]| gi|1787104 (AE000189) o380; 27% identical (27 gaps) to 332 residues from | |
| membrane fusion protein precursor, MTRC_NEIGO SW: P43505 (412 aa) |
| [Escherichia coli] Length = 380 |
| Score = 193 bits (485), Expect = 2e−48 |
| Identities = 120/345 (34%), Positives = 182/345 (51%), Gaps = 13/345 (3%) |
|
| Query: | 29 | PQAAYITETVRRGDISRTVSATGEISPSNLVSVGAQASGQIKKLYVKLGQQVKKGDLIAE | 88 | |
| | P Y T VR GD+ ++V ATG++ V VGAQ SGQ+K L V +G +VKK L+ |
| Sbjct: | 41 | PVPTYQTLIVRPGDLQQSVLATGKLDALRKVDVGAQVSGQLKTLSVAIGDKVKKDQLLGV | 100 |
|
| Query: | 89 | INSTTQTNTIDMEKSKLETYQAKLVSAQIALGSAEKKYKRQAALWKDDATSKEXXXXXXX | 148 |
| | I+ N I ++ L +A+ A+ L A Y RQ L+ A S++ |
| Sbjct: | 101 | IDPEQAENQIKEVEATLMELRAQRQQAEAELKLARVTYSROQRLAQTKAVSQQDLDTAAT | 160 |
|
| Query: | 149 | XXXXXXXXXXXXXXXIRQSKISINTAESDLGYTRITATMDGTVVAIPVEEGQTVNAAQST | 208 |
| | I++++ S++TA+++L YTRI A M G V I +GQTV AAQ |
| Sbjct: | 161 | EMAVKQAQIGTIDAQIKRNQASLDTAKTNLDYTRIVAPMAGEVTQITTLQGQTVIAAQQA | 220 |
|
| Query: | 209 | PTIVQLANLDMMLNKMQIAEGDITKVKAGQDISFTILSEPDTPIKAKLDSVDPGLTTMSS | 268 |
| | P I+ LA++ ML K Q++E D+ +K GQ FT+L +P T + ++ V P |
| Sbjct: | 221 | PNILTLADMSAMLVKAQVSEADVIHLKPGQKAWFTVLGDPLTRYEGQIKDVLP------- | 273 |
|
| Query: | 269 | GGYNSSTDTASNAVYYYARSFVPNPDGKLATGMTTQNTVEIDGVKNVLLIPSLTVKNRGG | 328 |
| | + + ++A++YYAR VPNP+G L MT Q +++ VKNVL IP + + G |
| Sbjct: | 274 | -----TPEKVNDAIFYYARFEVPNPNGLLRLDMTAQVHIQLTDVKNVLTIPLSALGDPVG | 328 |
|
| Query: | 329 | KAFVRV-LGADGKAVEREIRTGMKDSMNTEVKSGLKEGDKVVISE | 372 |
| | +V L +G+ ERE+ G ++ + E+ GL+ GD+VVI E |
| Sbjct: | 329 | DNRYKVKLLRNGETREREVTIGARNDTDVEIVKGLEAGDEVVIGE | 373 |
[1597]Based on this analysis, it was predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1598]ORF85-1 (40.4 kDa) was cloned in the pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 19A shows the results of affinity purification of the GST-fusion protein. Purified GST-fusion protein was used to immunise mice, whose sera were used for Western blot (FIG. 19B), FACS analysis (FIG. 19C), and ELISA (positive result). These experiments confirm that ORF85-1 is a surface-exposed protein, and that it is a useful immunogen.
Example 92
[1599]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 773>:
[0000]| 1 | ..ATTCCCGCCA CGATGACATT TGAACGCAGC GGCAATGCTT ACAAAATCGT | |
|
| 51 | TTCGACGATT AAAGTGCCGC TATACAATAT CCGTTTCGAG TCCGGCGGTA |
|
| 101 | CGGTTGTCGG CAATACCCTG CACCCTACCT ACTATAGAGA CATACGCAGG |
|
| 151 | GGCAAACTGT ATGCGGAAgc CAAATTCGCC GACgGcAGCG TAACTTACGG |
|
| 201 | CAAAGCGGGC GAGAGCAAAA CCGAGCAAAG CCCCAAGGCT ATGGATTTGT |
|
| 251 | TCACGCTTGC CTGGCAGTTG GCGGCAAATG ACGCGAAACT CCCCCCGGGG |
|
| 301 | CTGAAAATCA CCAACGGCAA AAAACTTTAT TCCGTCGGCG GTTTGAATAA |
|
| 351 | GGCGGGTACA GGAAAATACA GCATAGGCGG CGTGGAAACC GAAGTCGTCA |
|
| 401 | AATATCGGGT GCGGCGCGGC GACGATGCGG TAATGTATTT cTTCGCACCG |
|
| 451 | TCCCTGAACA ATATTCCGGC ACAAATCGGC TATACCGACG ACGGCAAAAC |
|
| 501 | CTATACGCTG AAACTCAAAT CGGTGCAGAT CAACGGCCAG GCAGCCAAAC |
|
| 551 | CGTAA |
[1600]This corresponds to the amino acid sequence <SEQ ID 774; ORF120>:
[0000]| 1 | ..IPATMTFERS GNAYKIVSTI KVPLYNIRFE SGGTVVGNTL HPTYYRDIRR | |
|
| 51 | GKLYAEAKFA DGSVTYGKAG ESKTEQSPKA MDLFTLAWQL AANDAKLPPG |
|
| 101 | LKITNGKKLY SVGGLNKAGT GKYSIGGVET EVVKYRVRRG DDAVMYFFAP |
|
| 151 | SLNNIPAQIG YTDDGKTYTL KLKSVQINGQ AAKP* |
[1601]Further work revealed the complete nucleotide sequence <SEQ ID 775>:
[0000]| 1 | ATGATGAAGA CTTTTAAAAA TATATTTTCC GCCGCCATTT TGTCCGCCGC | |
|
| 51 | CCTGCCGTGC GCGTATGCGG CAGGGCTGCC CCAATCCGCC GTGCTGCACT |
|
| 101 | ATTCCGGCAG CTACGGCATT CCCGCCACGA TGACATTTGA ACGCAGCGGC |
|
| 151 | AATGCTTACA AAATCGTTTC GACGATTAAA GTGCCGCTAT ACAATATCCG |
|
| 201 | TTTCGAGTCC GGCGGTACGG TTGTCGGCAA TACCCTGCAC CCTACCTACT |
|
| 251 | ATAGAGACAT ACGCAGGGGC AAACTGTATG CGGAAGCCAA ATTCGCCGAC |
|
| 301 | GGCAGCGTAA CTTACGGCAA AGCGGGCGAG AGCAAAACCG AGCAAAGCCC |
|
| 351 | CAAGGCTATG GATTTGTTCA CGCTTGCCTG GCAGTTGGCG GCAAATGACG |
|
| 401 | CGAAACTCCC CCCGGGGCTG AAAATCACCA ACGGCAAAAA ACTTTATTCC |
|
| 451 | GTCGGCGGTT TGAATAAGGC GGGTACAGGA AAATACAGCA TAGGCGGCGT |
|
| 501 | GGAAACCGAA GTCGTCAAAT ATCGGGTGCG GCGCGGCGAC GATGCGGTAA |
|
| 551 | TGTATTTCTT CGCACCGTCC CTGAACAATA TTCCGGCACA AATCGGCTAT |
|
| 601 | ACCGACGACG GCAAAACCTA TACGCTGAAA CTCAAATCGG TGCAGATCAA |
|
| 651 | CGGCCAGGCA GCCAAACCGT AA |
[1602]This corresponds to the amino acid sequence <SEQ ID 776; ORF120-1>:
[0000]| 1 | MMKTFKNIFS AAILSAALPC AYAAGLPQSA VLHYSGSYGI PATMTFERSG | |
|
| 51 | NAYKIVSTIK VPLYNIRFES GGTVVGNTLH PTYYRDIRRG KLYAEAKFAD |
|
| 101 | GSVTYGKAGE SKTEQSPKAM DLFTLAWQLA ANDAKLPPGL KITNGKKLYS |
|
| 151 | VGGLNKAGTG KYSIGGVETE VVKYRVRRGD DAVMYFFAPS LNNIPAQIGY |
|
| 201 | TDDGKTYTLK LKSVQINGQA AKP* |
[1603]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1604]ORF120 shows 92.4% identity over a 184aa overlap with an ORF (ORF120a) from strain A of N. meningitidis.
[0000]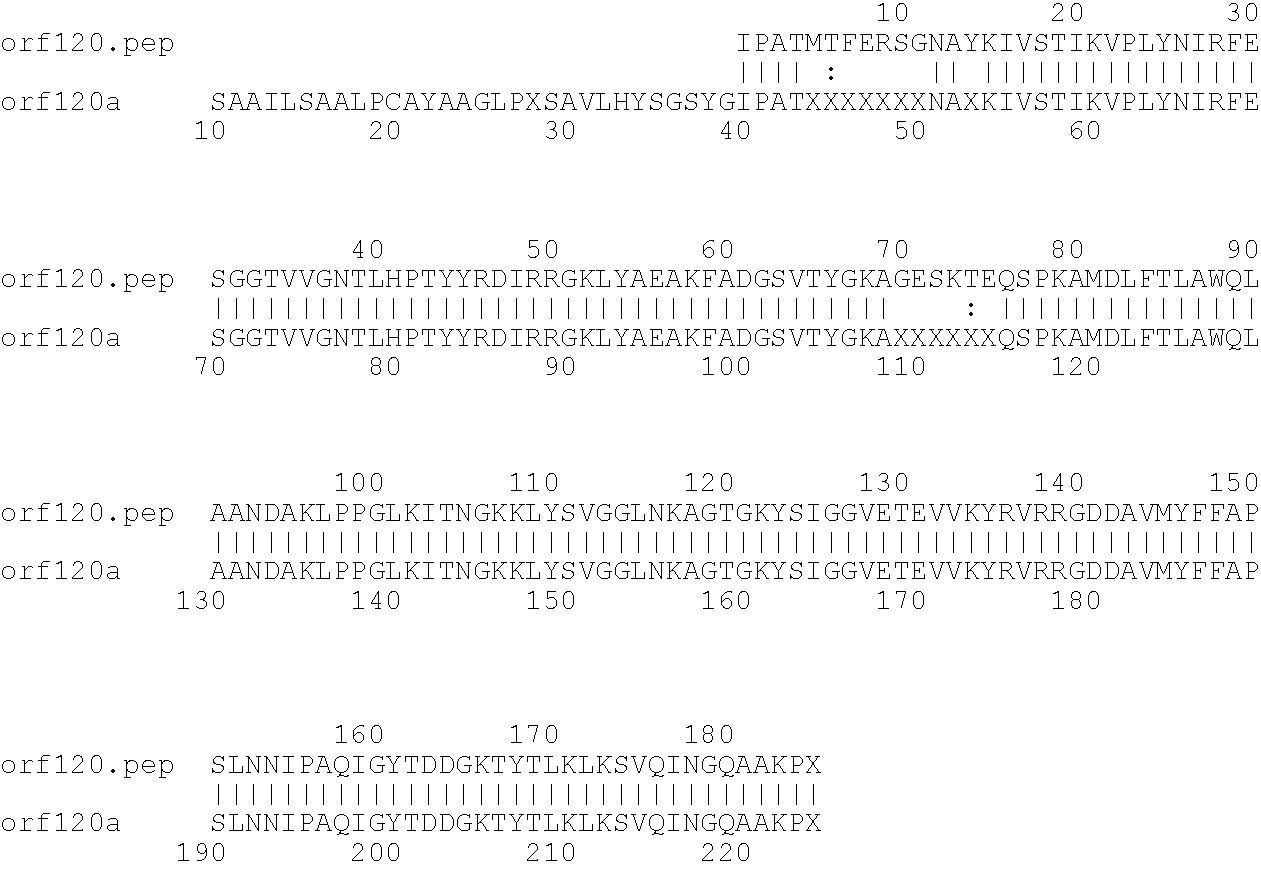
[1605]The complete length ORF120a nucleotide sequence <SEQ ID 777> is:
[0000]| 1 | ATGATGAAGA CTTTTAAAAA TATATTTTCC GCCGCCATTT TGTCCGCCGC | |
|
| 51 | CCTGCCGTGC GCGTATGCGG CAGGGCTGCC CNAATCCGCC GTGCTGCACT |
|
| 101 | ATTCCGGCAG CTACGGCATT CCCGCCACNA NNANNTNNGN ACNNNGNGNC |
|
| 151 | AATGCTTNCA AAATCGTTTC GACGATTAAA GTGCCGCTAT ACAATATCCG |
|
| 201 | TTTCGAGTCC GGCGGTACGG TTGTCGGCAA TACCCTGCAC CCTACCTACT |
|
| 251 | ATAGAGACAT ACGCAGGGGC AAACTGTATG CGGAAGCCAA ATTCGCCGAC |
|
| 301 | GGCAGCGTAA CCTACGGCAA AGCGGNNNNN ANCNNNNNNG NGCAAAGCCC |
|
| 351 | CAAGGCTATG GATTTGTTCA CGCTTGCNTG GCAGTTGGCG GCAAATGACG |
|
| 401 | CGAAACTCCC CCCGGGGCTG AAAATCACCA ACGGCAAAAA ACTTTATTCC |
|
| 451 | GTCGGCGGTT TGAATAAGGC GGGTACAGGA AAATACAGCA TAGGCGGCGT |
|
| 501 | GGAAACCGAA GTCGTCAAAT ATCGGGTGCG GCGCGGCGAC GATGCGGTAA |
|
| 551 | TGTATTTCTT CGCACCGTCC CTGAACAATA TTCCGGCACA AATCGGCTAT |
|
| 601 | ACCGACGACG GCAAAACCTA TACGCTGAAA CTCAAATCGG TGCAGATCAA |
|
| 651 | CGGCCAGGCA GCCAAACCGT AA |
[1606]This encodes a protein having amino acid sequence <SEQ ID 778>:
[0000]| 1 | MMKTFKNIFS AAILSAALPC AYAAGLPXSA VLHYSGSYGI PATXXXXXXX | |
|
| 51 | NAXKIVSTIK VPLYNIRFES GGTVVGNTLH PTYYRDIRRG KLYAEAKFAD |
|
| 101 | GSVTYGKAXX XXXXQSPKAM DLFTLAWQLA ANDAKLPPGL KITNGKKLYS |
|
| 151 | VGGLNKAGTG KYSIGGVETE VVKYRVRRGD DAVMYFFAPS LNNIPAQIGY |
|
| 201 | TDDGKTYTLK LKSVQINGQA AKP* |
[1607]ORF120a and ORF120-1 show 93.3% identity in 223 aa overlap:
[0000]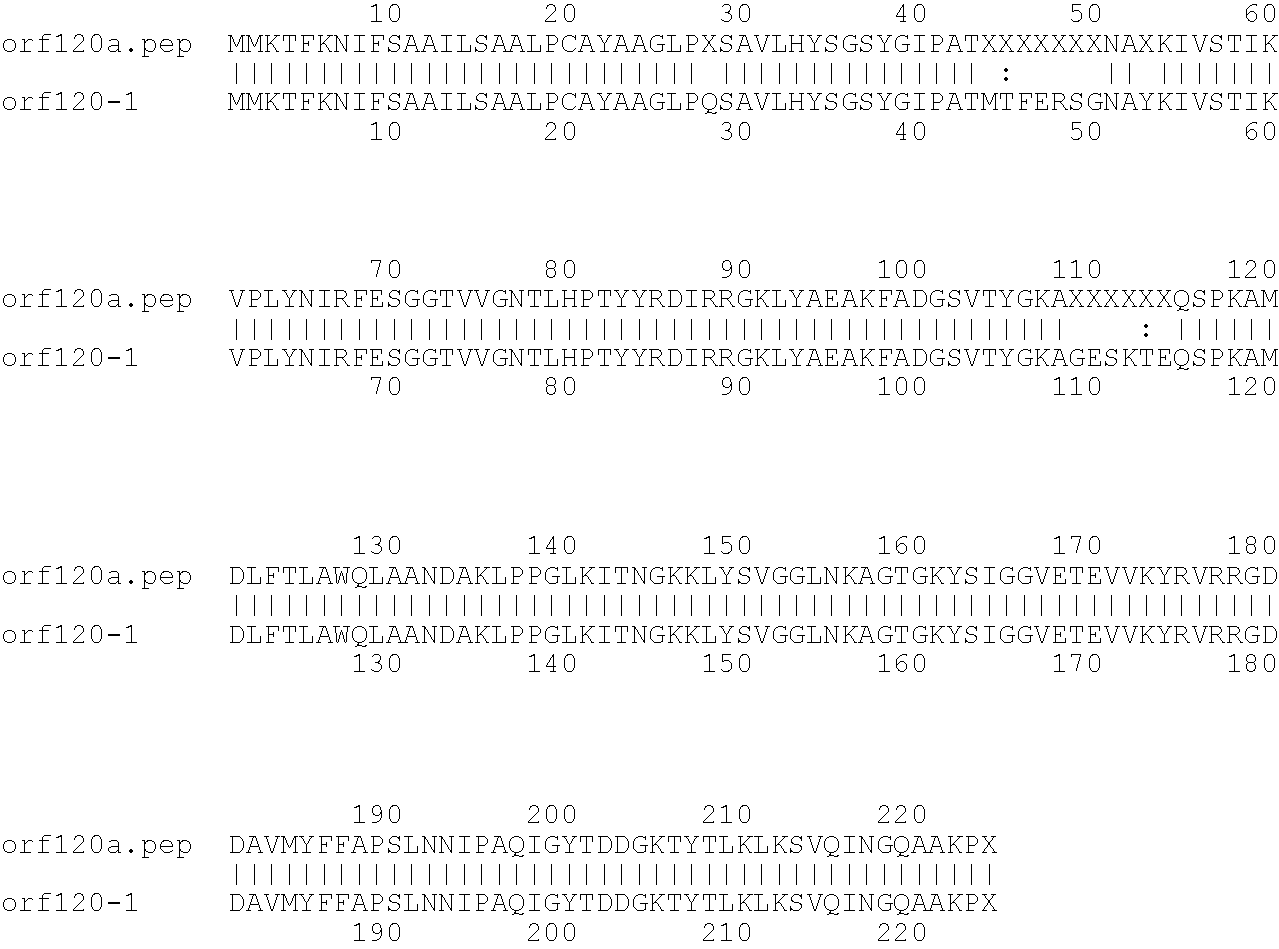
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1608]ORF120 shows 97.8% identity over 184 aa overlap with a predicted ORF (ORF120ng) from N. gonorrhoeae:
[0000]
[1609]The complete length ORF120ng nucleotide sequence <SEQ ID 779> is:
[0000]| 1 | ATGATGAAGA CTTTTAAAAA TATATTTTCC GCCGCCATTT TGTCCGCCGC | |
|
| 51 | CCTGCCGTGC GCGTATGCGG CAAGGCTACC CCAATCCGCC GTGCTGCACT |
|
| 101 | ATTCCGGCAG CTACGGCATT CCCGCCACGA TGACATTTGA ACGCAGCGGC |
|
| 151 | AATGCTTACA AAATCGTTTC GACGATTAAA GTGCCGCTAT ACAATATCCG |
|
| 201 | TTTCGAATCC GGCGGTACGG TTGTCGGCAA TACCCTGCAC CCTGCCTACT |
|
| 251 | ATAAAGACAT ACGCAGGGGC AAACTGTATG CGGAAGCCAA ATTCGCCGAC |
|
| 301 | GGCAGCGTAA CCTACGGCAA AGCGGGCGAG AGCAAAACCG AGCAAAGCCC |
|
| 351 | CAAGGCTATG GATTTGTTCA CGCTTGCCTG GCAGTTGGCG GCAAATGACG |
|
| 401 | CGAAACTCCC CCCGGGTCTG AAAATCACCA ACGGCAAAAA ACTTTATTCC |
|
| 451 | GTCGGCGGCC TGAATAAGGC GGGTACGGGA AAATACAGCA TaggCGGCGT |
|
| 501 | GGAAACCGAA GTCGTCAAAT ATCGGGTGCG GCGCGGCGAC GATACGGTAA |
|
| 551 | CGTATTTCTT CGCACCGTCC CTGAACAATA TTCCGGCACA AATCGGCTAT |
|
| 601 | ACCGACGACG GCAAAACCTA TACGCTGAAG CTCAAATCGG TGCAGATCAA |
|
| 651 | CGGACAGGCC GCCAAACCGT AA |
[1610]This encodes a protein having amino acid sequence <SEQ ID 780>:
[0000]| 1 | MMKTFKNIFS AAILSAALPC AYAARLPQSA VLHYSGSYGI PATMTFERSG | |
|
| 51 | NAYKIVSTIK VPLYNIRFES GGTVVGNTLH PAYYKDIRRG KLYAEAKFAD |
|
| 101 | GSVTYGKAGE SKTEQSPKAM DLFTLAWQLA ANDAKLPPGL KITNGKKLYS |
|
| 151 | VGGLNKAGTG KYSIGGVETE VVKYRVRRGD DTVTYFFAPS LNNIPAQIGY |
|
| 201 | TDDGKTYTLK LKSVQINGQA AKP* |
[1611]In comparison with ORF120-1, ORF120ng shows 97.8% identity in 223 aa overlap:
[0000]
[1612]This analysis, including the presence of a putative leader sequence in the gonococcal protein suggests that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 93
[1613]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 781>:
[0000]| 1 | ATGTATCGGA GGAAAGGGCG GGGCATCAAG CCGTGGATGG GTGCCGGTGC | |
|
| 51 | .GCGTTTGCC GCCTTGGTCT GGCTGGTTTT CGCGCTCGGC GATACTTTGA |
|
| 101 | CTCCGTTTGC GGTTGCGGCG GTGCTGGCGT ATGTATTGGA CCCTTTGGTC |
|
| 151 | GAATGGTTGC AGAAAAAGGG TTTGAACCGT GCATCCGCTT CGATGTCTGT |
|
| 201 | GATGGTGTTT TCCTTGATTT TGTTGTTGGC ATTATTGTTG ATTATCGTCC |
|
| 251 | CTATGCTGGT CGGGCAGTTC AACAATTTGG CATCGCGCCT GCCCCAATTA |
|
| 301 | ATCGGTTTTA TGCAGAACAC GCTGCTGCCG TGGTTGAAAA ATACAATCGG |
|
| 351 | CGGATATGTG GAAATCGATC AGGCATCTAT TATTGCGTGG CTTCAGGCGC |
|
| 401 | ATACGGGAGA GTTGAGCAAC GCGCTTAAGG CGTGGTTTCC CGTTTTGATG |
|
| 451 | AGGCAGGGCG GCAATATT.. |
[1614]This corresponds to the amino acid sequence <SEQ ID 782; ORF121>:
[0000]| 1 | MYRRKGRGIK PWMGAGXAFA ALVWLVFALG DTLTPFAVAA VLAYVLDPLV | |
|
| 51 | EWLQKKGLNR ASASMSVMVF SLILLLALLL IIVPMLVGQF NNLASRLPQL |
|
| 101 | IGFMQNTLLP WLKNTIGGYV EIDQASIIAW LQAHTGELSN ALKAWFPVLM |
|
| 151 | RQGGNI.. |
[1615]Further work revealed the complete nucleotide sequence <SEQ ID 783>:
[0000]| 1 | ATGTATCGGA GGAAAGGGCG GGGCATCAAG CCGTGGATGG GTGCCGGTGC | |
|
| 51 | GGCGTTTGCC GCCTTGGTCT GGCTGGTTTT CGCGCTCGGC GATACTTTGA |
|
| 101 | CTCCGTTTGC GGTTGCGGCG GTGCTGGCGT ATGTATTGGA CCCTTTGGTC |
|
| 151 | GAATGGTTGC AGAAAAAGGG TTTGAACCGT GCATCCGCTT CGATGTCTGT |
|
| 201 | GATGGTGTTT TCCTTGATTT TGTTGTTGGC ATTATTGTTG ATTATCGTCC |
|
| 251 | CTATGCTGGT CGGGCAGTTC AACAATTTGG CATCGCGCCT GCCCCAATTA |
|
| 301 | ATCGGTTTTA TGCAGAACAC GCTGCTGCCG TGGTTGAAAA ATACAATCGG |
|
| 351 | CGGATATGTG GAAATCGATC AGGCATCTAT TATTGCGTGG CTTCAGGCGC |
|
| 401 | ATACGGGAGA GTTGAGCAAC GCGCTTAAGG CGTGGTTTCC CGTTTTGATG |
|
| 451 | AGGCAGGGCG GCAATATTGT CAGCAGTATC GGCAACCTGC TGCTGCTTCC |
|
| 501 | CTTGCTGCTT TACTATTTCC TGCTGGATTG GCAGCGGTGG TCGTGCGGCA |
|
| 551 | TTGCCAAACT GGTTCCGAgG CGTTTTGCCG GTGCTTATAC GCGCATTACA |
|
| 601 | GGCAATTTGA ACGAGGTATT GGGCGAATTT TTGCGCGGGC AGCTTCTGGT |
|
| 651 | AATGCTGATT ATGGGCTTGG TTTACGGTTT GGGATTGGTG CTGGTCGGGC |
|
| 701 | TGGATTCGGG GTTTGCCATC GGTATGCTTG CCGGTATTTT GGTGTTTGTC |
|
| 751 | CCTTATCTCG GGGCGTTTAC GGGATTGCTG CTTGCCACCG TCGCCGCCTT |
|
| 801 | GCTCCAGTTC GGTTCGTGGA ACGGCATCCT ATCGGTTTGG GCGGTTTTTG |
|
| 851 | CCGTAGGACA GTTTCTCGAA AGTTTTTTCA TTACGCCGAA AATCGTGGGA |
|
| 901 | GACCGTATCG GGCTGTCGCC GTTTTGGGTT ATCTTTTCGC TGATGGCGTT |
|
| 951 | CGGGCAGCTG ATGGGCTTTG TCGGAATGTT GGCGGGATTG CCTTTGGCCG |
|
| 1001 | CCGTAACCTT GGTCTTGCTT CGCGAGGGCG TGCAGAAATA TTTTGCCGGC |
|
| 1051 | AGTTTTTACC GGGGCAGGTA G |
[1616]This corresponds to the amino acid sequence <SEQ ID 784; ORF121-1>:
[0000]| 1 | MYRRKGRGIK PWMGAGAAFA ALVWLVFALG DTLTPFAVAA VLAYVLDPLV | |
|
| 51 | EWLQKKGLNR ASASMSVMVF SLILLLALLL IIVPMLVGQF NNLASRLPQL |
|
| 101 | IGFMQNTLLP WLKNTIGGYV EIDQASIIAW LQAHTGELSN ALKAWFPVLM |
|
| 151 | RQGGNIVSSI GNLLLLPLLL YYFLLDWQRW SCGIAKLVPR RFAGAYTRIT |
|
| 201 | GNLNEVLGEF LRGQLLVMLI MGLVYGLGLV LVGLDSGFAI GMLAGILVFV |
|
| 251 | PYLGAFTGLL LATVAALLQF GSWNGILSVW AVFAVGQFLE SFFITPKIVG |
|
| 301 | DRIGLSPFWV IFSLMAFGQL MGFVGMLAGL PLAAVTLVLL REGVQKYFAG |
|
| 351 | SFYRGR* |
[1617]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1618]ORF121 shows 98.7% identity over a 156aa overlap with an ORF (ORF121a) from strain A of N. meningitidis.
[0000]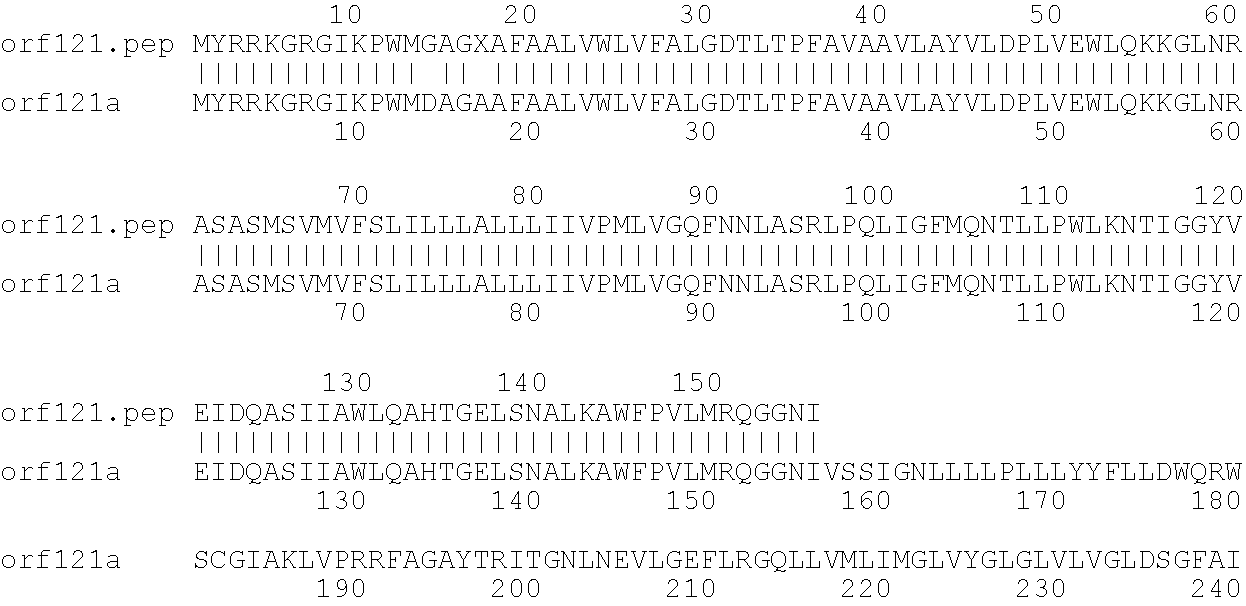
[1619]The complete length ORF121a nucleotide sequence <SEQ ID 785> is:
[0000]| 1 | ATGTATCGGA GGAAAGGGCG GGGCATCAAG CCGTGGATGG ATGCCGGTGC | |
|
| 51 | GGCGTTTGCC GCCTTGGTCT GGCTGGTTTT CGCGCTCGGC GATACTTTGA |
|
| 101 | CTCCGTTTGC GGTTGCGGCG GTGCTGGCGT ATGTATTGGA CCCTTTGGTC |
|
| 151 | GAATGGTTGC AGAAAAAGGG TTTGAACCGT GCATCCGCTT CGATGTCTGT |
|
| 201 | GATGGTGTTT TCCTTGATTT TGTTGTTGGC ATTATTGTTG ATTATTGTCC |
|
| 251 | CTATGCTGGT CGGGCAGTTC AACAATTTGG CATCGCGCCT GCCCCAATTA |
|
| 301 | ATCGGTTTTA TGCAGAACAC GCTGCTGCCG TGGTTGAAAA ATACAATCGG |
|
| 351 | CGGATATGTG GAAATCGATC AGGCATCTAT TATTGCGTGG CTTCAGGCGC |
|
| 401 | ATACGGGCGA GTTGAGCAAC GCGCTTAAGG CGTGGTTTCC CGTTTTGATG |
|
| 451 | AGGCAGGGCG GCAATATTGT CAGCAGTATC GGCAACCTGC TGCTGCTTCC |
|
| 501 | CTTGCTGCTT TACTATTTCC TGCTGGATTG GCAGCGGTGG TCGTGCGGCA |
|
| 551 | TTGCCAAACT GGTTCCGAGG CGTTTTGCCG GTGCTTATAC GCGCATTACA |
|
| 601 | GGCAATTTGA ACGAGGTATT GGGCGAATTT TTGCGCGGGC AGCTTCTGGT |
|
| 651 | GATGCTGATT ATGGGTTTGG TTTACGGCTT GGGGTTGGTG CTGGTCGGGC |
|
| 701 | TGGATTCGGG GTTTGCAATC GGTATGGTTG CCGGTATTTT GGTTTTTGTT |
|
| 751 | CCCTATTTGG GCGCGTTTAC AGGACTGCTG CTGGCAACCG TCGCCGCCTT |
|
| 801 | GCTCCAGTTC GGTTCGTGGA ACGGCATCTT GGCTGTTTGG GCGGTTTTTG |
|
| 851 | CCGTAGGACA GTTTCTCGAA AGTTTTTTCA TTACGCCGAA AATCGTGGGA |
|
| 901 | GACCGTATCG GCCTGTCGCC GTTTTGGGTT ATCTTTTCGC TGATGGCGTT |
|
| 951 | CGGGCAGCTG ATGGGCTTTG TCGGAATGTT GGCCGGATTG CCTTTGGCCG |
|
| 1001 | CCGTAACCTT GGTCTTGCTT CGCGAGGGCG TGCAGAAATA TTTTGCCGGC |
|
| 1051 | AGTTTTTACC GGGGCAGGTA G |
[1620]This encodes a protein having amino acid sequence <SEQ ID 786>:
[0000]| 1 | MYRRKGRGIK PWMDAGAAFA ALVWLVFALG DTLTPFAVAA VLAYVLDPLV | |
|
| 51 | EWLQKKGLNR ASASMSVMVF SLILLLALLL IIVPMLVGQF NNLASRLPQL |
|
| 101 | IGFMQNTLLP WLKNTIGGYV EIDQASIIAW LQAHTGELSN ALKAWFPVLM |
|
| 151 | RQGGNIVSSI GNLLLLPLLL YYFLLDWQRW SCGIAKLVPR RFAGAYTRIT |
|
| 201 | GNLNEVLGEF LRGQLLVMLI MGLVYGLGLV LVGLDSGFAI GMVAGILVFV |
|
| 251 | PYLGAFTGLL LATVAALLQF GSWNGILAVW AVFAVGQFLE SFFITPKIVG |
|
| 301 | DRIGLSPFWV IFSLMAFGQL MGFVGMLAGL PLAAVTLVLL REGVQKYFAG |
|
| 351 | SFYRGR* |
[1621]ORF121a and ORF121-1 show 99.2% identity in 356 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1622]ORF121 shows 97.4% identity over a 156 aa overlap with a predicted ORF (ORF121ng) from N. gonorrhoeae:
[0000]
[1623]An ORF121ng nucleotide sequence <SEQ ID 787> was predicted to encode a protein having amino acid sequence <SEQ ID 788>:
[0000]| 1 | MYRRKGRGIK PWMGAGAAFA ALVWLVYALG DTLTPFAVAA VLAYVLDPLV | |
|
| 51 | EWLQKKGLNR ASASMSVMVF SLILLLALLL IIVPMLVGQF NNLASRLPQL |
|
| 101 | IGFMQNTLLP WLKNTIGGYV EIDQASIIAW FQAHTGELSN ALKAWFPVLM |
|
| 151 | KQGGNIVSTI GNLLLPPLLL YYFLLDWHRW SCGIPKLVPR RFAGAYTRIT |
|
| 201 | GNLNKVWGKF LRGQLLGETE RGAVVCRVGR ECWEGGGARS RPSDDGWPRW |
|
| 251 | GGG* |
[1624]Further work revealed the following gonoccocal DNA sequence <SEQ ID 789>:
[0000]| 1 | ATGTATCGGA GAAAAGGACG GGGCATCAAG CCGTGGATGG GTGCCGGCGC | |
|
| 51 | GGCGTTTGCC GCCTTGGTCT GGCTGGTTTA CGCGCTCGGC GATACTTTGA |
|
| 101 | CTCCGTTTGC GGTTGCGGCG GTGCTGGCGT ATGTGTTGGA CCCTTTGGTC |
|
| 151 | GAATGGTTGC AGAAAAAGGG TTTGAACCGT GCATCCGCTT CGATGTCTGT |
|
| 201 | GATGGTGTTT TCCTTGATTT TGTTGTTGGC ATTATTGTTG ATTATTGTCC |
|
| 251 | CTATGCTGGT CGGGCAGTTC AATAATTTGG CATCTCGCCT GCCCCAATTA |
|
| 301 | ATCGGTTTTA TGCAGAACAC GCTGCTGCCG TGGTTGAAAA ATACAATCGG |
|
| 351 | CGGATATGTG GAAATCGATC AGGCATCTAT TATTGCGTGG TTTCAGGCGC |
|
| 401 | ATACGGGCGA GTTGAGCAAC GCGCTTAAGG CGTGGTTTCC CGTTTTGATG |
|
| 451 | AAACAGGGCG GCAATATTGT CAGCAGTATC GGCAACCTGC TGCTGCCGCC |
|
| 501 | CTTGCTGCTT TACTATTTCC TGCTGGATTG GCAGCGGTGG TCGTGCGGCA |
|
| 551 | TCGCCAAACT GGTTCCGAGG CGTTTTGCCG GTGCTTATAC GCGCATTACG |
|
| 601 | GGTAATTTGA ACGAGGTATT GGGCGAATTT TTGCGCGGTC AGCTTCTGGT |
|
| 651 | GATGCTGATT ATGGGCTTGG TTTACGGTTT GGGATTGATG CTAGTCGGAC |
|
| 701 | TGGATTCGGG ATTTGCCATC GGTATGGTTG CCGGTATTTT GGTGTTTGTC |
|
| 751 | CCCTATTTGG GTGCGTTTAC GGGATTGCTG CTTGCCACTG TTGCAGCCTT |
|
| 801 | GCTCCAGTTC GGTTCGTGGA ACGGAATCTT GGCTGTTTGG GCGGTTTTTG |
|
| 851 | CCGTCGGTCA GTTTCTCGAA AGTTTTTTCA TTACGCCGAA AATTGTAGGA |
|
| 901 | GACCGTATCG GCCTGTCGCC GTTTTGGGTT ATCTTTTCGC TGATGGCGTT |
|
| 951 | CGGAGAGCTG ATGGGCTTTG TCGGAATGTT GGCCGGATTG CCTTTGGCCG |
|
| 1001 | CCGTAACCTT GGTCTTGCTT CGCGAGGGCG CGCAGAAATA TTTTGCCGGC |
|
| 1051 | AGTTTTTACC GGGGCAGGTA G |
[1625]This corresponds to the amino acid sequence <SEQ ID 790; ORF121ng-1>:
[0000]| 1 | MYRRKGRGIK PWMGAGAAFA ALVWLVYALG DTLTPFAVAA VLAYVLDPLV | |
|
| 51 | EWLQKKGLNR ASASMSVMVF SLILLLALLL IIVPMLVGQF NNLASRLPQL |
|
| 101 | IGFMQNTLLP WLKNTIGGYV EIDQASIIAW FQAHTGELSN ALKAWFPVLM |
|
| 151 | KQGGNIVSSI GNLLLPPLLL YYFLLDWQRW SCGIAKLVPR RFAGAYTRIT |
|
| 201 | GNLNEVLGEF LRGQLLVMLI MGLVYGLGLM LVGLDSGFAI GMVAGILVFV |
|
| 251 | PYLGAFTGLL LATVAALLQF GSWNGILAVW AVFAVGQFLE SFFITPKIVG |
|
| 301 | DRIGLSPFWV IFSLMAFGEL MGFVGMLAGL PLAAVTLVLL REGAQKYFAG |
|
| 351 | SFYRGR* |
[1626]ORF121ng-1 and ORF121-1 show 97.5% identity in 356 aa overlap:
[0000]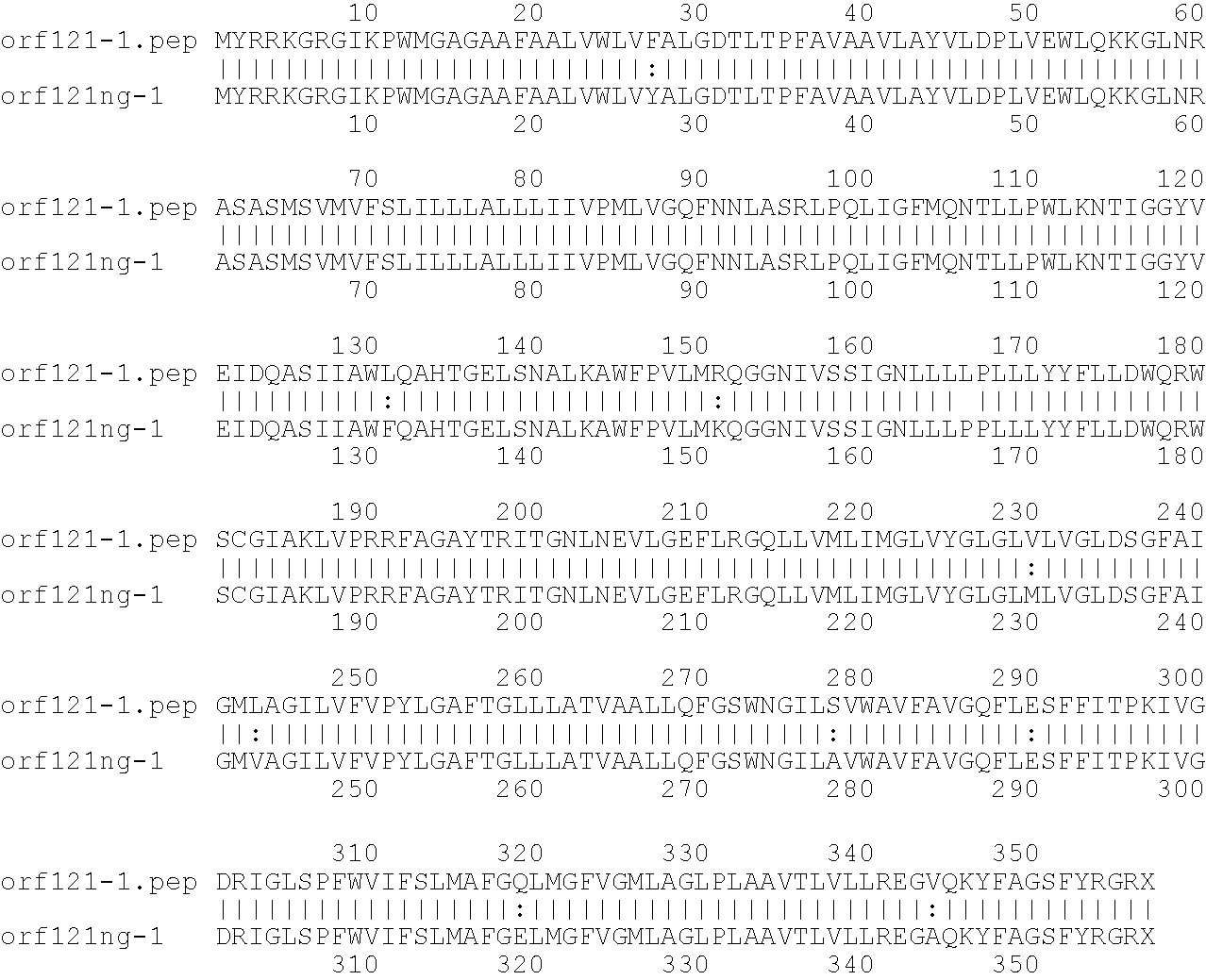
[1627]In addition, ORF121ng-1 shows homology to a permease from H. influenzae:
[0000]| sp|P43969|PERM_HAEIN PUTATIVE PERMEASE PERM HOMOLOG Length = 349 | |
| Score = 69.9 bits (168), Expect = 2e−11 |
| Identities = 67/317 (21%), Positives = 120/317 (37%), Gaps = 7/317 (2%) |
|
| Query: | 26 | VYALGDTLTPFAVAAVLAYVLDPLVEWL-QKKGLNRASASMSVMVFSXXXXXXXXXXXVP | 84 | |
| | +Y GD + P +A VL+Y+L+ + +L Q R A++ + VP |
| Sbjct: | 32 | IYFFGDLIAPLLIALVLSYLLEIPINFLNQYLKCPRMLATILIFGSFIGLAAVFFLVLVP | 91 |
|
| Query: | 85 | MLVGQFNNLASRLPQLIGFMQNTLLPWLKNTIGGYVE-IDQASIIAWFQAHTGELSNALK | 143 |
| | ML Q +L S LP + N WL N Y E ID + + + F + ++ + |
| Sbjct: | 92 | MLWNQTISLLSDLPAMF----NKSNEWLLNLPKNYPELIDYSMVDSIFNSVREKILGFGE | 147 |
|
| Query: | 144 | AWFPVLMKQGGNIVSSIGNXXXXXXXXXXXXXDWQRWSCGIAKLVPRRFAGAYTRITGNL | 203 |
| | + + + N+VS D G+++ +P+ A+ R + |
| Sbjct: | 148 | SAVKLSLASIMNLVSLGIYAFLVPLMMFFMLKDKSELLQGVSRFLPKNRNLAFXRWK-EM | 206 |
|
| Query: | 204 | NEVLGEFLRGQXXXXXXXXXXXXXXXXXXXXDSGFAIGMVAGILVFVPYXXXXXXXXXXX | 263 |
| | + + ++ G+ + + G+ V VPY |
| Sbjct: | 207 | QQQISNYINGKLLEILIVTLITYIIFLIFGLNYPLLLAFAVGLSVLVPYIGAVIVTIPVA | 266 |
|
| Query: | 264 | XXXXXQFGSWNGILAVWAVFAVGQFLESFFITPKIVGDRIGLSPFWVIFSLMAFGELMGF | 323 |
| | QFG + FAV Q L+ + P + + + L P +I S++ FG L GF |
| Sbjct: | 267 | LVALFQFGISPTFWYIIIAFAVSQLLDGNLLVPYLFSEAVNLHPLIIIISVLIFGGLWGF | 326 |
|
| Query: | 324 | VGMLAGLPLAAVTLVLL | 340 |
| | G+ +PLA + ++ |
| Sbjct: | 327 | WGVFFAIPLATLVKAVI | 343 |
[1628]Based on this analysis, including the presence of a putative leader sequence and transmembrane domains in the two proteins, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 94
[1629]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 791>:
[0000]| 1 | ..ACTGCTTTTT CGGCGGCGCT GCGCTTGAGT CCATCATGAC TCGTCATATT | |
|
| 51 | TTTGTCCTTT GGGAAACCGT ATCAACAAAC AGCCGCCATC TTAACATTTT |
|
| 101 | TTTGCACGTC CTGCCCGCCG CGTTCAAATG CGTACCAGCA ATACCGCCGC |
|
| 151 | CTGCGCCTCT ATGCCTTCCA TCCGCCCGAG ATAGCCGAGT TTTTCGTTGG |
|
| 201 | TTTTGCCTTT GATGTTGACG CACGAAATGT CTATGCCCAA ATCGGCGGCG |
|
| 251 | ATGTTGGCAC GCATTTGCGG AATGTGCGGC GCGAGTGTGG GTTTCTGTGC |
|
| 301 | AATCACGGTC GTATCGACAT TGACCGCCTG CCAACCCTGC GCCTGAACGC |
|
| 351 | TTTGATACGC CGCACGCAAA AGGACGCGGC TGTCCGCATC TTTGAACTCT |
|
| 401 | GCGGCGGTGT CGGGGAAATG GCTGCCGATA TCGCCCAAAC CTGCCGCACC |
|
| 451 | GAGCAGCGCG TCGGTAACGG CGTGCAGCAG CGCATCGGCA TCGGAGTGTC |
|
| 501 | CGAGCAGCCC TTTTTCAAAT GGGATTTCAA CTCCGCCAAG TATCAG.. |
[1630]This corresponds to the amino acid sequence <SEQ ID 792; ORF122>:
[0000]| 1 | ..TAFSAALRLS PSXLVIFLSF GKPYQQTAAI LTFFCTSCPP RSNAYQQYRR | |
|
| 51 | LRLYAFHPPE IAEFFVGFAF DVDARNVYAQ IGGDVGTHLR NVRRECGFLC |
|
| 101 | NHGRIDIDRL PTLRLNALIR RTQKDAAVRI FELCGGVGEM AADIAQTCRT |
|
| 151 | EQRVGNGVQQ RIGIGVSEQP FFKWDFNSAK YQ.. |
[1631]Further work revealed the complete nucleotide sequence <SEQ ID 793>:
[0000]| 1 | ATATCGTACT GGGCAAGCAG TTCGCCGGAT TTTTTGGAAG TAGATACCGC | |
|
| 51 | GCCTTTGATT TTTTTGCCGC TCTTACCCAA GGCTTCGATG AAAAAGTTGA |
|
| 101 | TGGTCGAGCC GGTACCGATG CCGATATATT CATTTTCGGG TACGAATTCG |
|
| 151 | ACTGCTTTTT CGGCGGCGAT GCGCTTGAGT TCGTCTTGTG TCGTCATATT |
|
| 201 | TTTGTCCTTT GGGAAACCGT ATCAACAAAC AGCCGCCATC TTAACATTTT |
|
| 251 | TTTGCACGTC CTGCCCGCCG CGTTCAAATG CGTACCAGCA ATACCGCCGC |
|
| 301 | CTGCGCCTCT ATGCCTTCCA TCCGCCCGAG ATAGCCGAGT TTTTCGTTGG |
|
| 351 | TTTTGCCTTT GATGTTGACG CACGAAATGT CTATGCCCAA ATCGGCGGCG |
|
| 401 | ATGTTGGCAC GCATTTGCGG AATGTGCGGC GCGAGTTTGG GTTTCTGTGC |
|
| 451 | AATCACGGTC GTATCGACAT TGACCGCCTG CCAACCCTGC GCCTGAACGC |
|
| 501 | TTTGATACGC CGCACGCAAA AGGACGCGGC TGTCCGCATC TTTGAACTCT |
|
| 551 | GCGGCGGTGT CGGGGAAATG GCTGCCGATA TCGCCCAAAC CTGCCGCACC |
|
| 601 | GAGCAGCGCG TCGGTAACGG CGTGCAGCAG CGCATCGGCA TCGGAGTGTC |
|
| 651 | CGAGCAGCCC TTTTTCAAAT GGGATTTCAA CTCCGCCAAG TATCAGCTTT |
|
| 701 | CTGCCTTCGG TCAGTTGGTG GACATCGTAG CCCTGTCCGA TACGGATGTT |
|
| 751 | CGTCATCGTT TGTGTTCCTG A |
[1632]This corresponds to the amino acid sequence <SEQ ID 794; ORF122-1>:
[0000]| 1 | ISYWASSSPD FLEVDTAPLI FLPLLPKASM KKLMVEPVPM PIYSFSGTNS | |
|
| 51 | TAFSAAMRLS SSCVVIFLSF GKPYQQTAAI LTFFCTSCPP RSNAYQQYRR |
|
| 101 | LRLYAFHPPE IAEFFVGFAF DVDARNVYAQ IGGDVGTHLR NVRREFGFLC |
|
| 151 | NHGRIDIDRL PTLRLNALIR RTQKDAAVRI FELCGGVGEM AADIAQTCRT |
|
| 201 | EQRVGNGVQQ RIGIGVSEQP FFKWDFNSAK YQLSAFGQLV DIVALSDTDV |
|
| 251 | RHALCS* |
[1633]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1634]ORF122 shows 94.0% identity over a 182aa overlap with an ORF (ORF122a) from strain A of N. meningitidis:
[0000]
[1635]The complete length ORF122a nucleotide sequence <SEQ ID 795> is:
[0000]| 1 | ATATCATATT GGGCAAGCAG TTCACTGGAT TTTTTGGAAG TAGATACCGC | |
|
| 51 | GCCTTTGATT TTTTTGCCGC TCTTACCCAA GGCTTCGATG AAAAAGTTGA |
|
| 101 | TGGTCGAACC GGTACCGATG CCGATGTATT CGTTTTCGGG TACGAATTCG |
|
| 151 | ACTGCNTTTT CGGCGGCGAT GCGCTTGAGT TCGTCTTGTG TCGTCATATT |
|
| 201 | TTTGTCCTTT GGGAAACCGT ATCAACAAAC AGCCGCCATC TTAACATTTT |
|
| 251 | TTNNNACGTC CTGCCCGCCG CGTTCAAATC CTTACCAGCA ATACCGCCGC |
|
| 301 | CTGCGACTCT ATGCCTTCCA TGCGCCCGAG ATAACCGAGT TTTTCGTTGG |
|
| 351 | TTTTGCCTTT GANGTTGACG CACGAAATGT CTATGCCCAA ATCGGCGGCG |
|
| 401 | ATGTTGGCAC GCATTTGCGG AATATGCGGC GCGAGTTTGG GTTTCTGTGC |
|
| 451 | AATCACGGTC GTATCGACAT TGACCGCCTG CCAACCCTGC GCCTGAACGC |
|
| 501 | TTTGATACGC CGCACGCAAA AGGACGCGGC TGTCCGCATC TTTGAACTCT |
|
| 551 | GCGGCGGTGT CGGGGAAATG GCTGCCGATA TCGCCCAAAC CTGCCGCACC |
|
| 601 | GAGCAGCGCG TCGGTAACGG CGTGCAGCAG CGCATCGGCA TCGGAGTGTC |
|
| 651 | CGAGCAGCCC TTTTTCAAAT GGGATTTCAA CTCCGCCAAG TATCAGCTTT |
|
| 701 | CTGCCTTCGG TCAGTTGGTG GACATCGTAG CCCTGTCCGA TACGGATGTT |
|
| 751 | CGTCATCGTT TGTGTTCCTG A |
[1636]This encodes a protein having amino acid sequence <SEQ ID 796>:
[0000]| 1 | ISYWASSSLD FLEVDTAPLI FLPLLPKASM KKLMVEPVPM PMYSFSGTNS | |
|
| 51 | TAFSAAMRLS SSCVVIFLSF GKPYQQTAAI LTFFXTSCPP RSNPYQQYRR |
|
| 101 | LRLYAFHAPE ITEFFVGFAF XVDARNVYAQ IGGDVGTHLR NMRREFGFLC |
|
| 151 | NHGRIDIDRL PTLRLNALIR RTQKDAAVRI FELCGGVGEM AADIAQTCRT |
|
| 201 | EQRVGNGVQQ RIGIGVSEQP FFKWDFNSAK YQLSAFGQLV DIVALSDTDV |
|
| 251 | RHRLCS* |
[1637]ORF122a and ORF122-1 show 96.9% identity in 256 aa overlap:
[0000]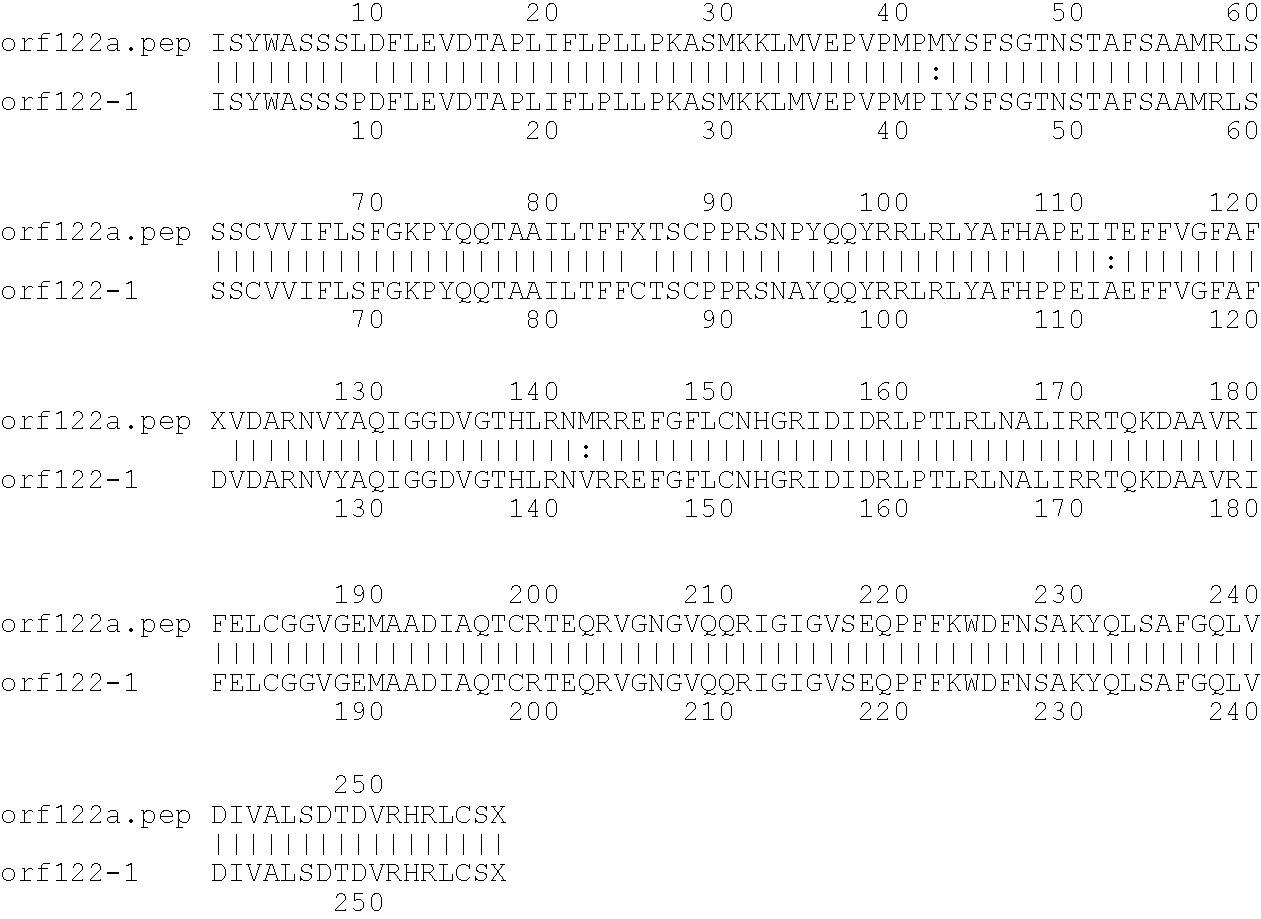
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1638]ORF122 shows 89.6% identity over a 182 aa overlap with a predicted ORF (ORF122ng) from N. gonorrhoeae:
[0000]
[1639]The complete length ORF122ng nucleotide sequence <SEQ ID 797> is:
[0000]| 1 | ATGTCGTACC GGGCAAGCAG TTCGCCGGAT TTTTTGGAGG TTGAAACCGC | |
|
| 51 | GCCTTTGATT TTTTTACCGC TTTTGCCCAA GGCTTCGATG AAGAAATTGa |
|
| 101 | tgGTCGAACC GgtaCCGATG CCGATGTATT CGTTTTCGGG TACGAATTCG |
|
| 151 | ACTGCTTTTT CGGCGGCGAT GCGCttgAgt TCgtcttgcg TcgTCATATT |
|
| 201 | TTTAtccttt gGGAAaccct atcaAcaAAc agccgccatC TTAACATTTT |
|
| 251 | TTTGCACGtc ctggccgccg cgttcaAATc cgtaccaGca ataccgccgc |
|
| 301 | ctgcgcctCT AtgcCTTCCA TCCGCCCGAG ATAGCCGAGT TTTTCGTTGG |
|
| 351 | TTTTGCCTTT GATatTGACG CACGAAATAT CGatacCCAa atcggcgGCG |
|
| 401 | ATGTTGGCAC GCATTTGCGG AATGTGCGGT GCGAGTTTGG GTTTCTGTGC |
|
| 451 | AATCACGGTC GTATCGACAT TGACCACCTG CCAACCCTGC GCCTGAACGC |
|
| 501 | TTTGATACGC CGCACGCAAA AGGACGCGGC TGTCCGCATC TTTGAACTCT |
|
| 551 | GCGGCGGTGT CGGGAAAATG GCTGCCGATG TCGCCCAAAC CTGCCGCACC |
|
| 601 | GAGCAGCgcg tcggtaaCGG CGTGCAGCAG cgcgTcgGCA TCCGAATGCC |
|
| 651 | CGAGCAGCCC TTTTTCAAAT GGGATTTCAA CTCCGCCAAG TATCAGCTTT |
|
| 701 | CTGCCTTCGG TCAATTGGTG GACATCGTAG CCCTGTCCGA TACGGATATT |
|
| 751 | CGTCATCGTT TGTGTTCCTG A |
[1640]This encodes a protein having amino acid sequence <SEQ ID 798>:
[0000]| 1 | MSYRASSSPD FLEVETAPLI FLPLLPKASM KKLMVEPVPM PMYSFSGTNS | |
|
| 51 | TAFSAAMRLS SSCVVIFLSF GKPYQQTAAI LTFFCTSWPP RSNPYQQYRR |
|
| 101 | LRLYAFHPPE IAEFFVGFAF DIDARNIDTQ IGGDVGTHLR NVRCEFGFLC |
|
| 151 | NHGRIDIDHL PTLRLNALIR RTQKDAAVRI FELCGGVGKM AADVAQTCRT |
|
| 201 | EQRVGNGVQQ RVGIRMPEQP FFKWDFNSAK YQLSAFGQLV DIVALSDTDI |
|
| 251 | RHRLCS* |
[1641]ORF122ng and ORF122-1 show 92.6% identity in 256 aa overlap:
[0000]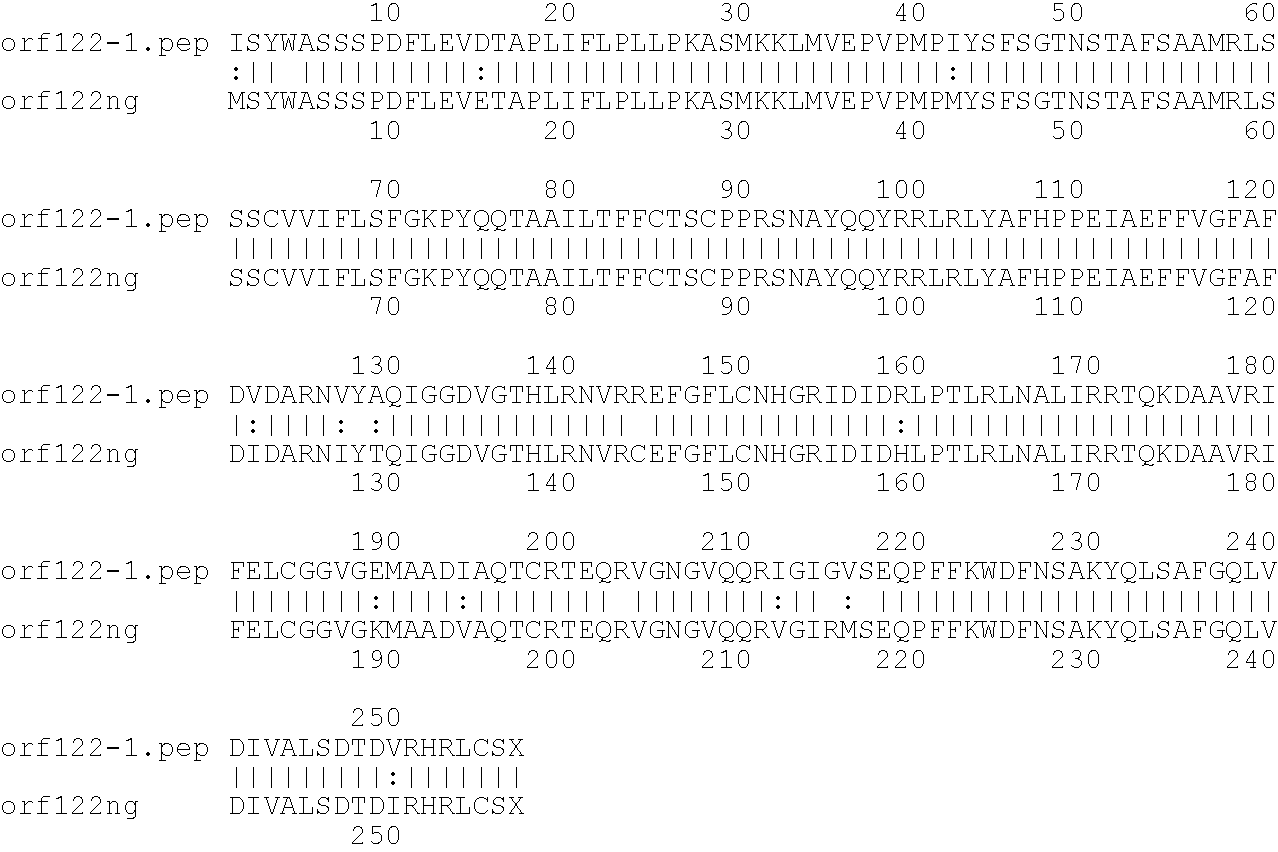
[1642]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 95
[1643]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 799>:
[0000]| 1 | ..GCCGGCGCGA GTGCGAACAA CATTTCCGCG CGTTTTGCGG AAACACCCGT | |
|
| 51 | CGCTGTCAGC GTTACCCTGA TCGGCACGGT ACTTGCCGTC ATGCTGCCCG |
|
| 101 | TTACCGAATA TGAAAACTTC CTGCTGCTTA TCGGCTCGGT ATTTGCGCCG |
|
| 151 | ATGGGGCGGA TTTTGATTGC CGACTTTTTC GTCTTGAAAC GGCGTGA |
[1644]This corresponds to the amino acid sequence <SEQ ID 800; ORF125>:
[0000]| 1 | ..AGASANNISA RFAETPVAVS VTLIGTVLAV MLPVTEYENF LLLIGSVFAP | |
|
| 51 | MGGFDCRLFR LETA* |
[1645]Further work revealed the complete nucleotide sequence <SEQ ID 801>:
[0000]| 1 | ATGTCGGGCA ATGCCTCCTC TCCTTCATCT TCCTCCGCCA TCGGGCTGAT | |
|
| 51 | TTGGTTCGGC GCGGCGGTAT CGATTGCCGA AATCAGCACG GGTACGCTGC |
|
| 101 | TTGCGCCTTT GGGCTGGCAG CGCGGTCTGG CGGCTCTACT TTTGGGTCAT |
|
| 151 | GCCGTCGGCG GCGCGCTGTT TTTTGCGGCG GCGTATATCG GCGCACTGAC |
|
| 201 | CGGACGCAGC TCGATGGAAA GCGTGCGCCT GTCGTTCGGC AAACGCGGTT |
|
| 251 | CAGTGCTGTT TTCCGTGGCG AATATGCTGC AACTGGCCGG CTGGACGGCG |
|
| 301 | GTGATGATTT ACGCCGGCGC AACGGTCAGC TCCGCTTTGG GCAAAGTGTT |
|
| 351 | GTGGGACGGC GAATCTTTTG TCTGGTGGGC ATTGGCAAAC GGCGCGCTGA |
|
| 401 | TTGTGCTGTG GCTGGTTTTC GGCGCACGCA AAACAGGCGG GCTGAAAACC |
|
| 451 | GTTTCGATGC TGCTGATGCT GTTGGCGGTT CTGTGGCTGA GTGCCGAAGT |
|
| 501 | CTTTTCCACG GCAGGCAGCA CCGCCGCACA GGTTTCAGAC GGCATGAGTT |
|
| 551 | TCGGAACGGC AGTCGAGCTG TCCGCCGTGA TGCCGCTTTC CTGGCTGCCG |
|
| 601 | CTTGCCGCCG ACTACACGCG CCACGCGCGC CGCCCGTTTG CGGCAACCCT |
|
| 651 | GACGGCAACG CTCGCCTACA CGCTGACCGG CTGCTGGATG TATGCCTTGG |
|
| 701 | GTTTGGCAGC GGCGTTGTTC ACCGGAGAAA CCGACGTGGC AAAAATCCTG |
|
| 751 | CTGGGCGCAG GTTTGGGTGC GGCAGGCATT TTGGCGGTCG TCCTCTCCAC |
|
| 801 | CGTTACCACA ACGTTTCTCG ATGCCTATTC CGCCGGCGCG AGTGCGAACA |
|
| 851 | ACATTTCCGC GCGTTTTGCG GAAACACCCG TCGCTGTCGG CGTTACCCTG |
|
| 901 | ATCGGCACGG TACTTGCCGT CATGCTGCCC GTTACCGAAT ATGAAAACTT |
|
| 951 | CCTGCTGCTT ATCGGCTCGG TATTTGCGCC GATGGCGGCG GTTTTGATTG |
|
| 1001 | CCGACTTTTT CGTCTTGAAA CGGCGTGAGG AGATTGAAGG CTTTGACTTT |
|
| 1051 | GCCGGACTGG TTCTGTGGCT TGCGGGCTTC ATCCTCTACC GCTTCCTGCT |
|
| 1101 | CTCGTCCGGC TGGGAAAGCA GCATCGGTCT GACCGCCCCC GTAATGTCTG |
|
| 1151 | CCGTTGCCAT TGCCACCGTA TCGGTACGCC TTTTCTTTAA AAAAACCCAA |
|
| 1201 | TCTTTACAAA GGAACCCGTC ATGA |
[1646]This corresponds to the amino acid sequence <SEQ ID 802; ORF125-1>:
[0000]| 1 | MSGNASSPSS SSAIGLIWFG AAVSIAEIST GTLLAPLGWQ RGLAALLLGH | |
|
| 51 | AVGGALFFAA AYIGALTGRS SMESVRLSFG KRGSVLFSVA NMLQLAGWTA |
|
| 101 | VMIYAGATVS SALGKVLWDG ESFVWWALAN GALIVLWLVF GARKTGGLKT |
|
| 151 | VSMLLMLLAV LWLSAEVFST AGSTAAQVSD GMSFGTAVEL SAVMPLSWLP |
|
| 201 | LAADYTRHAR RPFAATLTAT LAYTLTGCWM YALGLAAALF TGETDVAKIL |
|
| 251 | LGAGLGAAGI LAVVLSTVTT TFLDAYSAGA SANNISARFA ETPVAVGVTL |
|
| 301 | IGTVLAVMLP VTEYENFLLL IGSVFAPMAA VLIADFFVLK RREEIEGFDF |
|
| 351 | AGLVLWLAGF ILYRFLLSSG WESSIGLTAP VMSAVAIATV SVRLFFKKTQ |
|
| 401 | SLQRNPS* |
[1647]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1648]ORF125 shows 76.5% identity over a 51aa overlap with an ORF (ORF125a) from strain A of N. meningitidis:
[0000]
[1649]The ORF125a partial nucleotide sequence <SEQ ID 803> is:
[0000]| 1 | ATGTCGGGCA ATGCCTCCTC TCNTTCATCT TCCGCCGCCA TCGGGCTGAT | |
|
| 51 | TTGGTTCGGC GCGGCGGTAT CGATTGCCGA AATCAGCACG GGTACACTGC |
|
| 101 | TTGCGCCTTT GGGCTGGCAG CGCGGTCTGG CNGCTCTGCT TTTGGGTCAT |
|
| 151 | GCCGTCGGCG GCGCGCTGTT TTTTGCGGCG GCGTATATCG GCGCACTGAC |
|
| 201 | CGGACNCANC TCGATGGAAA GCGTGCGCCT GTCGTTCGGC AAACGCGGTT |
|
| 251 | CAGTGCTGTT TTCCGTGGCG AATATGCTGC AACTGGCCGG CTGGACGGCG |
|
| 301 | GTGATGATTT ACGCCGGCGC AACGGTCAGC TCCGCTTTGG GCAAAGTGTT |
|
| 351 | GTGGGACGGC GAATCTTTTG TCTGGTGGGC ATTGGCAAAC GGCGCGCTGA |
|
| 401 | TTGTGCTGTG GCTGGTTTTC GGCGCACGCA AAACAGGCGG GCTGAAAACC |
|
| 451 | GTTTCGATGC TGCTGATGCT GTTGGCGGTT CTGTGGCTGA GTGCCGAANT |
|
| 501 | NTTTTCCACG GCAGGCAGCA CCGCCGCANN GGTNNCAGAC GGCATGAGTT |
|
| 551 | TCGGAACGGC AGTCGAGCTG TCCGCCGTNA TGCCGCTTTC TTGGCTGCCG |
|
| 601 | CTGGCCGCCG ACTACACGCG CCACGCGCGC CGCCCGTTTG CGGCAACCCT |
|
| 651 | GACGGCAACG CTCGCCTACA CGCTGACCGG CTGCTGGATG TATGCCTTGG |
|
| 701 | GTTTGGCAGC GGCGTTGTTC ACCGGAGAAA CCGACGTGGC AAAAATCCTG |
|
| 751 | CTGGGCGCAG GTTTGGGTGC GGCAGGCATT TTGGCGGTCG TCCTGTCGAC |
|
| 801 | CGTTACCACC ACTTTTCTCG ATGCNTACTC CGCCGGCGTA AGTGCCAACA |
|
| 851 | ATATTTCCGC CAAACTTTCG GAAATACCNA TCGCCGTTGC CGTCGCCGTT |
|
| 901 | GTCGGCACAC TGCTTGCCGT CCTCCTGCCC GTTACCGAAT ATGAAAACTT |
|
| 951 | CCTGCTGCTT ATCGGCTCGG TATTTGCGCC GATGGCGGCG GTTTTGATTG |
|
| 1001 | CCGACTTTTT CGTCTTGAAA CGGCGTGAGG AGATTGAAGG C.. |
[1650]This encodes a protein having the partial amino acid sequence <SEQ ID 804>:
[0000]| 1 | MSGNASSXSS SAAIGLIWFG AAVSIAEIST GTLLAPLGWQ RGLAALLLGH | |
|
| 51 | AVGGALFFAA AYIGALTGXX SMESVRLSFG KRGSVLFSVA NMLQLAGWTA |
|
| 101 | VMIYAGATVS SALGKVLWDG ESFVWWALAN GALIVLWLVF GARKTGGLKT |
|
| 151 | VSMLLMLLAV LWLSAEXFST AGSTAAXVXD GMSFGTAVEL SAVMPLSWLP |
|
| 201 | LAADYTRHAR RPFAATLTAT LAYTLTGCWM YALGLAAALF TGETDVAKIL |
|
| 251 | LGAGLGAAGI LAVVLSTVTT TFLDAYSAGV SANNISAKLS EIPIAVAVAV |
|
| 301 | VGTLLAVLLP VTEYENFLLL IGSVFAPMAA VLIADFFVLK RREEIEG.. |
[1651]ORF125a and ORF125-1 show 94.5% identity in 347 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1652]ORF125 shows 86.2% identity over a 65aa overlap with a predicted ORF (ORF125ng) from N. gonorrhoeae:
[0000]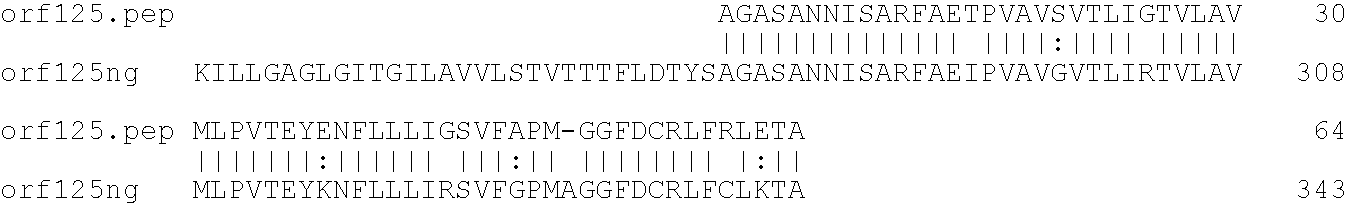
[1653]An ORF125ng nucleotide sequence <SEQ ID 805> was predicted to encode a protein having amino acid sequence <SEQ ID 806>:
[0000]| 1 | MSGNASSPSS SAAIGLVWFG AAVSIAEIST GTLLAPLGWQ RGLAALLLGH | |
|
| 51 | AVGGALFFAA AYIGALTGRS SMESVRLSFG KCGSVLFSVA NMLQLAGWTA |
|
| 101 | VMIYVGATVS SALGKVLWDG ESFVWWALAN GALIVLWLVF GARRTGGLKT |
|
| 151 | VSMLLMLLAV LWLSVEVFAS SGTNAAPAVS DGMTFGTAVE LSAVMPLSWL |
|
| 201 | PLAADYTRQA RRPFAATLTA TLAYTLTGCW MYALGLAAAL FTGETDVAKI |
|
| 251 | LLGAGLGITG ILAVVLSTVT TTFLDTYSAG ASANNISARF AEIPVAVGVT |
|
| 301 | LIRTVLAVML PVTEYKNFLL LIRSVFGPMA GGFDCRLFCL KTA* |
[1654]Further work revealed the following gonococcal DNA sequence <SEQ ID 807>:
[0000]| 1 | ATGTCGGGCA ATGCCTCCTC TCCTTCATCT TCCGCCGCCA TCGGGCTGGT | |
|
| 51 | TTGGTTCGGC GCGGCGGTAT CGATTGCCGA AATCAGCACG GGTACGCTGC |
|
| 101 | TCGCCCCCTT GGGCTGGCAG CGCGGTCTGG CGGCCCTGCT TTTGGGTCAT |
|
| 151 | GCCGTCGGCG GCGCGCTGTT TTTTGCGGCG GCGTATATCG GCGCACTGAC |
|
| 201 | CGGACGCAGC TCGATGGAAA GTGTGCGCCT GTCGTTCGGC AAATGCGGTT |
|
| 251 | CAGTGCTGTT TTCCGTGGCG AATATGCTGC AACTGGCCGG CTGGACGGCG |
|
| 301 | GTGATGATTT ACGTCGGCGC AACGGTCAGC TCCGCTTTGG GCAAAGTGTT |
|
| 351 | GTGGGACGGC GAATCCTTTG TCTGGTGGGC ATTGGCAAAC GGCGCACTGA |
|
| 401 | TCGTGCTGTG GCTGGTTTTC GGCGCACGCA GAACGGGCGG GCTGAAAACC |
|
| 451 | GTTTCGATGC TGCTGATGCT GCTTGCCGTG TTGTGGTTGA GCGTCGAAGT |
|
| 501 | GTTCGCTTCG TCCGGCACAA ACGCCGCGCC CGCCGTTTCA GACGGCATGA |
|
| 551 | CCTTCGGAAC GGCAGTCGAA CTGTCCGCCG TCATGCCGCT TTCCTGGCTG |
|
| 601 | CCGCTGGCCG CCGACTACAC GCGCCAAGCA CGCCGCCCGT TTGCGGCAAC |
|
| 651 | CCTGACGGCA ACGCTCGCCT ATACGCTGAC GGGCTGCTGG ATGTATGCCT |
|
| 701 | TGGGTTTGGC GGCGGCTCTG TTTACCGGAG AAACCGACGT GGCGAAAATC |
|
| 751 | CTGTTGGGCG CGGGCTTGGG CATAACGGGC ATTCTGGCAG TCGTCCTCTC |
|
| 801 | CACCGTTACC ACAACGTTTC TCGATACCTA TTCCGCCGGC GCGAGTGCGA |
|
| 851 | ACAACATTTC CGCGCGTTTT GCGGAAATAC CCGTCGCTGT CGGCGTTACC |
|
| 901 | CTGATCGGCA CGGTGCTTGC CGTCATGCTG CCCGTTACCG AATATAAAAA |
|
| 951 | CTTCCTGCTG CTTATCGGCT CGGTATTTGC GCCGATGGCG GCGGTTTTGA |
|
| 1001 | TTGCCGACTT TTTCGTCTTA AAACGGCGTG AGGAGATTGA AGGCTTTGAC |
|
| 1051 | TTTGCCGGAC TGGTTCTGTG GCTGGCAGGC TTCATCCTCT ACCGCTTCCT |
|
| 1101 | GCTCTCGTCC GGTTGGGAAA GCAGCATCGG TCTGACCGCC CCCGTAATGT |
|
| 1151 | CTGCCGTTGC CATTGCCACC GTATCGGTAC GCCTTTTCTT TAAAAAAACC |
|
| 1201 | CAATCTTTAC AAAGGAACCC GTCATGA |
[1655]This corresponds to the amino acid sequence <SEQ ID 808; ORF125ng-1>:
[0000]| 1 | MSGNASSPSS SAAIGLVWFG AAVSIAEIST GTLLAPLGWQ RGLAALLLGH | |
|
| 51 | AVGGALFFAA AYIGALTGRS SMESVRLSFG KCGSVLFSVA NMLQLAGWTA |
|
| 101 | VMIYVGATVS SALGKVLWDG ESFVWWALAN GALIVLWLVF GARRTGGLKT |
|
| 151 | VSMLLMLLAV LWLSVEVFAS SGTNAAPAVS DGMTFGTAVE LSAVMPLSWL |
|
| 201 | PLAADYTRQA RRPFAATLTA TLAYTLTGCW MYALGLAAAL FTGETDVAKI |
|
| 251 | LLGAGLGITG ILAVVLSTVT TTFLDTYSAG ASANNISARF AEIPVAVGVT |
|
| 301 | LIGTVLAVML PVTEYKNFLL LIGSVFAPMA AVLIADFFVL KRREEIEGFD |
|
| 351 | FAGLVLWLAG FILYRFLLSS GWESSIGLTA PVMSAVAIAT VSVRLFFKKT |
|
| 401 | QSLQRNPS* |
[1656]ORF125ng-1 and ORF125-1 show 95.1% identity in 408 aa overlap:
[0000]
[1657]Based on this analysis, including the presence of putative leader sequence and transmembrane domains in the gonococcal protein, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 96
[1658]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 809>:
[0000]| 1 | ATGACCCGTA TCGCCATCCT CGGCGGCGGC CTCTCGGGAA GGCTGACCGC | |
|
| 51 | GTTGCAGCTT GCAGAACAAG GTTATCAGAT TGCACTTTTC GATAAAAGCT |
|
| 101 | GCCGCCGGGG CGAACACGCC GCCGCCTATG TAGCCGCCGC CATGCTCGCG |
|
| 151 | CCTGCAGCGG A.ACGGTCGA AGCCACGCCC GAAGTGGTCA GGCTGGGCAG |
|
| 201 | GCAGAGCATC CCGCTTTGGC GCGGCATCCG ATGCCGTCTG AACACGCACA |
|
| 251 | CGATGATGCA GGAAAACGGC AGCCTGATTG TATGGCACGG GCAGGACAAG |
|
| 301 | CCATTATCCA GCGAGTTCGT CCGCCATCTC AAACGCGGCG GCGT.ACGGA |
|
| 351 | TGACGAAATC GTCCGTTGGC GCGCCGACGA CATCGCCGAA CGCGAACCGC |
|
| 401 | AACTCGGCGG ACGTTTTTAA GACGGCATCT ACCTGCCGAC CGAAGC.CAG |
|
| 451 | CTCGACGGGC GGCAATTATA GTCTGCACTT GCCGACGCTT TGGACGAACT |
|
| 501 | GAACGTCCCC TGCCATTGGG AACACGAATG CGTCCCCGAA GCCTGCAAG.. |
[1659]This corresponds to the amino acid sequence <SEQ ID 810; ORF126>:
[0000]| 1 | MTRIAILGGG LSGRLTALQL AEQGYQIALF DKSCRRGEHA AAYVAAAMLA | |
|
| 51 | PAAXTVEATP EVVRLGRQSI PLWRGIRCRL NTHTMMQENG SLIVWHGQDK |
|
| 101 | PLSSEFVRHL KRGGXTDDEI VRWRADDIAE REPQLGGRFX DGIYLPTEXQ |
|
| 151 | LDGRQLXSAL ADALDELNVP CHWEHECVPE ACK... |
[1660]Further work revealed the complete nucleotide sequence <SEQ ID 811>:
[0000]| 1 | ATGACCCGTA TCGCCATCCT CGGCGGCGGC CTCTCGGGAA GGCTGACCGC | |
|
| 51 | GTTGCAGCTT GCAGAACAAG GTTATCAGAT TGCACTTTTC GATAAAGGCT |
|
| 101 | GCCGCCGGGG CGAACACGCC GCCGCCTATG TTGCCGCCGC CATGCTCGCG |
|
| 151 | CCTGCGGCGG AAGCGGTCGA AGCCACGCCC GAAGTGGTCA GGCTGGGCAG |
|
| 201 | GCAGAGCATC CCGCTTTGGC GCGGCATCCG ATGCCGTCTG AACACGCACA |
|
| 251 | CGATGATGCA GGAAAACGGC AGCCTGATTG TGTGGCACGG GCAGGACAAG |
|
| 301 | CCATTATCCA GCGAGTTCGT CCGCCATCTC AAACGCGGCG GCGTAGCGGA |
|
| 351 | TGACGAAATC GTCCGTTGGC GCGCCGACGA CATCGCCGAA CGCGAACCGC |
|
| 401 | AACTCGGCGG ACGTTTTTCA GACGGCATCT ACCTGCCGAC CGAAGGCCAG |
|
| 451 | CTCGACGGGC GGCAAATATT GTCTGCACTT GCCGACGCTT TGGACGAACT |
|
| 501 | GAACGTCCCC TGCCATTGGG AACACGAATG CGTCCCCGAA GGCCTGCAAG |
|
| 551 | CCCAATACGA CTGGCTGATC GACTGCCGCG GCTACGGCGC AAAAACCGCG |
|
| 601 | TGGAACCAAT CCCCCGAGCA CACCAGCACC CTGCGCGGCA TACGCGGCGA |
|
| 651 | AGTGGCGCGG GTTTACACAC CCGAAATCAC GCTCAACCGC CCCGTGCGTC |
|
| 701 | TGCTCCATCC GCGTTATCCG CTCTACATCG CCCCGAAAGA AAACCACGTC |
|
| 751 | TTCGTCATCG GCGCGACCCA AATCGAAAGC GAAAGCCAAG CCCCCGCCAG |
|
| 801 | CGTGCGTTCA GGGTTGGAAC TCTTGTCCGC ACTCTATGCC ATCCACCCCG |
|
| 851 | CCTTCGGCGA AGCCGACATC CTCGAAATCG CCACCGGCCT GCGCCCCACG |
|
| 901 | CTCAACCACC ACAACCCCGA AATCCGTTAC AACCGCGCCC GACGCCTGAT |
|
| 951 | TGAAATCAAC GGCCTTTTCC GCCACGGTTT CATGATCTCC CCCGCCGTAA |
|
| 1001 | CCGCCGCCGC CGCCAGATTG GCAGTGGCAC TGTTTGACGG AAAAGACGCG |
|
| 1051 | CCCGAACGCG ATAAAGAAAG CGGTTTGGCG TATATCCGAA GACAAGATTA |
|
| 1101 | A |
[1661]This corresponds to the amino acid sequence <SEQ ID 812; ORF126-1>:
[0000]| 1 | MTRIAILGGG LSGRLTALQL AEQGYQIALF DKGCRRGEHA AAYVAAAMLA | |
|
| 51 | PAAEAVEATP EVVRLGRQSI PLWRGIRCRL NTHTMMQENG SLIVWHGQDK |
|
| 101 | PLSSEFVRHL KRGGVADDEI VRWRADDIAE REPQLGGRFS DGIYLPTEGQ |
|
| 151 | LDGRQILSAL ADALDELNVP CHWEHECVPE GLQAQYDWLI DCRGYGAKTA |
|
| 201 | WNQSPEHTST LRGIRGEVAR VYTPEITLNR PVRLLHPRYP LYIAPKENHV |
|
| 251 | FVIGATQIES ESQAPASVRS GLELLSALYA IHPAFGEADI LEIATGLRPT |
|
| 301 | LNHHNPEIRY NRARRLIEIN GLFRHGFMIS PAVTAAAARL AVALFDGKDA |
|
| 351 | PERDKESGLA YIRRQD* |
[1662]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1663]ORF126 shows 90.0% identity over a 180aa overlap with an ORF (ORF126a) from strain A of N. meningitidis:
[0000]
[1664]The complete length ORF126a nucleotide sequence <SEQ ID 813> is:
[0000]| 1 | ATGACCCGTA TCGCCATCCT CGGCGGCGGC CTCTCNGGAA GGCTGACCGC | |
|
| 51 | ACTGCAGCTT GCAGAACAAG GTTATCAGAT TGCACTTTTC GATAAAGGCT |
|
| 101 | GCCGCCGGGG CGAACACGCC GCCGCCTATG TTGCCGCCGC CATGCTCGCG |
|
| 151 | CCTGCGGCGG AAGCGGTCGA AGCCACGCCT GAAGTGGTCA GGCTGGGCAG |
|
| 201 | GCAGANCATC CCGCTTTGGC GCGGCATCCG ATGCCATCTG AAAACGCCTG |
|
| 251 | CCATGATGCA NGAAAACGGC AGCCTGATTG TGTGGCACGG GCAGGACAAA |
|
| 301 | CCTTTATCCA ACGAGTTCGT CCGCCATCTC AAACGCGGCG GCGTAGCGGA |
|
| 351 | TGACNAAATC GTCCGTTGGC GCGCCGACGA CATCGCCGAA CGCGAACCGC |
|
| 401 | AACTCGGCGG ACGTTTTTCA GACGGCATCT ACCTGCCGAC CGAAGGCCAG |
|
| 451 | CTCGACGGGC GGCAAATATT GTCTGCACTT GCCGACGCTT TGGACGAACT |
|
| 501 | GAACGTCCCC TGCCATTGGG AACACGAATG TGCCCCCGAA GACTTGCAAG |
|
| 551 | CCCAATACGA CTGGCTGATC GACTGCCGCG GCTACGGCGC AAAAACCGCG |
|
| 601 | TGGAACCAAT CCCCCGANNA NACCAGCACC CTGCGCGGCA TACGCGGCGA |
|
| 651 | AGTGGCGCGG GTTTACACAC CCGAAATCAC GCTCAACCGC CCCGTGCGCC |
|
| 701 | TGCTACACCC GCGCTATCCG CTNTACATCG CCCCGAAAGA AAACCNCGTC |
|
| 751 | TTCGTCATCG GCGCGACCCA AATCGAAAGC GAAAGCCAAG CACCTGCCAG |
|
| 801 | CGTGCGTTCC GGGCTGGAAC TCTTATCCGC ACTCTATGCC GTCCACCCCG |
|
| 851 | CCTTCGGCGA AGCCGACATC CTCGAAATCG CCACCGGCCT GCGCCCCACG |
|
| 901 | CTCAATCACC ACAACCCCGA AATCCGTTAC AACCGCGCCC GACGCCTGAT |
|
| 951 | TGAAATCAAC GGCCTTTTCC GCCACGGTTT CATGATCTCC CCCGCCGTAA |
|
| 1001 | CCGCCGCCGC CGTCAGATTG GCAGTGGCAC TGTTTGACGG AAAAGANGCG |
|
| 1051 | CCCGAACGCG ATGAAGAAAG CGGTTTGGCG TATATCCGAA GACAAGATTA |
|
| 1101 | A |
[1665]This encodes a protein having amino acid sequence <SEQ ID 814>:
[0000]| 1 | MTRIAILGGG LSGRLTALQL AEQGYQIALF DKGCRRGEHA AAYVAAAMLA | |
|
| 51 | PAAEAVEATP EVVRLGRQXI PLWRGIRCHL KTPAMMXENG SLIVWHGQDK |
|
| 101 | PLSNEFVRHL KRGGVADDXI VRWRADDIAE REPQLGGRFS DGIYLPTEGQ |
|
| 151 | LDGRQILSAL ADALDELNVP CHWEHECAPE DLQAQYDWLI DCRGYGAKTA |
|
| 201 | WNQSPXXTST LRGIRGEVAR VYTPEITLNR PVRLLHPRYP LYIAPKENXV |
|
| 251 | FVIGATQIES ESQAPASVRS GLELLSALYA VHPAFGEADI LEIATGLRPT |
|
| 301 | LNHHNPEIRY NRARRLIEIN GLFRHGFMIS PAVTAAAVRL AVALFDGKXA |
|
| 351 | PERDEESGLA YIRRQD* |
[1666]ORF126a and ORF126-1 show 95.4% identity in 366 aa overlap:
[0000]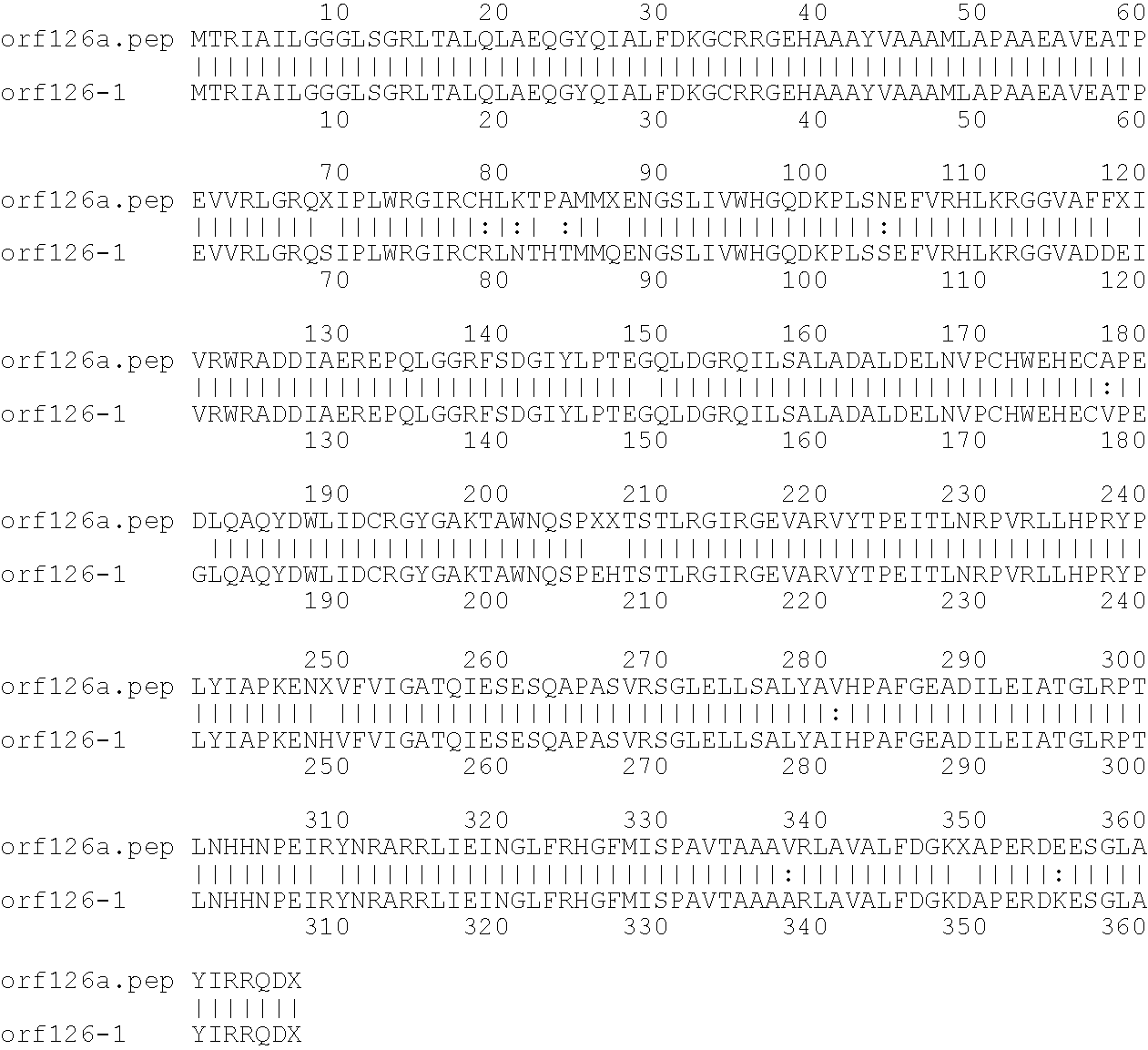
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1667]ORF126 shows 90% identity over a 180 aa overlap with a predicted ORF (ORF126ng) from N. gonorrhoeae:
[0000]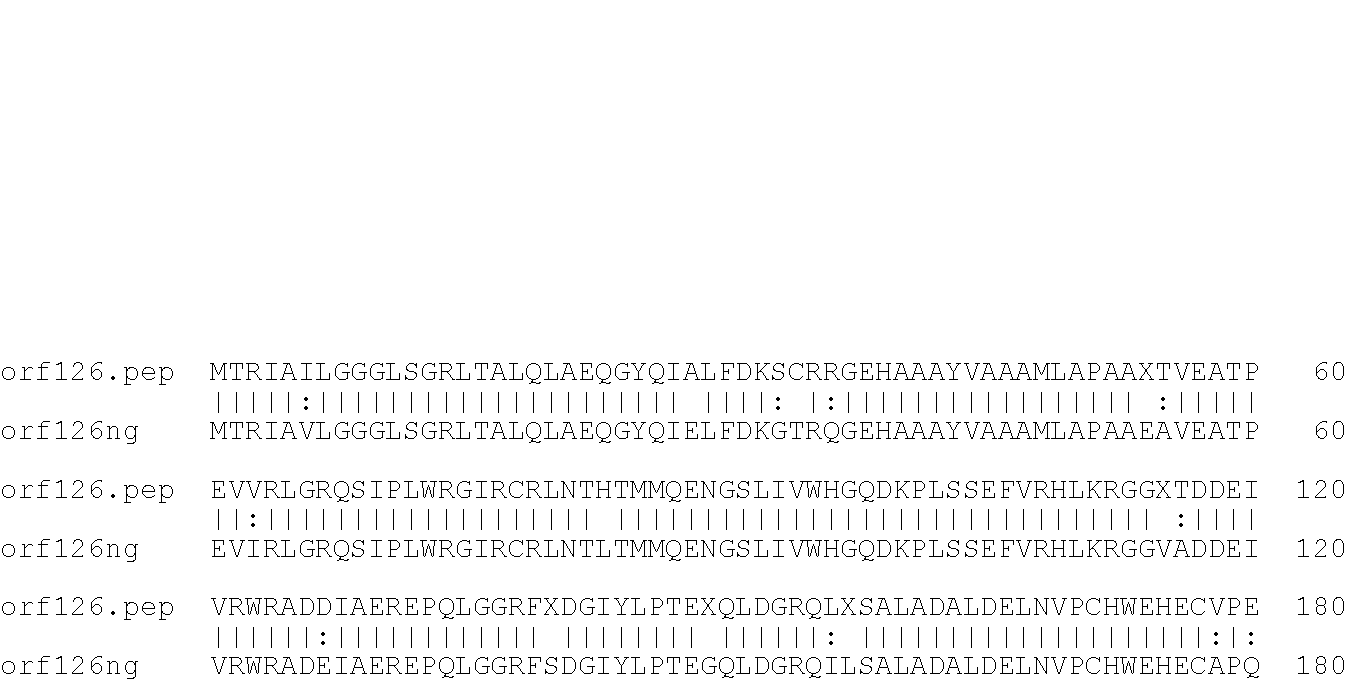
[1668]An ORF126ng nucleotide sequence <SEQ ID 815> was predicted to encode a protein having amino acid sequence <SEQ ID 816>:
[0000]| 1 | MTRIAVLGGG LSGRLTALQL AEQGYQIELF DKGTRQGEHA AAYVAAAMLA | |
|
| 51 | PAAEAVEATP EVIRLGRQSI PLWRGIRCRL NTLTMMQENG SLIVWHGQDK |
|
| 101 | PLSSEFVRHL KRGGVADDEI VRWRADEIAE REPQLGGRFS DGIYLPTEGQ |
|
| 151 | LDGRQILSAL ADALDELNVP CHWEHECAPQ DLQAQYDWVI DCRGYGAKTA |
|
| 201 | WNQSPEHTST LRGIRGEVRG FTRPKSRSTA PCACCTRAIR STSPRKKTTS |
|
| 251 | SSSARPKSKA KAKPPPAYVP GWNSYPRSMP STPPSAKPTS SKWRPGLRPT |
|
| 301 | LNHHNPEIRY SRERRLIEIN GLFRHGFMIS PAVTAAAVRL AVALFDGKDA |
|
| 351 | PERDEESGLA YIGRQD* |
[1669]Further work revealed the following gonococcal DNA sequence <SEQ ID 817>:
[0000]| 1 | ATGACCCGTA TCGCCGTCCT CGGAGGCGGC CTTTCCGGAA GGCTGACCGC | |
|
| 51 | ATTGCAGCTT GCAGAACAAG GTTATCAGAT TGAACTTTTC GACAAGGGCA |
|
| 101 | CCCGCCAAGG CGAACACGCC GCCGCCTATG TTGCCGCCGC GATGCTCGCG |
|
| 151 | CCTGCGGCGG AAGCGGTCGA GGCAACGCCC GAAGTCATCA GGCTGGGCAG |
|
| 201 | GCAGAGCATT CCGCTTTGGC GCGGCATCCG ATGCCGTCTG AACACGCTCA |
|
| 251 | CGATGATGCA GGAAAACGGC AGCCTGATTG TGTGGCACGG GCAGGACAAG |
|
| 301 | CCATTATCCA GCGAGTTCGT CCGCCATCTC AAACGCGGCG GCGTAGCGGA |
|
| 351 | TGACGAAATC GTCCGTTGGC GCGCCGATGA AATCGCCGAA CGCGAACCGC |
|
| 401 | AACTCGGCGG ACGTTTTTCA GACGGCATCT ACCTGCCGAC CGAAGGCCAG |
|
| 451 | CTCGACGGGC GGCAAATATT GTCTGCACTT GCCGACGCTT TGGACGAACT |
|
| 501 | GAACGTCCCT TGCCATTGGG AACACGAATG CGCCCCCCAA GACCTGCAAG |
|
| 551 | CCCAATACGA CTGGGTAATC GACTGCCGGG GCTACGGCGC GAAAACCGCG |
|
| 601 | TGGAACCAAT CCCCCGAGCA CACCAGCACC TTGCGCGGCA TACGCGGCGA |
|
| 651 | AGTGGCGCGG GTTTACACGC CCGAAATCAC GCTCAACCGC CCCGTGCGCC |
|
| 701 | TGCTGCACCC GCGCTATCCG CTCTACATCG CCCCGAAAGA AAACCACGTC |
|
| 751 | TTCGTCATCG GCGCGACCCA AATCGAAAGC GAAAGCCAAG CCCCCGCCAG |
|
| 801 | CGTACGTTCC GGGCTGGAAC TCTTATCCGC GCTCTATGCC GTCCACCCCG |
|
| 851 | CCTTCGGCGA AGCCGACATC CTCGAAATCG CCGCCGGCCT GCGCCCCACG |
|
| 901 | CTCAACCACC ACAACCCCGA AATCCGCTAC AGCCGCGAAC GCCGCCTCAT |
|
| 951 | CGAAATCAAC GGCCTTTTCC GGCACGGCTT TATGATTTCC CCCGCCGTAA |
|
| 1001 | CCGCCGCCGC CGTCAGATTG GCAGTGGCAC TGTTTGACGG AAAAGACGCG |
|
| 1051 | CCCGAACGTG ATGAAGAAAG CGGTTTGGCG TATATCGGAA GACAAGATTA |
|
| 1101 | A |
[1670]This corresponds to the amino acid sequence <SEQ ID 818; ORF126ng-1>:
[0000]| 1 | MTRIAVLGGG LSGRLTALQL AEQGYQIELF DKGTRQGEHA AAYVAAAMLA | |
|
| 51 | PAAEAVEATP EVIRLGRQSI PLWRGIRCRL NTLTMMQENG SLIVWHGQDK |
|
| 101 | PLSSEFVRHL KRGGVADDEI VRWRADEIAE REPQLGGRFS DGIYLPTEGQ |
|
| 151 | LDGRQILSAL ADALDELNVP CHWEHECAPQ DLQAQYDWVI DCRGYGAKTA |
|
| 201 | WNQSPEHTST LRGIRGEVAR VYTPEITLNR PVRLLHPRYP LYIAPKENHV |
|
| 251 | FVIGATQIES ESQAPASVRS GLELLSALYA VHPAFGEADI LEIAAGLRPT |
|
| 301 | LNHHNPEIRY SRERRLIEIN GLFRHGFMIS PAVTAAAVRL AVALFDGKDA |
|
| 351 | PERDEESGLA YIGRQD* |
[1671]ORF126ng-1 and ORF126-1 show 95.1% identity in 366 aa overlap:
[0000]
[1672]Furthermore, ORF126ng-1 shows homology to a putative Rhizobium oxidase flavoprotein:
[0000]| gi|2627327 (AF004408) putative amino acid oxidase flavoprotein | |
| [Rhizobium etli] |
| Length = 327 |
| Score = 169 bits (423), Expect = 3e−41 |
| Identities = 112/329 (34%), Positives = 163/329 (49%), Gaps = 25/329 (7%) |
|
| Query: | 3 | RIAVLGGGLSGRLTALQLAEQGYQIELFDKGTRQGEHXXXXXXXXXXXXXXXXXXXXXXX | 62 | |
| | RI V G G++G A QL G+++ L ++ G |
| Sbjct: | 2 | RILVNGAGVAGLTVAWQLYRHGFRVTLAERAGTVGA-GASGFAGGMLAPWCERESAEEPV | 60 |
|
| Query: | 63 | IRLGRQSIPLWRGIRCRLNTLTMMQENGSLIVWHGQDKPLSSEFVRHLKRGGVADDEIVR | 122 |
| | + LGR + W + G+L+V G+D F R G DE+ |
| Sbjct: | 61 | LTLGRLAADWWEAA-----LPGHVHRRGTLVVAGGRDTGELDRFSRRTS-GWEWLDEVA- | 113 |
|
| Query: | 123 | WRADEIAEREPQLGGRFSDGIYLPTEGQLDGRQILSALADALDELNVPCHWEHECAPQDL | 182 |
| | IA EP L GRF ++ E LD RQ L+ALA L++ + + |
| Sbjct: | 114 | -----IAALEPDLAGRFRRALFFRQEAHLDPRQALAALAAGLEDARMRLTLG---VVGES | 165 |
|
| Query: | 183 | QAQYDWVIDCRGYGAKTAWNQSPEHTSTLRGIRGEVARVYTPEITLNRPVRLLHPRYPLY | 242 |
| | +D V+DC G LRG+RGE+ V T E++L+RPVRLLHPR+P+Y |
| Sbjct: | 166 | DVDHDRVVDCTGAA-------QIGRLPGLRGVRGEMLCVETTEVSLSRPVRLLHPRHPIY | 218 |
|
| Query: | 243 | IAPKENHVFVIGATQIESESQAPASVRSGLELLSALYAVHPAFGEADILEIAAGLRPTLN | 302 |
| | I P++ + F++GAT IES+ P + RS +ELL+A YA+HPAFGEA + E AG+RP |
| Sbjct: | 219 | IVPRDKNRFMVGATMIESDDGGPITARSLMELLNAAYAMHPAFGEARVTETGAGVRPAYP | 278 |
|
| Query: | 303 | HHNPEIRYSRERRLIEINGLFRHGFMISP | 331 |
| | + P R ++E R + +NGL+RHGF+++P |
| Sbjct: | 279 | DNLP--RVTQEGRTLHVNGLYRHGFLLAP | 305 |
[1673]This analysis suggests that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 97
[1674]The following DNA sequence, believed to be complete, was identified in N. meningitidis <SEQ ID 819>:
[0000]| 1 | ATGACTGATA ATCGGGGGTT TACGCTGGTT GAATTAATAT CAGTGGTCTT | |
|
| 51 | GATATTGTCT GTACTTGCTT TAATTGTTTA TCCGAGCTAT CGCAATTATG |
|
| 101 | TTGAGAAAGC AAAGATAAAT GCAGTGCGGG CAGCCTTGTT AGAAAATGCA |
|
| 151 | CATTTTATGG AAAAGTTTTA TCTGCAGAAT GGGAGGTTTA AACAAACATC |
|
| 201 | TACCAAGTGG CCAAGTTTGC CGATTAAAGA GGCAGAAGGC TTTTGTATCC |
|
| 251 | GTTTGAATGG AATCGtCGCG CGGG..GCTT TAGACAGTAA ATTCATGTTG |
|
| 301 | AAGGCGGTAG CCATAGATAA AGATAAAAAT CCTTTTATTA TTAAGATGAA |
|
| 351 | TGAAAATCTA GTAACCTTTA aTTTGCAAGA AGTCCGCCAG TTCGTGTAGT |
|
| 401 | GACGGGCTGG ATTATTTTAA AGGAAATGAT AAGGACTGCA AGTTACTTAA |
|
| 451 | GTAG |
[1675]This corresponds to the amino acid sequence <SEQ ID 820; ORF127>:
[0000]| 1 | MTDNRGFTLV ELISVVLILS VLALIVYPSY RNYVEKAKIN AVRAALLENA | |
|
| 51 | HFMEKFYLQN GRFKQTSTKW PSLPIKEAEG FCIRLNGIVA RXALDSKFML |
|
| 101 | KAVAIDKDKN PFIIKMNENL VTFICKKSAS SCSDGLDYFK GNDKDCKLLK |
|
| 151 | * |
[1676]Further work revealed the following DNA sequence <SEQ ID 821>:
[0000]| 1 | ATGACTGATA ATCGGGGGTT TACGCTGGTT GAATTAATAT CAGTGGTCTT | |
|
| 51 | GATATTGTCT GTACTTGCTT TAATTGTTTA TCCGAGCTAT CGCAATTATG |
|
| 101 | TTGAGAAAGC AAAGATAAAT GCAGTGCGGG CAGCCTTGTT AGAAAATGCA |
|
| 151 | CATTTTATGG AAAAGTTTTA TCTGCAGAAT GGGAGGTTTA AACAAACATC |
|
| 201 | TACCAAGTGG CCAAGTTTGC CGATTAAAGA GGCAGAAGGC TTTTGTATCC |
|
| 251 | GTTTGAATGG AATCGCGCGC GGGGCTTTAG ACAGTAAATT CATGTTGAAG |
|
| 301 | GCGGTAGCCA TAGATAAAGA TAAAAATCCT TTTATTATTA AGATGAATGA |
|
| 351 | AAATCTAGTA ACCTTTATTT GCAAGAAGTC CGCCAGTTCG TGTAGTGACG |
|
| 401 | GGCTGGATTA TTTTAAAGGA AATGATAAGG ACTGCAAGTT ACTTAAGTAG |
[1677]This corresponds to the amino acid sequence <SEQ ID 822; ORF127-1>:
[0000]| 1 | MTDNRGFTLV ELISVVLILS VLALIVYPSY RNYVEKAKIN AVRAALLENA | |
|
| 51 | HFMEKFYLQN GRFKQTSTKW PSLPIKEAEG FCIRLNGIAR GALDSKFMLK |
|
| 101 | AVAIDKDKNP FIIKMNENLV TFICKKSASS CSDGLDYFKG NDKDCKLLK* |
[1678]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1679]ORF127 shows 98.0% identity over a 150aa overlap with an ORF (ORF127a) from strain A of N. meningitidis.
[0000]
[1680]The complete length ORF127a nucleotide sequence <SEQ ID 823> is:
[0000]| 1 | ATGACTGATA ATCGGGGGTT TACGCTGGTT GAATTAATAT CAGTGGTCTT | |
|
| 51 | GATATTGTCT GTACTTGCTT TAATTGTTTA TCCGAGCTAT CGCAATTATG |
|
| 101 | TTGAGAAAGC AAAGATAAAT ACAGTGCGGG CAGCCTTGTT AGAAAATGCA |
|
| 151 | CATTTTATGG AAAAGTTTTA TCTGCAGAAT GGGAGATTTA AACAAACATC |
|
| 201 | TACCAAATGG CCAAGTTTGC CGATTAAAGA GGCAGAAGGC TTTTGTATCC |
|
| 251 | GTTTGAATGG AATCGCGCGC GGGGCCTTAG ACAGTAAATT CATGTTGAAG |
|
| 301 | GCGGTAGCCA TAGATAAAGA TAAAAATCCT TTTATTATTA AGATGAATGA |
|
| 351 | AAATCTAGTA ACCTTTATTT GCAAGAAGTC CGCCAGTTCG TGTAGTGACG |
|
| 401 | GGCTGGATTA TTTTAAAGGA AATGATAAGG ACTGCAAGTT ACTTAAGTAG |
[1681]This encodes a protein having amino acid sequence <SEQ ID 824>:
[0000]| 1 | MTDNRGFTLV ELISVVLILS VLALIVYPSY RNYVEKAKIN TVRAALLENA | |
|
| 51 | HFMEKFYLQN GRFKQTSTKW PSLPIKEAEG FCIRLNGIAR GALDSKFMLK |
|
| 101 | AVAIDKDKNP FIIKMNENLV TFICKKSASS CSDGLDYFKG NDKDCKLLK* |
[1682]ORF127a and ORF127-1 show 99.3% identity in 149 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1683]ORF127 shows 97.3% identity over a 150 aa overlap with a predicted ORF (ORF127ng) from N. gonorrhoeae:
[0000]
[1684]The complete length ORF127ng nucleotide sequence <SEQ ID 825> is:
[0000]| 1 | ATGACTGATA ATCGGGGGTT TACACTGGTT GAATTAATAT CAGTGGTCTT | |
|
| 51 | GATATTGTCT GTACTTGCTT TAATTGTTTA TCCGAGCTAT CGCAATTATG |
|
| 101 | TTGAGAAAGC AAAGATAAAT GCAGTGCGGG CAGCCTTGTT AGAAAATGCA |
|
| 151 | CATTTTATGG AAAAGTTTTA TCTGCAGAAT GGGAGATTTA AACAAACATC |
|
| 201 | TACCAAATGG CCAAGTTTGC CGATTAAAGA GGCAGAAGGC TTTTGTATCC |
|
| 251 | GTTTGAATGG AATCGCGCGC GGGGCTTTAG ACAGTAAATT CATGTTGAAG |
|
| 301 | GCGGTAGCCA TAGATAAAGA TAAAAATCCT TTTATTATTA AGATGAATGA |
|
| 351 | AAATCTAGTA ACCTTTATTT GCAAGAAGTC CGCCAGTTCG TGTAGTGACG |
|
| 401 | GGCTGGATTA TTTTAAAGGA AATGATAAGG ACTGCAAGTT ACTTAAGTAG |
[1685]This encodes a protein having amino acid sequence <SEQ ID 826>:
[0000]| 1 | MTDNRGFTLV ELISVVLILS VLALIVYPSY RNYVEKAKIN AVRAAFLENA | |
|
| 51 | HFMEKFYLQN GRFKQTSTKW PSLPIKEAEG FCIRLNGIAR GALDSKFMLK |
|
| 101 | AVAIDKDKNP FIIKMNENLV TFICKKSASS CSDRLDYFKG NDKDCKLLK* |
[1686]ORF127ng and ORF127-1 show 100.0% identity in 149 aa overlap:
[0000]
[1687]This analysis, including the fact that the predicted transmembrane domain is shared by the meningococcal and gonococcal proteins, suggests that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 98
[1688]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 827>
[0000]| 1 | ..GTGTCGCTGG CTTCGGTGAT TGCCTCTCAA ATCTTCCTTT ACGAAGATTT | |
|
| 51 | CAACCAAATG CGGAAAACCC GTGGAGCTAT CTGCGGTTTT CTTGTCCAAT |
|
| 101 | ATTTATCTGG GGTTTCAGCA GGGGTATTTC GATTTGAGTG CCGACGAGAA |
|
| 151 | CCCCGTACTG CATATCTGGT CTTTGGCAGT AGAGGAACAG TATTACCTCC |
|
| 201 | TGTATCCCCT TTTGCTGATA TTTTGCTGCA AAAAAACCAA ATCGCTACGG |
|
| 251 | GTGCTGCGTA ACATCAGCAT CATCCTGTTT TTGATTTTGA CTGCCTCATC |
|
| 301 | GTTTTTGCCA AGCGGGTTTT ATACCGACAT CCTCAACCAA CCCAATACTT |
|
| 351 | ATTACCTTTC GACACTGAGG TTTCCCGAGC TGTTGGCAGG TTCGCTGCTG |
|
| 401 | GCGGTTTACG GGCAAACGCA AAACGGCAGA CGGCAAACAG CAAATGGAAA |
|
| 451 | ACGGCAGTTG CTTTCATCAC TCTGCTTCGG CGCATTGCTT GCCTGCCTGT |
|
| 501 | TCGTGATTGA CAAACACAAT CCGTTTATCC CGGGAATGAC CCTGCTCCTT |
|
| 551 | CCCTGCCTGC TGACGGCACT GCTTATCCGG AGTATGCAAT ACGGGACACT |
|
| 601 | TCCGACCCGC ATCCTGTCGG CAAGCCCCAT CGTATTTGTC GGCAAAATCT |
|
| 651 | CTTATTCCCT ATACCTGTAC CATTGGATTT TTATTGCTTT CGCTCCGCTC |
|
| 701 | ATTAGAGGCG GGAAACAGCT CGGACTGCCT GCCG.. |
[1689]This corresponds to the amino acid sequence <SEQ ID 828; ORF128>:
[0000]| 1 | ..VSLASVIASQ IFLYEDFNQM RKTVELSAVF LSNIYLGFQQ GYFDLSADEN | |
|
| 51 | PVLHIWSLAV EEQYYLLYPL LLIFCCKKTK SLRVLRNISI ILFLILTASS |
|
| 101 | FLPSGFYTDI LNQPNTYYLS TLRFPELLAG SLLAVYGQTQ NGRRQTANGK |
|
| 151 | RQLLSSLCFG ALLACLFVID KHNPFIPGMT LLLPCLLTAL LIRSMQYGTL |
|
| 201 | PTRILSASPI VFVGKISYSL YLYHWIFIAF APLIRGGKQL GLPA.. |
[1690]Further work revealed the complete nucleotide sequence <SEQ ID 829>:
[0000]| 1 | ATGCAAGCTG TCCGATACAG ACCGGAAATT GACGGATTGC GGGCCGTCGC | |
|
| 51 | CGTGCTATCC GTCATGATTT TCCACCTGAA TAACCGCTGG CTGCCCGGAG |
|
| 101 | GATTCCTGGG GGTGGACATT TTCTTTGTCA TCTCAGGATT CCTCATTACC |
|
| 151 | GGCATCATTC TTTCTGAAAT ACAGAACGGT TCTTTTTCTT TCCGGGATTT |
|
| 201 | TTATACCCGC AGGATTAAGC GGATTTATCC TGCCTTTATT GCGGCCGTGT |
|
| 251 | CGCTGGCTTC GGTGATTGCC TCTCAAATCT TCCTTTACGA AGATTTCAAC |
|
| 301 | CAAATGCGGA AAACCGTGGA GCTTTCTGCG GTTTTCTTGT CCAATATTTA |
|
| 351 | TCTGGGGTTT CAGCAGGGGT ATTTCGATTT GAGTGCCGAC GAGAACCCCG |
|
| 401 | TACTGCATAT CTGGTCTTTG GCAGTAGAGG AACAGTATTA CCTCCTGTAT |
|
| 451 | CCCCTTTTGC TGATATTTTG CTGCAAAAAA ACCAAATCGC TACGGGTGCT |
|
| 501 | GCGTAACATC AGCATCATCC TGTTTTTGAT TTTGACTGCC TCATCGTTTT |
|
| 551 | TGCCAAGCGG GTTTTATACC GACATCCTCA ACCAACCCAA TACTTATTAC |
|
| 601 | CTTTCGACAC TGAGGTTTCC CGAGCTGTTG GCAGGTTCGC TGCTGGCGGT |
|
| 651 | TTACGGGCAA ACGCAAAACG GCAGACGGCA AACAGCAAAT GGAAAACGGC |
|
| 701 | AGTTGCTTTC ATCACTCTGC TTCGGCGCAT TGCTTGCCTG CCTGTTCGTG |
|
| 751 | ATTGACAAAC ACAATCCGTT TATCCCGGGA ATGACCCTGC TCCTTCCCTG |
|
| 801 | CCTGCTGACG GCACTGCTTA TCCGGAGTAT GCAATACGGG ACACTTCCGA |
|
| 851 | CCCGCATCCT GTCGGCAAGC CCCATCGTAT TTGTCGGCAA AATCTCTTAT |
|
| 901 | TCCCTATACC TGTACCATTG GATTTTTATT GCTTTCGCCC ATTACATTAC |
|
| 951 | AGGCGACAAA CAGCTCGGAC TGCCTGCCGT ATCGGCGGTT GCCGCGTTGA |
|
| 1001 | CGGCCGGATT TTCCCTGTTG AGTTATTATT TGATTGAACA GCCGCTTAGA |
|
| 1051 | AAACGGAAGA TGACCTTCAA AAAGGCATTT TTCTGCCTCT ATCTCGCCCC |
|
| 1101 | GTCCCTGATA CTTGTCGGTT ACAACCTGTA CGCAAGGGGG ATATTGAAAC |
|
| 1151 | AGGAACACCT CCGCCCGTTG CCCGGCGCGC CCCTTGCTGC GGAAAATCAT |
|
| 1201 | TTTCCGGAAA CCGTCCTGAC CCTCGGCGAC TCGCACGCCG GACACCTGAG |
|
| 1251 | GGGGTTTCTG GATTATGTCG GCAGCCGGGA AGGGTGGAAA GCCAAAATCC |
|
| 1301 | TGTCCCTCGA TTCGGAGTGT TTGGTTTGGG TAGATGAGAA GCTGGCAGAC |
|
| 1351 | AACCCGTTAT GTCGAAAATA CCGGGATGAA GTTGAAAAAG CCGAAGCCGT |
|
| 1401 | TTTCATTGCC CAATTCTATG ATTTGAGGAT GGGCGGCCAG CCTGTGCCGA |
|
| 1451 | GATTTGAAGC GCAATCCTTC CTAATACCCG GGTTCCCAGC CCGATTCAGG |
|
| 1501 | GAAACCGTCA AAAGGATAGC CGCCGTCAAA CCCGTCTATG TTTTTGCAAA |
|
| 1551 | CAACACATCA ATCAGCCGTT CGCCCCTGAG GGAGGAAAAA TTGAAAAGAT |
|
| 1601 | TTGCCGCAAA CCAATATCTC CGCCCCATTC AGGCTATGGG CGACATCGGC |
|
| 1651 | AAGAGCAATC AGGCGGTCTT TGATTTGATT AAAGATATTC CCAATGTGCA |
|
| 1701 | TTGGGTGGAC GCACAAAAAT ACCTGCCCAA AAACACGGTC GAAATATACG |
|
| 1751 | GCCGCTATCT TTACGGCGAC CAAGACCACC TGACCTATTT CGGTTCTTAT |
|
| 1801 | TATATGGGGC GGGAATTCCA CAAACACGAA CGCCTGCTTA AATCTTCCCA |
|
| 1851 | CGGCGGCGCA TTGCAGTAG |
[1691]This corresponds to the amino acid sequence <SEQ ID 830; ORF128-1>:
[0000]| 1 | MQAVRYRPEI DGLRAVAVLS VMIFHLNNRW LPGGFLGVDI FFVISGFLIT | |
|
| 51 | GIILSEIQNG SFSFRDFYTR RIKRIYPAFI AAVSLASVIA SQIFLYEDFN |
|
| 101 | QMRKTVELSA VFLSNIYLGF QQGYFDLSAD ENPVLHIWSL AVEEQYYLLY |
|
| 151 | PLLLIFCCKK TKSLRVLRNI SIILFLILTA SSFLPSGFYT DILNQPNTYY |
|
| 201 | LSTLRFPELL AGSLLAVYGQ TQNGRRQTAN GKRQLLSSLC FGALLACLFV |
|
| 251 | IDKHNPFIPG MTLLLPCLLT ALLIRSMQYG TLPTRILSAS PIVFVGKISY |
|
| 301 | SLYLYHWIFI AFAHYITGDK QLGLPAVSAV AALTAGFSLL SYYLIEQPLR |
|
| 351 | KRKMTFKKAF FCLYLAPSLI LVGYNLYARG ILKQEHLRPL PGAPLAAENH |
|
| 401 | FPETVLTLGD SHAGHLRGFL DYVGSREGWK AKILSLDSEC LVWVDEKLAD |
|
| 451 | NPLCRKYRDE VEKAEAVFIA QFYDLRMGGQ PVPRFEAQSF LIPGFPARFR |
|
| 501 | ETVKRIAAVK PVYVFANNTS ISRSPLREEK LKRFAANQYL RPIQAMGDIG |
|
| 551 | KSNQAVFDLI KDIPNVHWVD AQKYLPKNTV EIYGRYLYGD QDHLTYFGSY |
|
| 601 | YMGREFHKHE RLLKSSHGGA LQ* |
[1692]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with Hypothetical Integral Membrane Protein HI0392 of H. influenzae (Accession Number U32723)
[1693]ORF128 and HI0392 show 52% aa identity in 180aa overlap:
[0000]| Orf128: | 1 | VSLASVIASQIFLYEDFNQMRKTVELSAVFLSNIYLGFQQGYFDLSADENPVLHIWSLAV | 60 | |
| | ++L S IAS IF+Y DFN++RKT+EL+ FLSN YLG QGYFDLSA+ENPVLHIWSLAV | |
| HI0392: | 46 | MALVSFIASAIFIYNDFNKLRKTIELAIAFLSNFYLGLTQGYFDLSANENPVLHIWSLAV | 105 |
|
| Orf128: | 61 | EEQXXXXXXXXXIFCCKKTKSLRVLRNISIILFLILTASSFLPSGFYTDILNQPNTYYLS | 120 |
| | E Q I KK + ++VL I++ILF IL A+SF+ + FY ++L+QPN YYLS | |
| HI0392: | 106 | EGQYYLIFPLILILAYKKFREVKVLFIITLILFFILLATSFVSANFYKEVLHQPNIYYLS | 165 |
|
| Orf128: | 121 | TLRFPELLAGSLLAVYGQTQNGRRQTANGKRQLLSSLCFGALLACLFVIDKHNPFIPGMT | 180 |
| | LRFPELL GSLLA+Y N + Q + +L+ L L +CLF+++ + FIPG+T | |
| HI0392: | 166 | NLRFPELLVGSLLAIYHNLSN-KVQLSKQVNNILAILSTLLLFSCLFLMNNNIAFIPGIT | 224 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1694]ORF128 shows 98.0% identity over a 244aa overlap with an ORF (ORF128a) from strain A of N. meningitidis.
[0000]
[1695]The complete length ORF128a nucleotide sequence <SEQ ID 831> is:
[0000]| 1 | ATGCAAGCTG TCCGATACAG ACCGGAAATT GACGGATTGC GGGCCGTCGC | |
|
| 51 | CGTGCTATCC GTCATGATTT TCCACCTGAA TAACCGCTGG CTGCCCGGAG |
|
| 101 | GATTCCTGGG GGTGGACATT TTCTTTGTCA TCTCAGGATT CCTCATTACC |
|
| 151 | GGCATCATTC TTTCTGAAAT ACAGAACGGT TCTTTTTCTT TCCGGGATTT |
|
| 201 | TTATACCCGC AGGATTAAGC GGATTTATCC TGCTTTTATT GCGGCCGTGT |
|
| 251 | CGCTGGCTTC GGTGATTGCC TCTCAAATCT TCCTTTACGA AGATTTCAAC |
|
| 301 | CAAATGCGGA AAACCGTGGA GCTTTCTGCG GTTTTCTTGT CCAATATTTA |
|
| 351 | TCTGGGGTTT CAGCAGGGGT ATTTCGATTT GAGTGCCGAC GAGAACCCCG |
|
| 401 | TACTGCATAT CTGGTCTTTG GCAGTAGAGG AACAGTATTA CCTCCTGTAT |
|
| 451 | CCTCTTTTGC TGATATTTTG CTGCAAAAAA ACAAAATCGC TACGGGTGCT |
|
| 501 | GCGTAACATC AGCATCATCC TATTTCTGAT TTTGACTGCC ACATCGTTTT |
|
| 551 | TGCCAAGCGG GTTTTATACC GATATTCTCA ACCAACCCAA TACTTATTAC |
|
| 601 | CTTTCGACAC TGAGGTTTCC CGAGCTGTTG GCAGGTTCGC TGCTGGCGGT |
|
| 651 | TTACGGGCAA ACGCAAAACG GCAGACGGCA AACAGCAAAT GGAAAACGGC |
|
| 701 | AGTTGCTTTC ATCACTCTGC TTCGGCGCAT TGCTTGCCTG CCTGTTCGTG |
|
| 751 | ATTGACAAAC ACAATCCGTT TATCCCGGGA ATGACCCTGC TCCTTCCCTG |
|
| 801 | CCTGCTGACG GCACTGCTTA TCCGGAGTAT GCAATACGGG ACACTTCCGA |
|
| 851 | CCCGCATCCT GTCGGCAAGC CCCATCGTAT TTGTCGGCAA AATCTCTTAT |
|
| 901 | TCCCTATACC TGTACCATTG GATTTTTATT GCTTTCGCCC ATTACATTAC |
|
| 951 | AGGCGACAAA CAGCTCGGAC TGCCTGCCGT ATCGGCGGTT GCCGCGTTGA |
|
| 1001 | CGGCCGGATT TTCCCTGTTG AGTTATTATT TGATTGAACA GCCGCTTAGA |
|
| 1051 | AAACGGAAGA TGACCTTCAA AAAGGCATTT TTCTGCCTCT ATCTCGCCCC |
|
| 1101 | GTCCCTGATA CTTGTCGGTT ACAACCTGTA CGCAAGGGGG ATATTGAAAC |
|
| 1151 | AGGAACACCT CCGCCCGTTG CCCGGCGCGC CCCTTGCTGC GGAAAATCAT |
|
| 1201 | TTTCCGGAAA CCGTCCTGAC CCTCGGCGAC TCGCACGCCG GACACCTGCG |
|
| 1251 | GGGGTTTCTG GATTATGTCG GCAGCCGGGA AGGGTGGAAA GCCAAAATCC |
|
| 1301 | TGTCCCTCGA TTCGGAGTGT TTGGTTTGGG TAGATGAGAA GCTGGCAGAC |
|
| 1351 | AACCCGTTAT GTCGAAAATA CCGGGATGAA GTTGAAAAAG CCGAAGCCGT |
|
| 1401 | TTTCATTGCC CAATTCTATG ATTTGAGGAT GGGCGGCCAG CCCGTGCCGA |
|
| 1451 | GATTTGAAGC GCAATCCTTC CTAATACCCG GGTTCCCAGC CCGATTCAGG |
|
| 1501 | GAAACCGTCA AAAGGATAGC CGCCGTCAAA CCCGTCTATG TTTTTGCAAA |
|
| 1551 | CAACACATCA ATCAGCCGTT CGCCCCTGAG GGAGGAAAAA TTGAAAAGAT |
|
| 1601 | TTGCCGCAAA CCAATATCTC CGCCCCATTC AGGCTATGGG CGACATCGGC |
|
| 1651 | AAGAGCAATC AGGCGGTCTT TGATTTGATT AAAGATATTC CCAATGTGCA |
|
| 1701 | TTGGGTGGAC GCACAAAAAT ACCTGCCCAA AAACACGGTC GAAATATACG |
|
| 1751 | GCCGCTATCT TTACGGCGAC CAAGACCACC TGACCTATTT CGGTTCTTAT |
|
| 1801 | TATATGGGGC GGGAATTTCA CAAACACGAA CGCCTGCTTA AATCTTCTCG |
|
| 1851 | CGACGGCGCA TTGCAGTAG |
[1696]This encodes a protein having amino acid sequence <SEQ ID 832>:
[0000]| 1 | MQAVRYRPEI DGLRAVAVLS VMIFHLNNRW LPGGFLGVDI FFVISGFLIT | |
|
| 51 | GIILSEIQNG SFSFRDFYTR RIKRIYPAFI AAVSLASVIA SQIFLYEDFN |
|
| 101 | QMRKTVELSA VFLSNIYLGF QQGYFDLSAD ENPVLHIWSL AVEEQYYLLY |
|
| 151 | PLLLIFCCKK TKSLRVLRNI SIILFLILTA TSFLPSGFYT DILNQPNTYY |
|
| 201 | LSTLRFPELL AGSLLAVYGQ TQNGRRQTAN GKRQLLSSLC FGALLACLFV |
|
| 251 | IDKHNPFIPG MTLLLPCLLT ALLIRSMQYG TLPTRILSAS PIVFVGKISY |
|
| 301 | SLYLYHWIFI AFAHYITGDK QLGLPAVSAV AALTAGFSLL SYYLIEQPLR |
|
| 351 | KRKMTFKKAF FCLYLAPSLI LVGYNLYARG ILKQEHLRPL PGAPLAAENH |
|
| 401 | FPETVLTLGD SHAGHLRGFL DYVGSREGWK AKILSLDSEC LVWVDEKLAD |
|
| 451 | NPLCRKYRDE VEKAEAVFIA QFYDLRMGGQ PVPRFEAQSF LIPGFPARFR |
|
| 501 | ETVKRIAAVK PVYVFANNTS ISRSPLREEK LKRFAANQYL RPIQAMGDIG |
|
| 551 | KSNQAVFDLI KDIPNVHWVD AQKYLPKNTV EIYGRYLYGD QDHLTYFGSY |
|
| 601 | YMGREFHKHE RLLKSSRDGA LQ* |
[1697]ORF128a and ORF128-1 show 99.5% identity in 622 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1698]ORF128 shows 93.4% identity over 244 aa overlap with a predicted ORF (ORF128ng) from N. gonorrhoeae:
[0000]
[1699]The complete length ORF128ng nucleotide sequence <SEQ ID 833> is:
[0000]| 1 | ATGCAAGCTG TCCGATACAG GCCTGAAATT GACGGATTGC GGGCCGTCGC | |
|
| 51 | CGTGCTATCC GTCATTATTT TCCACCTGAA TAACCGCTGG CTGCCCGGAG |
|
| 101 | GATTCCTGGG GGTGGACATT TTCTTTGTCA TCTCGGGATT CCTCATTACC |
|
| 151 | AACATCATTC TTTCTGAAAT ACAGAACGGT TCTTTTTCTT TCCGGGATTT |
|
| 201 | TTATACCCGC AGGATTAAGC GGATTTATCC TGCTTTTATT GCGGCCGTGT |
|
| 251 | CCCTGGCTTC GGTGATTGCT TCTCAAATCT TCCTTTACGA AGATTTCAAC |
|
| 301 | CAAATGAGGA AAACCATAGA GCTTTCTACG GTTTTTTTGT CCAATATTTA |
|
| 351 | TTTGGGGTTC CGATTGGGGT ATTTCGATTT GAGTGCCGAC GAGAACCCCG |
|
| 401 | TACTGCATAT CTGGTCTTTG GCGGTAGAGG AACAGTATTA CCTCCTGTAT |
|
| 451 | CCTCTTTTGC TGATATTCTG TTACAAAAAA ACCAAATCAC TACGGGTGCT |
|
| 501 | GCGTAATATC AGCATCATCC TGTTTCTGAT TTTGACCGCA TCATCGTTTT |
|
| 551 | TGCCGGCCGG GTTTTATACC GACATCCTCA ACCAACCcaa TACTTATTAC |
|
| 601 | CTTTCGACAC TGAGGTTTCC CGAGCTGTTG GTGGGTTCGC TGTTGGCGGT |
|
| 651 | TTACGGGCAA ACGCAAAACG GCAGACGGCA AACAGAAAAT GGAAAACGGC |
|
| 701 | AGTTGCTTTC ATTACTCTGT TTCGGCGCat tgCTTGTCTG CCTGTTCGTG |
|
| 751 | ATCGACAAAC ACGATCCGTT TATCCCGGGA ATAACCCTGC TCCTTCCCTG |
|
| 801 | CCTGCTGACG GCGCTGCTTA TCCGGAGTAT GCAATACGGG ACACTTCCGA |
|
| 851 | CCCGCATCCT GTCGGCAAGC CCCATCGTAT TTGTCGGCAA AATCTCTTAT |
|
| 901 | TCCCTATACC TGTACCATTG GATTTTTATT GCCTTCGCCC ATTACATTAC |
|
| 951 | AGGCGACAAA CAGCTCGGAC TGCCTGCCGT ATCGGCGGTT GCCGCGTTGA |
|
| 1001 | CGGCCGGATT TTCCCTGTTG AGCTATTATT TGATTGAACA GCCGCTTAGA |
|
| 1051 | AAACGGAAGA TGACCTTCAA AAAGGCATTT TTCTGCCTTT ATCTCGCCCC |
|
| 1101 | GTCCCTGATG CTTGTCGGTT ACAACCTGTA TTCAAGAGGG ATATTGAAAC |
|
| 1151 | AGGAACACCT CCGCCCGCTG CCCGGCACGC CCGTTGCTGC GGAAAATAAT |
|
| 1201 | TTTCCGGAAA CCGTCTTGAC CCTCGGCGAC TCGCACGCCG GACACCTGCG |
|
| 1251 | GGGGTTTCTG GATTATGTCG GCGGCAGGGA AGGGTGGAAA GCTAAAATCC |
|
| 1301 | TGTCCCTCGA TTCGGAGTGT TTGGTTTGGG TGGATGAGAA GCTGGCAGAC |
|
| 1351 | AACCCGTTGT GCCGAAAATA CCGGGATGAA GTTGAAAAAG CCGAAGCTGT |
|
| 1401 | TTTCATTGCC CAATTCTATG ATTTGAGGAT GGGCGGCCAG CCCGTGCCGA |
|
| 1451 | GATTTGAAGC GCAATCCTTC CTGATACCCG GGTTCAAAGC CCGATTCAGG |
|
| 1501 | GAAACCGTCA AGAGGATAGC CGCCGTCAAA CCTGTATATG TTTTTGCAAA |
|
| 1551 | CAATACATCA ATCAGCCGTT CTCCCTTGAG GGAGGAAAAA TTGAAAAGAT |
|
| 1601 | TTGCTATAAA CCAATACCTC CGGCCTATTC GGGCTATGGG CGACATCGGC |
|
| 1651 | AAGAGCAATC AGGCGGTCTT TGATTTGGTT AAAGATATTC CCAATGTGCA |
|
| 1701 | TTGGGTGGAC GCACAAAAAT ACCTGCCCAA AAACACGGTC GAAATACACG |
|
| 1751 | GACGCTATCT TTACGGCGAC CAAGACCACC TGACCTATTT CGGTTCTTAT |
|
| 1801 | TATATGGGGC GGGAATTTCA CAAACACGAA CGCCTGCTCA AGCATTCCCG |
|
| 1851 | AGGCGGCGCA TTGCAGTAG |
[1700]This encodes a protein having amino acid sequence <SEQ ID 834>:
[0000]| 1 | MQAVRYRPEI DGLRAVAVLS VIIFHLNNRW LPGGFLGVDI FFVISGFLIT | |
|
| 51 | NIILSEIQNG SFSFRDFYTR RIKRIYPAFI AAVSLASVIA SQIFLYEDFN |
|
| 101 | QMRKTIELST VFLSNIYLGF RLGYFDLSAD ENPVLHIWSL AVEEQYYLLY |
|
| 151 | PLLLIFCYKK TKSLRVLRNI SIILFLILTA SSFLPAGFYT DILNQPNTYY |
|
| 201 | LSTLRFPELL VGSLLAVYGQ TQNGRRQTEN GKRQLLSLLC FGALLVCLFV |
|
| 251 | IDKHDPFIPG ITLLLPCLLT ALLIRSMQYG TLPTRILSAS PIVFVGKISY |
|
| 301 | SLYLYHWIFI AFAHYITGDK QLGLPAVSAV AALTAGFSLL SYYLIEQPLR |
|
| 351 | KRKMTFKKAF FCLYLAPSLM LVGYNLYSRG ILKQEHLRPL PGTPVAAENN |
|
| 401 | FPETVLTLGD SHAGHLRGFL DYVGGREGWK AKILSLDSEC LVWVDEKLAD |
|
| 451 | NPLCRKYRDE VEKAEAVFIA QFYDLRMGGQ PVPRFEAQSF LIPGFKARFR |
|
| 501 | ETVKRIAAVK PVYVFANNTS ISRSPLREEK LKRFAINQYL RPIRAMGDIG |
|
| 551 | KSNQAVFDLV KDIPNVHWVD AQKYLPKNTV EIHGRYLYGD QDHLTYFGSY |
|
| 601 | YMGREFHKHE RLLKHSRGGA LQ* |
[1701]ORF128ng and ORF128-1 show 95.7% identity in 622 aa overlap:
[0000]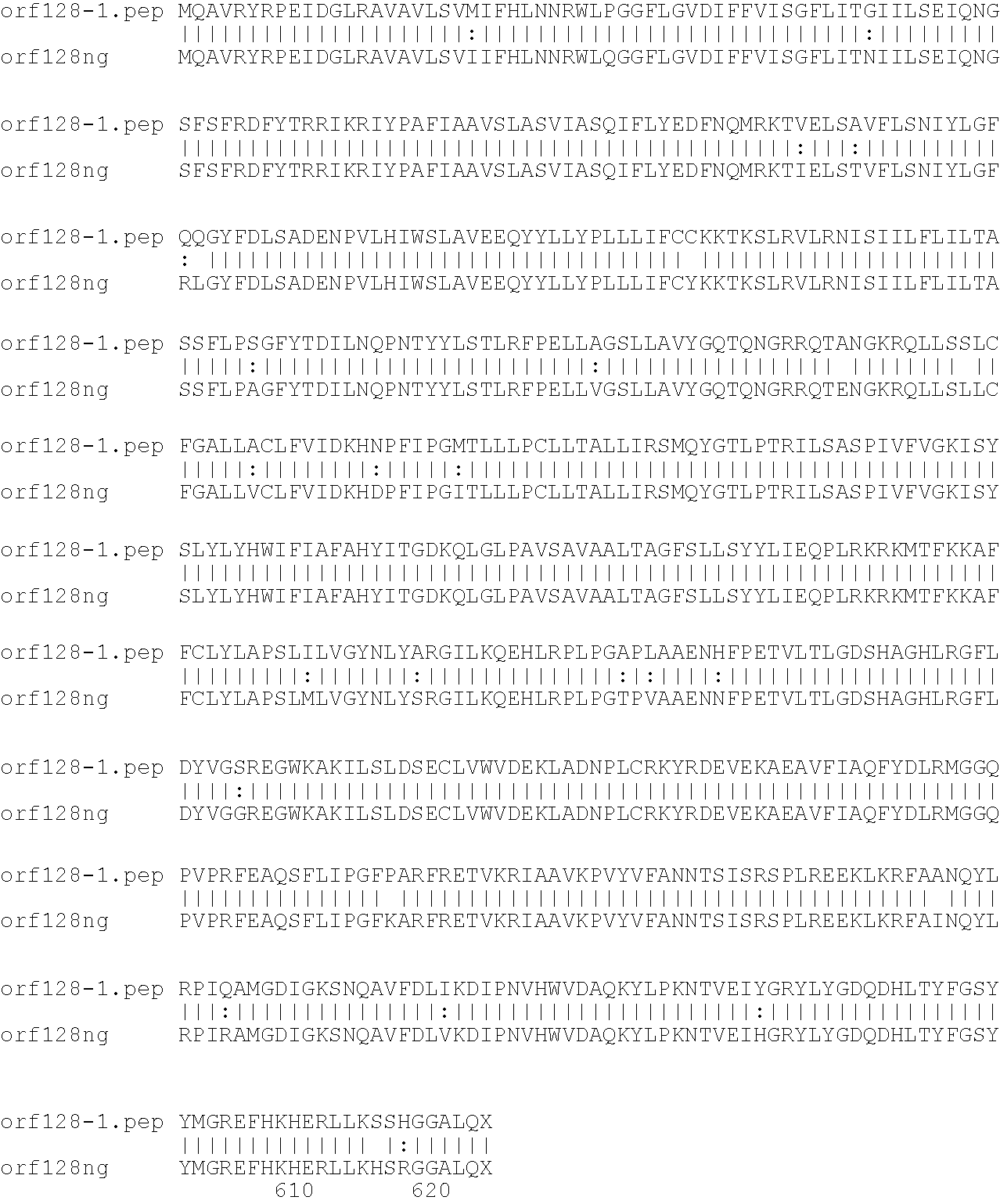
[1702]In addition, ORF218ng shows homology to a hypothetical H. influenzae protein:
[0000]| sp|P43993|Y392_HAEIN HYPOTHETICAL PROTEIN HI0392 >gi|1074385|pir||B64007 | |
| hypothetical protein HI0392 - Haemophilus influenzae (strain Rd KW20) |
| >gi|1573364 (U32723) H. influenzae predicted coding region HI0392 [Haemophilus |
| influenzae] Length = 245 |
| Score = 239 bits (604), Expect = 3e−62 |
| Identities = 124/225 (55%), Positives = 152/225 (67%), Gaps = 1/225 (0%) |
|
| Query: | 38 | VDIFFVISGFLITNIILSEIQNGSFSFRDFYTRRIKRIYPXXXXXXXXXXXXXXXXFLYE | 97 | |
| | +DIFFVISGFLIT II++EIQ SFS + FYTRRIKRIYP F+Y | |
| Sbjct: | 1 | MDIFFVISGFLITGIIITEIQQNSFSLKQFYTRRIKRIYPAFITVMALVSFIASAIFIYN | 60 |
|
| Query: | 98 | DFNQMRKTIELSTVFLSNIYLGFRLGYFDLSADENPVLHIWSLAVEEQXXXXXXXXXIFC | 157 |
| | DFN++RKTIEL+ FLSN YLG GYFDLSA+ENPVLHIWSLAVE Q I | |
| Sbjct: | 61 | DFNKLRKTIELAIAFLSNFYLGLTQGYFDLSANENPVLHIWSLAVEGQYYLIFPLILILA | 120 |
|
| Query: | 158 | YKKTKSLRVLRNISIILFLILTASSFLPAGFYTDILNQPNTYYLSTLRFPELLVGSLLAV | 217 |
| | YKK + ++VL I++ILF IL A+SF+ A FY ++L+QPN YYLS LRFPELLVGSLLA+ | |
| Sbjct: | 121 | YKKFREVKVLFIITLILFFILLATSFVSANFYKEVLHQPNIYYLSNLRFPELLVGSLLAI | 180 |
|
| Query: | 218 | YGQTQNGRRQTENGKRQLLSLLCFGALLVCLFVIDKHDPFIPGIT | 262 |
| | Y N + Q +L++L L CLF+++ + FIPGIT | |
| Sbjct: | 181 | YHNLSN-KVQLSKQVNNILAILSTLLLFSCLFLMNNNIAFIPGIT | 224 |
[1703]This analysis, including the identification of several putative transmembrane domains, suggests that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 99
[1704]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 835>:
[0000]| 1 | ..ATTATTTACG AATACCGCTG GATGTTTCTT TACGGCGCAC TGACGACCTT | |
|
| 51 | GGGGCTGACG GTCGTGGCAA C.GCGGGCGG TTCGGTATTG GGTCTGTTGT |
|
| 101 | TGGCGTTGGC GCGCCTGATT CACTTGGAAA AAGCCGGTGC GCCGATGCGC |
|
| 151 | GTGCTGGCGT GGGCGTTGCG TAAAGTTTCG CTGCTGTATG TTACGCTGTT |
|
| 201 | CCGGGGTACG CCGCTGTTTG TGCAGATTGT GATTTGGGCG TATGTGTGGT |
|
| 251 | TTCCGTTTTT CGTC.. |
[1705]This corresponds to the amino acid sequence <SEQ ID 836; ORF129>:
[0000]| 1 | ..IIYEYRWMFL YGALTTLGLT VVAXAGGSVL GLLLALARLI HLEKAGAPMR | |
|
| 51 | VLAWALRKVS LLYVTLFRGT PLFVQIVIWA YVWFPFFV.. |
[1706]Further work revealed the complete nucleotide sequence <SEQ ID 837>:
[0000]| 1 | ATGGATTTTC GTTTTGACAT TATTTACGAA TACCGCTGGA TGTTTCTTTA | |
|
| 51 | CGGCGCACTG ACGACCTTGG GGCTGACGGT CGTGGCAACG GCGGGCGGTT |
|
| 101 | CGGTATTGGG TCTGTTGTTG GCGTTGGCGC GCCTGATTCA CTTGGAAAAA |
|
| 151 | GCCGGTGCGC CGATGCGCGT GCTGGCGTGG GCGTTGCGTA AAGTTTCGCT |
|
| 201 | GCTGTATGTT ACGCTGTTCC GGGGTACGCC GCTGTTTGTG CAGATTGTGA |
|
| 251 | TTTGGGCGTA TGTGTGGTTT CCGTTTTTCG TCCATCCTTC AGACGGCATT |
|
| 301 | TTGGTCAGCG GCGAGGCGGC AATCGCGCTG CGTCGCGGAT ACGGGCCGCT |
|
| 351 | GATTGCCGGT TCTTTGGCAC TGATCGCCAA CTCGGGGGCG TATATCTGTG |
|
| 401 | AGATTTTCCG CGCGGGCATC CAGTCTATAG ACAAAGGACA GATGGAGGCG |
|
| 451 | GCGCGTTCTT TGGGGCTGAC CTATCCGCAG GCGATGCGCT ATGTGATTCT |
|
| 501 | GCCGCAGGCA TTGCGCCGCA TGCTGCCGCC TTTGGCGAGC GAGTTCATCA |
|
| 551 | CGCTCTTGAA AGACAGCTCG CTGCTGTCGG TCATTGCTGT GGCGGAGTTG |
|
| 601 | GCGTATGTTC AGAATACGAT TACGGGCCGG TATTCGGTTT ATGAAGAACC |
|
| 651 | GCTTTACACC GTCGCCCTGA TTTATCTGTT GATGACGACT TTCTTAGGCT |
|
| 701 | GGATATTCCT GCGTTTGGAA AAACGTTACA ATCCGCAACA CCGCTGA |
[1707]This corresponds to the amino acid sequence <SEQ ID 838; ORF129-1>:
[0000]| 1 | MDFRFDIIYE YRWMFLYGAL TTLGLTVVAT AGGSVLGLLL ALARLIHLEK | |
|
| 51 | AGAPMRVLAW ALRKVSLLYV TLFRGTPLFV QIVIWAYVWF PFFVHPSDGI |
|
| 101 | LVSGEAAIAL RRGYGPLIAG SLALIANSGA YICEIFRAGI QSIDKGQMEA |
|
| 151 | ARSLGLTYPQ AMRYVILPQA LRRMLPPLAS EFITLLKDSS LLSVIAVAEL |
|
| 201 | AYVQNTITGR YSVYEEPLYT VALIYLLMTT FLGWIFLRLE KRYNPQHR* |
[1708]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1709]ORF129 shows 98.9% identity over a 88aa overlap with an ORF (ORF129a) from strain A of N. meningitidis:
[0000]
[1710]The complete length ORF129a nucleotide sequence <SEQ ID 839> is:
[0000]| 1 | ATGGATTTTC GTTTTGACAT TATTTACGAA TACCGCTGGA TGTTTCTTTA | |
|
| 51 | CGGCGCACTG ACGACCTTGG GGCTGACGGT CGTGGCGACG GCGGGCGGTT |
|
| 101 | CGGTATTGGG TCTGTTGTTG GCGTTGGCGC GCCTGATTCA CTTGGAAAAA |
|
| 151 | GCCGGTGCGC CGATGCGCGT GCTGGCGTGG GCGTTGCGTA AGGTTTCGCT |
|
| 201 | GCTGTATGTT ACGCTGTTCC GGGGTACGCC GCTGTTTGTG CAGATTGTGA |
|
| 251 | TTTGGGCGTA TGTGTGGTTT CCGTTTTTCG TCCATCCTTC AGACGGCATT |
|
| 301 | TTGGTTAGCG GCGAGGCGGC AATCGCGCTG CGTCGCGGAT ACGGGCCGCT |
|
| 351 | GATTGCCGGT TCTTTGGCAC TGATCGCCAA CTCGGGGGCG TATATCTGTG |
|
| 401 | AGATTTTCCG CGCGGGCATC CAGTCTATAG ACAAAGGACA GATGGAGGCG |
|
| 451 | GCGCGTTCTT TGGGGCTGAC CTATCCGCAG GCGATGCGCT ATGTGATTCT |
|
| 501 | GCCGCAGGCA TTGCGCCGTA TGCTGCCGCC TTTGGCGAGC GAGTTCATCA |
|
| 551 | CGCTCTTGAA AGACAGCTCG CTGCTGTCGG TCATTGCTGT GGCGGAGTTG |
|
| 601 | GCGTATGTTC AGAATACGAT TACGGGCCGG TATTCGGTTT ATGAAGAACC |
|
| 651 | GCTTTACACC GTCGCCCTGA TTTATCTGTT GATGACGACT TTCTTAGGCT |
|
| 701 | GGATATTCCT GCGTTTGGAA AAACGTTACA ATCCGCAACA CCGCTGA |
[1711]This encodes a protein having amino acid sequence <SEQ ID 840>:
[0000]| 1 | MDFRFDIIYE YRWMFLYGAL TTLGLTVVAT AGGSVLGLLL ALARLIHLEK | |
|
| 51 | AGAPMRVLAW ALRKVSLLYV TLFRGTPLFV QIVIWAYVWF PFFVHPSDGI |
|
| 101 | LVSGEAAIAL RRGYGPLIAG SLALIANSGA YICEIFRAGI QSIDKGQMEA |
|
| 151 | ARSLGLTYPQ AMRYVILPQA LRRMLPPLAS EFITLLKDSS LLSVIAVAEL |
|
| 201 | AYVQNTITGR YSVYEEPLYT VALIYLLMTT FLGWIFLRLE KRYNPQHR* |
[1712]ORF129a and ORF129-1 show 100.0% identity in 248 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1713]ORF129 shows 98.9% identity over a 88 aa overlap with a predicted ORF (ORF129ng) from N. gonorrhoeae:
[0000]
[1714]An ORF129ng nucleotide sequence <SEQ ID 841> was predicted to encode a protein having amino acid sequence <SEQ ID 842>:
[0000]| 1 | MDFRFDIIYE YRWMFLYGAL TTLGLTVVAT AGGSVLGLLL ALARLIHLEK | |
|
| 51 | AGAPMRVLAW ALRKVSLLYV TLFRGTPLFV QIVIWAYVWF PFFVILHTAF |
|
| 101 | LGNAMRQSRR VPDKGRWIAG SLELNCQPRG RKTRGEFPPG ESNLGTEPRN |
|
| 151 | PLSMGQRRFP GCENWYPPQN FIKK* |
[1715]Further work revealed the following gonococcal sequence <SEQ ID 843>:
[0000]| 1 | ATGGATTTTc gtTTTGACAT TATTTAcgaA TACCGCTGGA TGTTTCTTTA | |
|
| 51 | CGGCGCACTG Acgaccttgg ggctgacggt cgtggcgacg gCGGGCGGTT |
|
| 101 | CGGtattggG TCTGTTGTTG GCGTTGGCGC GCCTGATTCA CTTGGAAAAA |
|
| 151 | GCCGGTGCGC CGATGCGCGT GCTGGCGTGG GCGTTGCGTA AGGTTTCGCT |
|
| 201 | GCTGTACGTT ACCCTGTTCC GGGGTACGCC GCTGTTTGTG CAGATTGTGA |
|
| 251 | TTTGGGCGTA TGTGTGGTTT CCGTTTTTCG TCCATCCTTC AGACGGCATT |
|
| 301 | TTGGTCAGCG GCGAGGCGGC AATCGCGCTG CGTCGCGGAT ACGGGCCGCT |
|
| 351 | GATTGCCGGT TCTTTGGCAC TGATCGCCAA CTCGGGGGCG TATATCTGTG |
|
| 401 | AGATTTTCCG CGCGGGCATC CAGTCTATAG ACAAAGGACA GATGGAGGCG |
|
| 451 | GCGTGTTCTT TGGGACTGAC CTATCCGCAG GCGATGCGCT ATGTGATTCT |
|
| 501 | GCCGCAGGCA TTGCGCCGTA TGCTGCCGCC TTTGGCGAGC GAGTTCATCA |
|
| 551 | CGCTCTTGAA AGACAGCTCG CTGCTGTCGG TCATTGCTGT GGCGGAGTTG |
|
| 601 | GCGTATGTTC AGAATACGAT TACGGGCCGG TATTCGGTTT ATGAAGAACC |
|
| 651 | GCTTTACACC GCCGCCCTGA TTTATCTGTT GATGACGACT TTCTTAGGCT |
|
| 701 | GGATATTCCT GCGTTTGGAA AAACGTTACA ATCCGCAACA CCGCTGA |
[1716]This corresponds to the amino acid sequence <SEQ ID 844; ORF129ng-1>:
[0000]| 1 | MDFRFDIIYE YRWMFLYGAL TTLGLTVVAT AGGSVLGLLL ALARLIHLEK | |
|
| 51 | AGAPMRVLAW ALRKVSLLYV TLFRGTPLFV QIVIWAYVWF PFFVHPSDGI |
|
| 101 | LVSGEAAIAL RRGYGPLIAG SLALIANSGA YICEIFRAGI QSIDKGQMEA |
|
| 151 | ARSLGLTYPQ AMRYVILPQA LRRMLPPLAS EFITLLKDSS LLSVIAVAEL |
|
| 201 | AYVQNTITGR YSVYEEPLYT VALIYLLMTT FLGWIFLRLE KRYNPQHR* |
[1717]ORF129ng-1 and ORF129-1 show 99.2% identity in 248 aa overlap:
[0000]
[1718]In addition, ORF129ng-1 is homologous to an ABC transporter from A. fulgidus:
[0000]| 2650409(AE001090) glutamine ABC transporter, permease protein (glnP) | |
| [Archaeoglobus fulgidus]Length = 224 |
| Score = 132 bits (329), Expect = 2e−30 |
| Identities = 86/178 (48%), Positives = 103/178 (57%), Gaps = 18/178 (10%) |
|
| Query: | 65 | VSLLYVTLFRGTPLFVQIVIWAYVWFPFFVHPSDGILVSGEAAIALRRGYGPLIAGSLAL | 124 | |
| | +S YV + RGTPL VQI+I +F P+ GI + E A G +AL |
| Sbjct: | 58 | ISTAYVEVIRGTPLLVQILI------VYFGLPAIGINLQPEPA------------GIIAL | 99 |
|
| Query: | 125 | IANSGAYICEIFRAGIQSIDKGQMEAACSLGLTYPQAMRYVILPQALRRMLPPLASEFIT | 184 |
| | SGAYI EI RAGI+SI GQMEAA SLG+TY QAMRYVI PQA R +LP L +EFI |
| Sbjct: | 100 | SICSGAYIAEIVRAGIESIPIGQMEAARSLGMTYLQAMRYVIFPQAFRNILPALGNEFIA | 159 |
|
| Query: | 185 | LLKDSSLLSVIAVAELAYVQNTITGRYSVYEEPLYTAALIYLLMTTFLGWIFLRLEKR | 242 |
| | LLKDSSLLSVI++ EL V I P AL YL+MT L + +K+ |
| Sbjct: | 160 | LLKDSSLLSVISIVELTRVGRQIVNTTFNAWTPFLGVALFYLMMTIPLSRLVAYSQKK | 217 |
[1719]This analysis, including the identification of transmembrane domains in the two proteins, suggests that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 100
[1720]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 845>:
[0000]| 1 | ..CTGAAAGAAT GCCGTCTGAA AGACCCTGTT TTTATTCCAA ATATCGTTTA | |
|
| 51 | TAAGAACATC GCCATTACTT TCCTGCTCTT GCACGCCGCC GCCGAACTTT |
|
| 101 | GGCTGCCCGC GCAAACCGCC GGTTTTACCG CGCTCGCCGT CGGCTTCATC |
|
| 151 | CTGCTCGCCA AGCTGCGTGA gCTTCACCAT CACGAACTCT TACGTAAACA |
|
| 201 | cTACGTCCGC ACTTATTACy TGCTCCAACT CTTTGCCGCC GCAGgcTAgT |
|
| 251 | TTGTGGACAG GCGCGGCGwA ATTACAAAAC CTGCCCGCyT CCGCGCCCCT |
|
| 301 | GCACCTGATT ACCCTCGGCG GCATGATGGG CGGCGTGATG ATGGTGTGGc |
|
| 351 | TGACCGCCGG ACTGTGGCAC AGCGGCTTTA CCAAACTCGA CTACCCCAAA |
|
| 401 | CTCTGCCGCA TTGCCGTCCC CATCCTTTTC GCCGCCGCCG TCTCGCGCGC |
|
| 451 | TTTCTTGrTG AACGTGAACC CGrTATTTTT CATTACCGTT CCTGCGATTC |
|
| 501 | TGACCGCCGC CGTATTCGTA CTGTATCTTT TCrCGTTTAT ACCGATATTT |
|
| 551 | CGGGCGAATG CGTTTACAGA CGATCCGGAr TAr |
[1721]This corresponds to the amino acid sequence <SEQ ID 846; ORF130>:
[0000]| 1 | ..LKECRLKDPV FIPNIVYKNI AITFLLLHAA AELWLPAQTA GFTALAVGFI | |
|
| 51 | LLAKLRELHH HELLRKHYVR TYYLLQLFAA AGSLWTGAAX LQNLPASAPL |
|
| 101 | HLITLGGMMG GVMMVWLTAG LWHSGFTKLD YPKLCRIAVP ILFAAAVSRA |
|
| 151 | FLXNVNPXFF ITVPAILTAA VFVLYLFXFI PIFRANAFTD DPE* |
[1722]Further work revealed the complete nucleotide sequence <SEQ ID 847>:
[0000]| 1 | ATGCGGCCGT TTTTCGTCGG CGCGGCGGTG CTTGCCATAC TCGGTGCGCT | |
|
| 51 | GGTGTTTTTC ATCAACCCCG GTGCCATCGT CCTGCACCGC CAAATTTTCT |
|
| 101 | TGGAACTTAT GCTGCCGGCG GCATACGGCG GTTTTTTGAC TGCGGCTTTG |
|
| 151 | TTGGACTGGA CGGGTTTTTC GGGTAACCTG AAACCTGTCG CGACTTTGAT |
|
| 201 | GGCGGCATTA TTGCTCGCCG CATCCGCTAT ACTGCCCTTT TCGCCGCAAA |
|
| 251 | CTGCCTCGTT TTTCGTCGCC GCCTATTGGC TGGTGTTGCT GCTGTTCTGC |
|
| 301 | GCCCGGCTGA TTTGGCTAGA CCGAAACACC GACAACTTCG CCCTGCTAAT |
|
| 351 | GTTACTTGCC GCGTTCACTG TTTTTCAGAC GGCATATGCC GTCAGCGGCG |
|
| 401 | ATTTGAACCT GTTGCGCGCG CAAGTGCATC TAAATATGGC GGCGGTGATG |
|
| 451 | TTCGTATCCG TGCGCGTCAG TATTCTTTTG GGCGCGGAAG CCCTGAAAGA |
|
| 501 | ATGCCGTCTG AAAGACCCTG TTTTTATTCC AAATATCGTT TATAAAAACA |
|
| 551 | TCGCCATTAC TTTCCTGCTC TTGCACGCCG CCGCCGAACT TTGGCTGCCC |
|
| 601 | GCGCAAACCG CCGGTTTTAC CGCGCTCGCC GTCGGCTTCA TCCTGCTCGC |
|
| 651 | CAAGCTGCGT GAGCTTCACC ATCACGAACT CTTACGTAAA CACTACGTCC |
|
| 701 | GCACTTATTA CCTGCTCCAA CTCTTTGCCG CCGCAGGCTA TTTGTGGACA |
|
| 751 | GGCGCGGCGA AATTACAAAA CCTGCCCGCC TCCGCGCCCC TGCACCTGAT |
|
| 801 | TACCCTCGGC GGCATGATGG GCGGCGTGAT GATGGTGTGG CTGACCGCCG |
|
| 851 | GACTGTGGCA CAGCGGCTTT ACCAAACTCG ACTACCCCAA ACTCTGCCGC |
|
| 901 | ATTGCCGTCC CCATCCTTTT CGCCGCCGCC GTCTCGCGCG CTTTCTTGAT |
|
| 951 | GAACGTGAAC CCGATATTTT TCATTACCGT TCCTGCGATT CTGACCGCCG |
|
| 1001 | CCGTATTCGT ACTGTATCTT TTCACGTTTA TACCGATATT TCGGGCGAAT |
|
| 1051 | GCGTTTACAG ACGATCCGGA ATAA |
[1723]This corresponds to the amino acid sequence <SEQ ID 848; ORF130-1>:
[0000]| 1 | MRPFFVGAAV LAILGALVFF INPGAIVLHR QIFLELMLPA AYGGFLTAAL | |
|
| 51 | LDWTGFSGNL KPVATLMAAL LLAASAILPF SPQTASFFVA AYWLVLLLFC |
|
| 101 | ARLIWLDRNT DNFALLMLLA AFTVFQTAYA VSGDLNLLRA QVHLNMAAVM |
|
| 151 | FVSVRVSILL GAEALKECRL KDPVFIPNIV YKNIAITFLL LHAAAELWLP |
|
| 201 | AQTAGFTALA VGFILLAKLR ELHHHELLRK HYVRTYYLLQ LFAAAGYLWT |
|
| 251 | GAAKLQNLPA SAPLHLITLG GMMGGVMMVW LTAGLWHSGF TKLDYPKLCR |
|
| 301 | IAVPILFAAA VSRAFLMNVN PIFFITVPAI LTAAVFVLYL FTFIPIFRAN |
|
| 351 | AFTDDPE* |
[1724]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1725]ORF130 shows 94.3% identity over a 193aa overlap with an ORF (ORF130a) from strain A of N. meningitidis:
[0000]
[1726]The complete length ORF130a nucleotide sequence <SEQ ID 849> is:
[0000]| 1 | ATGCGGCCGT TTTTCGTCGG CGCGGCGGTG CTTGCCATAC TCGGTGCGCT | |
|
| 51 | GGTGTTTTTC ATCAACCCCG GTGCCATCGT CCTGCACCGC CAAATTTTCT |
|
| 101 | TGGAACTTAT GCTGCCGGCG GCATACGGCG GTTTTTTGAC TGCGGCTTTG |
|
| 151 | TTGGACTGGA CGGGTTTTTC GGGTAACCTG AAACCTGTCG CGACTTTGAT |
|
| 201 | GGCGGCATTA TTGCTCGCCG CATCCGCTAT ACTGCCCTTT TCGCCGCAAA |
|
| 251 | CTGCCTCGTT TTTCGTCGCC GCCTATTGGC TGGTGTTGCT GCTGTTCTGC |
|
| 301 | GCCCGGCTGA TTTGGCTAGA CCGAAACACC GACAACTTCG CCCTGCTAAT |
|
| 351 | GTTACTTGCC GCGTTCACTG TTTTTCAGAC GGCATATGCC GTCAGCGGCG |
|
| 401 | ATTTGAACCT GTTGCGCGCG CAAGTGCATC TAAATATGGC GGCGGTGATG |
|
| 451 | TTCGTATCCG TGCGCGTCAG TATTCTTTTG GGCGCGGAAG CCCTGAAAGA |
|
| 501 | ATGCCGTCTG AAAGACCCAG TATTCATCCC CAATGTCGTC TATAAAAACA |
|
| 551 | TCGCCATTAC CTTCCTGCTC CTGCACGCCG CCGCCGAACT TTGGCTGCCT |
|
| 601 | GCGCAAACCG CCGGTTTTAC CTCGCTCGCC GTCGGCTTTA TCCTGCTTGC |
|
| 651 | CAAGCTGCGT GAGCTTCACC ATCACGAACT CCTGCGCAAA CACTACGTCC |
|
| 701 | GCACTTATTA CCTGCTCCAA CTCTTTGCCG CCGCAGGCTA TTTGTGGACA |
|
| 751 | GGCGCGGCGA AATTACAAAA CCTGCCCGCC TCCGCGCCCC TGCACCTGAT |
|
| 801 | TACCCTCGGT GGCATGATGG GCAGCGTGAT GATGGTGTGG CTGACTGCCG |
|
| 851 | GACTGTGGCA CAGCGGCTTT ACCAAGCTCG ACTACCCGAA ACTCTGCCGC |
|
| 901 | ATCGCCGTCC CCATCCTNTT CGCCGCCGCC GTTTCGCGCG CTGTTTTAAT |
|
| 951 | GAACGTAAAC CCGATATTCT TCATCACCGT CCCCGCAATT CTGACCGCCG |
|
| 1001 | CCGTGTTCGT GCTTTACCTG CTGACATTCG TACCGATCTT TCGGGCGAAC |
|
| 1051 | GCGTTTACAG ACGATCCGGA ATAA |
[1727]This encodes a protein having amino acid sequence <SEQ ID 850>:
[0000]| 1 | MRPFFVGAAV LAILGALVFF INPGAIVLHR QIFLELMLPA AYGGFLTAAL | |
|
| 51 | LDWTGFSGNL KPVATLMAAL LLAASAILPF SPQTASFFVA AYWLVLLLFC |
|
| 101 | ARLIWLDRNT DNFALLMLLA AFTVFQTAYA VSGDLNLLRA QVHLNMAAVM |
|
| 151 | FVSVRVSILL GAEALKECRL KDPVFIPNVV YKNIAITFLL LHAAAELWLP |
|
| 201 | AQTAGFTSLA VGFILLAKLR ELHHHELLRK HYVRTYYLLQ LFAAAGYLWT |
|
| 251 | GAAKLQNLPA SAPLHLITLG GMMGSVMMVW LTAGLWHSGF TKLDYPKLCR |
|
| 301 | IAVPILFAAA VSRAVLMNVN PIFFITVPAI LTAAVFVLYL LTFVPIFRAN |
|
| 351 | AFTDDPE* |
[1728]ORF130a and ORF130-1 show 98.3% identity in 357 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1729]ORF130 shows 91.7% identity over a 193 aa overlap with a predicted ORF (ORF130ng) from N. gonorrhoeae:
[0000]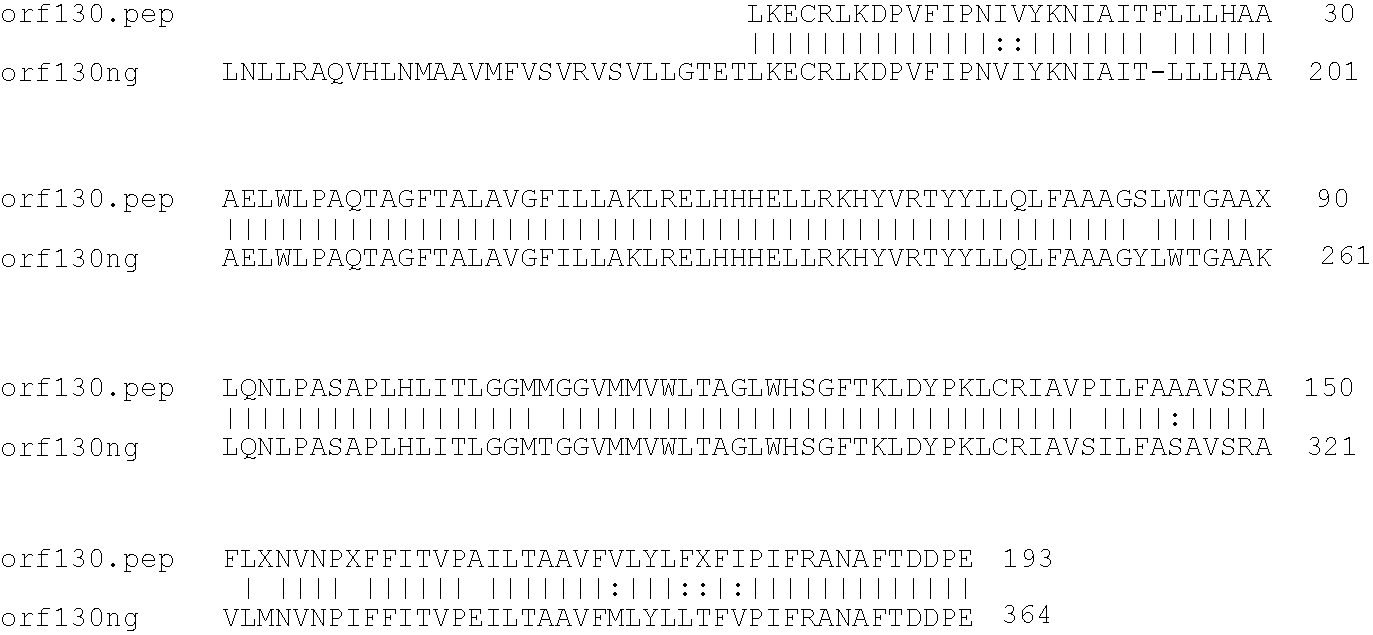
[1730]An ORF130ng nucleotide sequence <SEQ ID 851> was predicted to encode a protein having amino acid sequence <SEQ ID 852>:
[0000]| 1 | MNKFFTHPMR PFFVGAAVLA ILGALVFFHQ PRRYHPAPPN FLGTYAAGCI | |
|
| 51 | RRFFDYRFVG PDGFFRQPET CRYFDGGVVA CCGCFIAVFT ATCRIFRRRL |
|
| 101 | LAGVAAVLRL ADLARRQHRT LRSVDVTAAF TVFQTAYAVS GDLNLLRAQV |
|
| 151 | HLNMAAVMFV SVRVSVLLGT ETLKECRLKD PVFIPNVIYK NIAITLLLHA |
|
| 201 | AAELWLPAQT AGFTALAVGF ILLAKLRELH HHELLRKHYV RTYYLLQLFA |
|
| 251 | AAGYLWTGAA KLQNLPASAP LHLITLGGMT GGVMMVWLTA GLWHSGFTKL |
|
| 301 | DYPKLCRIAV SILFASAVSR AVLMNVNPIF FITVPEILTA AVFMLYLLTF |
|
| 351 | VPIFRANAFT DDPE* |
[1731]Further work revealed the following gonococcal DNA sequence <SEQ ID 853>:
[0000]| 1 | ATGCGCCCGT TTTTCGTCGG TGCGGCAGTA CTTGCCATAC TCGGTGCGTT | |
|
| 51 | GGTGTTTTTT ATCAACCCCG GCGCTATCAT CCTGCACCGC CAAATTTTCT |
|
| 101 | TGGAACTTAT GCTGCCGGCT GCATACGGCG GTTTTTTGAC TACCGCTTTG |
|
| 151 | TTGGACCGGA CGGGTTTTTC AGGCAACCTG AAACCTGCCG CTACTTTGAT |
|
| 201 | GGCGGTGTTG TTGCTTGTTG CGGCTGTTTT ATTGCCGTTT TTACCGCAAC |
|
| 251 | TTGCCGCATT TTTCGTCGCC GCCTATTGGC TGGTGTTGCT GCTGTTCTGC |
|
| 301 | GCCTGGCTGA TTTGGCTCGA CCGCAACACC GACAACTTCG CTCTGTTGAT |
|
| 351 | GTTACTTGCC GCATTTACCG TTTTTCAGAC GGCCTATGCC GTCAGCGGCG |
|
| 401 | ATTTGAACTT ACTGCGCGCG CAAGTGCATT TGAATATGGC GGCGGTCATG |
|
| 451 | TTCGTATCCG TCCGCGTCAG CGTCCTTTTG GGCACGGAAA CCCTGAAAGA |
|
| 501 | ATGCCGTCTG AAAGACCCCG TATTCATCCC CAACGTTATC TATAAAAACA |
|
| 551 | TCGCCATCAC CCTGCTGCTG CACGCCGCCG CCGAACTTTG GCTGCCCGCG |
|
| 601 | CAAACCGCCG GTTTTACTGC GCTTGCCGTC GGCTTCATCC TGCTCGCCAA |
|
| 651 | GCTGCGCGAA CTGCACCATC ACGAACTCTT ACGCAAACAC TACGTCCGCA |
|
| 701 | CTTATTACCT GCTCCAGCTC TTTGCCGCCG CAGGTTATCT GTGGACAGGC |
|
| 751 | GCGGCGAAAC TGCAAAACCT GCCCGCCTCC GCGCCCCTGC ACCTGATTAC |
|
| 801 | CCTCGGCGGC ATGACGGGTG GCGTGATGAT GGTGTGGCTG ACTGCCGGAC |
|
| 851 | TGTGGCACAG CGGCTTTACC AAACTCGACT ACCCGAAACT CTGCCGCATC |
|
| 901 | GCCGTCTCCA TCCTTTTCGC CTCCGCCGTT TCGCGCGCTG TTTTAATGAA |
|
| 951 | CGTGAATCCG ATATTCTTCA TCACCGTTCC CGAGATTCTG ACCGCCGCCG |
|
| 1001 | TGTTCATGCT TTACCTGCTG ACGTTCGTAC CGATTTTTCG AGCGAACGCG |
|
| 1051 | TTTACAGACG ATCCGGAATA A |
[1732]This corresponds to the amino acid sequence <SEQ ID 854; ORF130ng-1>:
[0000]| 1 | MRPFFVGAAV LAILGALVFF INPGAIILHR QIFLELMLPA AYGGFLTTAL | |
|
| 51 | LDRTGFSGNL KPAATLMAVL LLVAAVLLPF LPQLAAFFVA AYWLVLLLFC |
|
| 101 | AWLIWLDRNT DNFALLMLLA AFTVFQTAYA VSGDLNLLRA QVHLNMAAVM |
|
| 151 | FVSVRVSVLL GTETLKECRL KDPVFIPNVI YKNIAITLLL HAAAELWLPA |
|
| 201 | QTAGFTALAV GFILLAKLRE LHHHELLRKH YVRTYYLLQL FAAAGYLWTG |
|
| 251 | AAKLQNLPAS APLHLITLGG MTGGVMMVWL TAGLWHSGFT KLDYPKLCRI |
|
| 301 | AVSILFASAV SRAVLMNVNP IFFITVPEIL TAAVFMLYLL TFVPIFRANA |
|
| 351 | FTDDPE* |
[1733]ORF130ng-1 and ORF130-1 show 92.4% identity in 357 aa overlap:
[0000]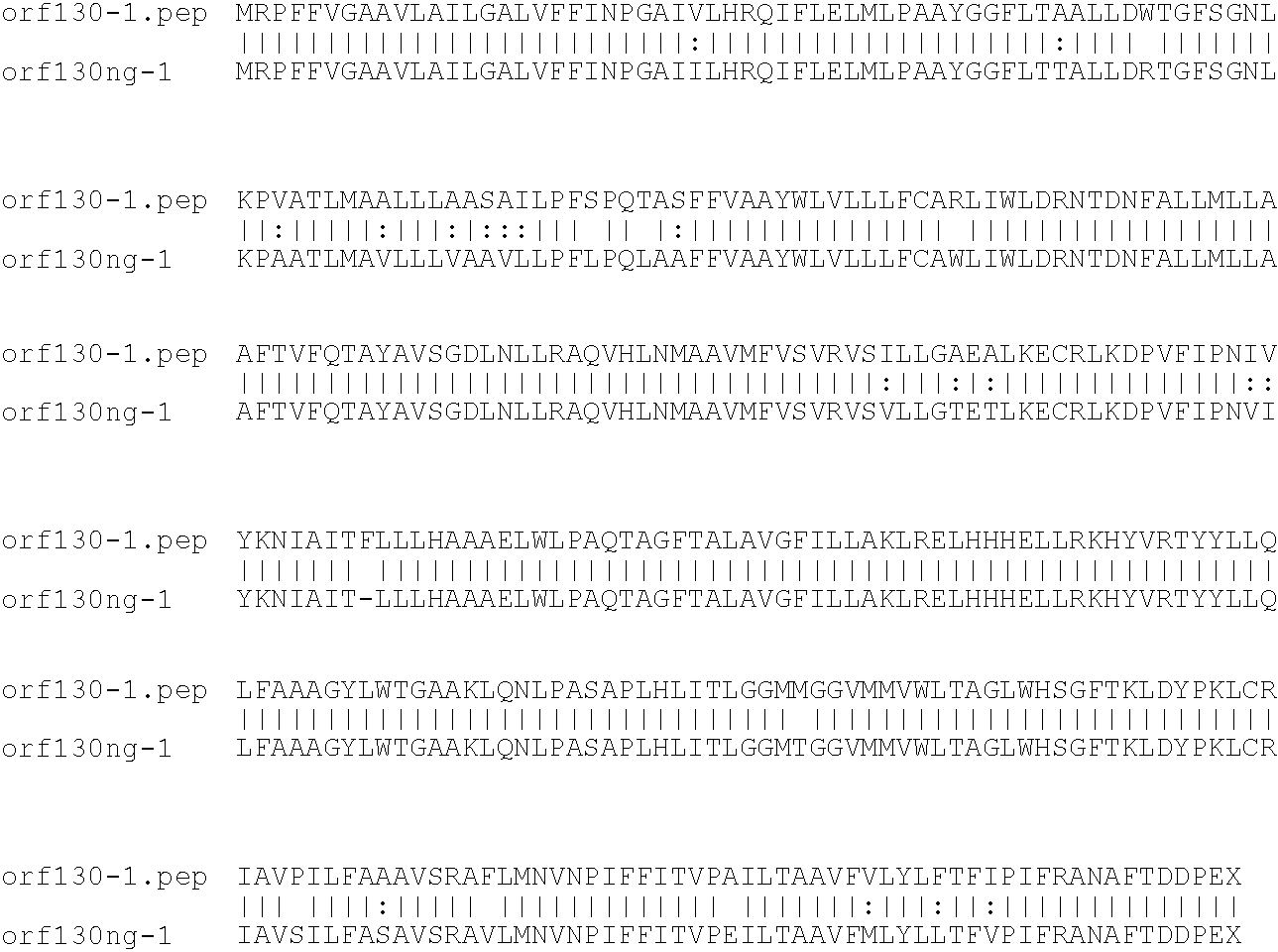
[1734]Based on this analysis, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 101
[1735]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 855>:
[0000]| 1 | ATGGAAATTC GGGCAATAAA ATATACGGCA ATGGCTGCGT TGCTTGCATT | |
|
| 51 | TACGGTTGCA GGCTGCCGGC TGGCGGGGTG GTATGAGTGT TCGTCCCTCA |
|
| 101 | CCGGCTGGTG TAAGCCGAGA AAACCGGCTG CCATCGATTT TTGGGATATT |
|
| 151 | GGCGGCGAGA GTCCGCCGTC TTTAGGGGAC TACGAGATAC CGCTTTCAGA |
|
| 201 | CGGCAATAGT TCCGTCAGGG CAAACGAATA TGAATCCGCA CAACAATCTT |
|
| 251 | ACTTTTACAG GAAAATAGGG AAGTTTGAAG C.TGCGGGCT GGATTGGCGT |
|
| 301 | ACGCGTGACG GCAAACCTTT GATTGAGACG TTCAAACAGG GAGGATTTGA |
|
| 351 | CTGCTTGGAA AAG.. |
[1736]This corresponds to the amino acid sequence <SEQ ID 856; ORF131>:
[0000]| 1 | MEIRAIKYTA MAALLAFTVA GCRLAGWYEC SSLTGWCKPR KPAAIDFWDI | |
|
| 51 | GGESPPSLGD YEIPLSDGNS SVRANEYESA QQSYFYRKIG KFEXCGLDWR |
|
| 101 | TRDGKPLIET FKQGGFDCLE K.. |
[1737]Further work revealed the complete nucleotide sequence <SEQ ID 857>:
[0000]| 1 | ATGGAAATTC GGGCAATAAA ATATACGGCA ATGGCTGCGT TGCTTGCATT | |
|
| 51 | TACGGTTGCA GGCTGCCGGC TGGCGGGGTG GTATGAGTGT TCGTCCCTCA |
|
| 101 | CCGGCTGGTG TAAGCCGAGA AAACCGGCTG CCATCGATTT TTGGGATATT |
|
| 151 | GGCGGCGAGA GTCCGCCGTC TTTAGGGGAC TACGAGATAC CGCTTTCAGA |
|
| 201 | CGGCAATCGT TCCGTCAGGG CAAACGAATA TGAATCCGCA CAACAATCTT |
|
| 251 | ACTTTTACAG GAAAATAGGG AAGTTTGAAG CCTGCGGGCT GGATTGGCGT |
|
| 301 | ACGCGTGACG GCAAACCTTT GATTGAGACG TTCAAACAGG GAGGATTTGA |
|
| 351 | CTGCTTGGAA AAGCAGGGGT TGCGGCGCAA CGGTCTGTCC GAGCGCGTCC |
|
| 401 | GATGGTAA |
[1738]This corresponds to the amino acid sequence <SEQ ID 858; ORF131-1>:
[0000]| 1 | MEIRAIKYTA MAALLAFTVA GCRLAGWYEC SSLTGWCKPR KPAAIDFWDI | |
|
| 51 | GGESPPSLGD YEIPLSDGNR SVRANEYESA QQSYFYRKIG KFEACGLDWR |
|
| 101 | TRDGKPLIET FKQGGFDCLE KQGLRRNGLS ERVRW* |
[1739]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1740]ORF131 shows 95.0% identity over a 121 aa overlap with an ORF (ORF131a) from strain A of N. meningitidis:
[0000]
[1741]The complete length ORF131a nucleotide sequence <SEQ ID 859> is:
[0000]| 1 | ATGGAAATTC GGGCAATAAA ATATACGGCA ATGGCTGCGT TGCTTGCATT | |
|
| 51 | TACGGTTGCA GGCTGCCGGT TGGCAGGTTG GTATGAGTGT TCGTCCCTGT |
|
| 101 | CCGGCTGGTG TAAGCCGAGA AAACCTGCCG CCATCGATTT TTGGGATATT |
|
| 151 | GGCGGCGAGA GTCCTCCGTC TTTAGAGGAC TACGAGATAC CGCTTTCAGA |
|
| 201 | CGGCAATCGT TCCGTCAGGG CAAACGAATA TGAATCCGCA CAACAATCTT |
|
| 251 | ACTTTTACAG GAAAATAGGG AAGTTTGAAG CCTGCGGGTT GGATTGGCGT |
|
| 301 | ACGCGTGACG GCAAACCTTT GATTGAGACG TTCAAACAGG AAGGTTTTGA |
|
| 351 | TTGTTTGAAA AAGCAGGGGT TGCGGCGCAA CGGTCTGTCC GAGCGCGTCC |
|
| 401 | GATGGTAA |
[1742]This encodes a protein having amino acid sequence <SEQ ID 860>:
[0000]| 1 | MEIRAIKYTA MAALLAFTVA GCRLAGWYEC SSLSGWCKPR KPAAIDFWDI | |
|
| 51 | GGESPPSLED YEIPLSDGNR SVRANEYESA QQSYFYRKIG KFEACGLDWR |
|
| 101 | TRDGKPLIET FKQEGFDCLK KQGLRRNGLS ERVRW* |
[1743]ORF131a and ORF131-1 show 97.0% identity in 135 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1744]ORF131 shows 89.3% identity over 121 aa overlap with a predicted ORF (ORF131ng) from N. gonorrhoeae:
[0000]
[1745]A complete length ORF131ng nucleotide sequence <SEQ ID 861> was predicted to encode a protein having amino acid sequence <SEQ ID 862>:
[0000]| 1 | MEIRVIKYTA TAALFAFTVA GCRLAGWYEC LSLSGWCKPR KPAAIDFWDI | |
|
| 51 | GGESPLSLED YEIPLSDGNR SVRANEYESA QKSYFYRKIG KFEACGLDWR |
|
| 101 | TRDGKPLVER FKQEGFDCLE KQGLRRNGLS ERVRW* |
[1746]Further work revealed the following gonococcal DNA sequence <SEQ ID 863>:
[0000]| 1 | ATGGAAATTC GGGTAATAAA ATATACGGCA ACGGCTGCGT TGTTTGCATT | |
|
| 51 | TACGGTTGCA GGCTGCCGGC TGGCGGGGTG GTATGAGTGT TCGTCCTTGT |
|
| 101 | CCGGCTGGTG TAAGCCGAGA AAACCTGCCG CCATCGATTT TTGGGATATT |
|
| 151 | GGCGGCGAGA GtccgctGTC TTTAGAGGAC TACGAGATAC CGCTTTCAGA |
|
| 201 | CGGCAATCGT TCCGTCAGGG CAAACGAATA TGAATCCGCG CAAAAATCTT |
|
| 251 | ACTTTTATAG GAAAATAGGG AAGTTTGAAG CCTGCGGGTT GGATTGGCGT |
|
| 301 | ACGCGTGACG GCAAACCTTT GGTTGAGAGG TTCAAACAGG AAGGTTTCGA |
|
| 351 | CTGTTTGGAA AAGCAGGGGT TGCGGCGCAA CGGCCTGTCC GAGCGCGTCC |
|
| 401 | GATGGTAA |
[1747]This corresponds to the amino acid sequence <SEQ ID 864; ORF131ng-1>:
[0000]| 1 | MEIRVIKYTA TAALFAFTVA GCRLAGWYEC SSLSGWCKPR KPAAIDFWDI | |
|
| 51 | GGESPLSLED YEIPLSDGNR SVRANEYESA QKSYFYRKIG KFEACGLDWR |
|
| 101 | TRDGKPLVER FKQEGFDCLE KQGLRRNGLS ERVRW* |
[1748]ORF131ng-1 and ORF131-1 show 92.6% identity in 135 aa overlap:
[0000]
[1749]Based on the presence of a predicted prokaryotic membrane lipoprotein lipid attachment site, it is predicted that the proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 102
[1750]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 865>
[0000]| 1 | ATGAAACACA TCCATATTAT CGGTATCGGC GGCACGTTTA TGGGCGGGCT | |
|
| 51 | TGCCGCCATT GCCAAAGAAG CGGGGTTTGA AGTCAGCGGT TGCGACGCGA |
|
| 101 | AGATGTATCC GCCGATGAGC ACCCAGCTCG AAGCCTTGGG TATAGACGTG |
|
| 151 | TATGAAGGCT TCGATGCCGC TCAGTTGGAC GAATTTAAAG CCGACGTTTA |
|
| 201 | CGTTATCGGC AATGTCGCCA AGCGCGGGAT GGATGTGGTT GAAGCGATTT |
|
| 251 | TGAACCTCGG CCTGCCtTAT ATtTcCGGCC CGCAATGGCT GTCGGAAAAC |
|
| 301 | GTGCTGCACC ATCATTGGGT ACTCGGTGTG GCGGGGACgC ACGGCAAAAC |
|
| 351 | GACCACCGCC TCCATGCTCG CATGGGTCTT GGAATATgCC GGCCTCGCGC |
|
| 401 | CGGGCTTCCT TATtGGCGGC GTACC.GGAA AATttCGGCG TTTCCGCCCG |
|
| 451 | CCTGCCGCAA ACGCCGCGCC AAGACCCGAA CAGCCAATCG CCGTTTTTcG |
|
| 501 | TCATCGAAGC CGACGAATAC GACACCGCCT TTtTCGACAA ACGTTCTAAA |
|
| 551 | TtCGTGCATT ACCGTCCGCG TACCGCCGTG TTGAACAATC TGGAATTCGA |
|
| 601 | CCACGCCGAC ATCTTTGCCG ACTTGGGCGC GATACAGACc CAGTTCCACT |
|
| 651 | ACCTCGTGCG TACCGTGCCG TCTGAAGGCT TAATCGTCTG CAACGGACGG |
|
| 701 | CAGCAAAGCC TGCAAGATAC TTTGGACAAA GGCTGCTGGA CGCCGGTGGA |
|
| 751 | AAAATTCGGC ACGGAACACG GCTGGCA.. |
[1751]This corresponds to the amino acid sequence <SEQ ID 866; ORF132>:
[0000]| 1 | MKHIHIIGIG GTFMGGLAAI AKEAGFEVSG CDAKMYPPMS TQLEALGIDV | |
|
| 51 | YEGFDAAQLD EFKADVYVIG NVAKRGMDVV EAILNLGLPY ISGPQWLSEN |
|
| 101 | VLHHHWVLGV AGTHGKTTTA SMLAWVLEYA GLAPGFLIGG VXGKFRRFRP |
|
| 151 | PAANAAPRPE QPIAVFRHRS RRIRHRLFRQ TFXIRALPSA YRRVEQSGIR |
|
| 201 | PRRHLCRLGR DTDPVPLPRA YRAVXRLNRL QRTAAKPARY FGQRLLDAGG |
|
| 251 | KIRHGTRLA.. |
[1752]Further work revealed the complete nucleotide sequence <SEQ ID 867>:
[0000]| 1 | ATGAAACACA TCCATATTAT CGGTATCGGC GGCACGTTTA TGGGCGGGCT | |
|
| 51 | TGCCGCCATT GCCAAAGAAG CGGGGTTTGA AGTCAGCGGT TGCGACGCGA |
|
| 101 | AGATGTATCC GCCGATGAGC ACCCAGCTCG AAGCCTTGGG TATAGACGTG |
|
| 151 | TATGAAGGCT TCGATGCCGC TCAGTTGGAC GAATTTAAAG CCGACGTTTA |
|
| 201 | CGTTATCGGC AATGTCGCCA AGCGCGGGAT GGATGTGGTT GAAGCGATTT |
|
| 251 | TGAACCTCGG CCTGCCTTAT ATTTCCGGCC CGCAATGGCT GTCGGAAAAC |
|
| 301 | GTGCTGCACC ATCATTGGGT ACTCGGTGTG GCGGGGACGC ACGGCAAAAC |
|
| 351 | GACCACCGCC TCCATGCTCG CATGGGTCTT GGAATATGCC GGCCTCGCGC |
|
| 401 | CGGGCTTCCT TATTGGCGGC GTACCGGAAA ATTTCGGCGT TTCCGCCCGC |
|
| 451 | CTGCCGCAAA CGCCGCGCCA AGACCCGAAC AGCCAATCGC CGTTTTTCGT |
|
| 501 | CATCGAAGCC GACGAATACG ACACCGCCTT TTTCGACAAA CGTTCTAAAT |
|
| 551 | TCGTGCATTA CCGTCCGCGT ACCGCCGTGT TGAACAATCT GGAATTCGAC |
|
| 601 | CACGCCGACA TCTTTGCCGA CTTGGGCGCG ATACAGACCC AGTTCCACTA |
|
| 651 | CCTCGTGCGT ACCGTGCCGT CTGAAGGCTT AATCGTCTGC AACGGACGGC |
|
| 701 | AGCAAAGCCT GCAAGATACT TTGGACAAAG GCTGCTGGAC GCCGGTGGAA |
|
| 751 | AAATTCGGCA CGGAACACGG CTGGCAGGCC GGCGAAGCCA ATGCCGACGG |
|
| 801 | CTCGTTCGAC GTGTTGCTCG ACGGCAAAAC CGCCGGACGC GTCAAATGGG |
|
| 851 | ATTTGATGGG CAGGCACAAC CGCATGAACG CGCTCGCCGT CATTGCCGCC |
|
| 901 | GCGCGTCATG TCGGTGTCGA TATTCAGACC GCCTGCGAAG CCTTGGGCGC |
|
| 951 | GTTTAAAAAC GTCAAACGCC GGATGGAAAT CAAAGGCACG GCAAACGGCA |
|
| 1001 | TCACCGTTTA CGACGACTTC GCCCACCACC CGACCGCCAT CGAAACCACG |
|
| 1051 | ATTCAAGGTT TGCGCCAACG CGTCGGCGGC GCGCGCATCC TCGCCGTCCT |
|
| 1101 | CGAACCGCGT TCCAACACGA TGAAGCTGGG CACGATGAAG TCCGCCCTGC |
|
| 1151 | CTGTAAGCCT CAAAGAAGCC GACCAAGTGT TCTGCTACGC CGGCGGCGTG |
|
| 1201 | GACTGGGACG TCGCCGAAGC CCTCGCGCCT TTGGGCGGCA GGCTGAACGT |
|
| 1251 | CGGCAAAGAC TTCGATGCCT TCGTTGCCGA AATCGTGAAA AACGCCGAAG |
|
| 1301 | TAGGCGACCA TATTTTGGTG ATGAGCAACG GCGGTTTCGG CGGAATACAC |
|
| 1351 | GGAAAGCTGC TGGAAGCTTT GAGATAG |
[1753]This corresponds to the amino acid sequence <SEQ ID 868; ORF132-1>:
[0000]| 1 | MKHIHIIGIG GTFMGGLAAI AKEAGFEVSG CDAKMYPPMS TQLEALGIDV | |
|
| 51 | YEGFDAAQLD EFKADVYVIG NVAKRGMDVV EAILNLGLPY ISGPQWLSEN |
|
| 101 | VLHHHWVLGV AGTHGKTTTA SMLAWVLEYA GLAPGFLIGG VPENFGVSAR |
|
| 151 | LPQTPRQDPN SQSPFFVIEA DEYDTAFFDK RSKFVHYRPR TAVLNNLEFD |
|
| 201 | HADIFADLGA IQTQFHYLVR TVPSEGLIVC NGRQQSLQDT LDKGCWTPVE |
|
| 251 | KFGTEHGWQA GEANADGSFD VLLDGKTAGR VKWDLMGRHN RMNALAVIAA |
|
| 301 | ARHVGVDIQT ACEALGAFKN VKRRMEIKGT ANGITVYDDF AHHPTAIETT |
|
| 351 | IQGLRQRVGG ARILAVLEPR SNTMKLGTMK SALPVSLKEA DQVFCYAGGV |
|
| 401 | DWDVAEALAP LGGRLNVGKD FDAFVAEIVK NAEVGDHILV MSNGGFGGIH |
|
| 451 | GKLLEALR* |
[1754]Computer analysis of this amino acid sequence gave the following results:
[0000]Homology with the Hypothetical o457 Protein of E. coli (Accession Number U14003)
[1755]ORF132 and o457 show 58% aa identity in 140 aa overlap:
[0000]| Orf132: | 4 | IHIIGIGGTFMGGLAAIAKEAGFEVSGCDAKMYPPMSTQLEALGIDVYEGFDAAQLDEFK | 63 | |
| | IHI+GI GTFMGGLA +A++ G EV+G DA +YPPMST LE GI++ +G+DA+QL+ + |
| o457: | 3 | IHILGICGTFMGGLAMLARQLGHEVTGSDANVYPPMSTLLEKQGIELIQGYDASQLEP-Q | 61 |
|
| Orf132: | 64 | ADVYVIGNVAKRGMDVVEAILNLGLPYISGPQWLSENVLHHHWVLGVAGTHGKTTTASML | 123 |
| | D+ +IGN RG VEA+L +PY+SGPQWL + VL WVL VAGTHGKTTTA M |
| o457: | 62 | PDLVIIGNAMTRGNPCVEAVLEKNIPYMSGPQWLHDFVLRDRWVLAVAGTHGKTTTAGMA | 121 |
|
| Orf132: | 124 | AWVLEYAGLAPGFLIGGVXG | 143 |
| | W+LE G PGF+IGGV G |
| o457: | 122 | TWILEQCGYKPGFVIGGVPG | 141 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1756]ORF132 shows 74.6% identity over a 189aa overlap with an ORF (ORF132a) from strain A of N. meningitidis:
[0000]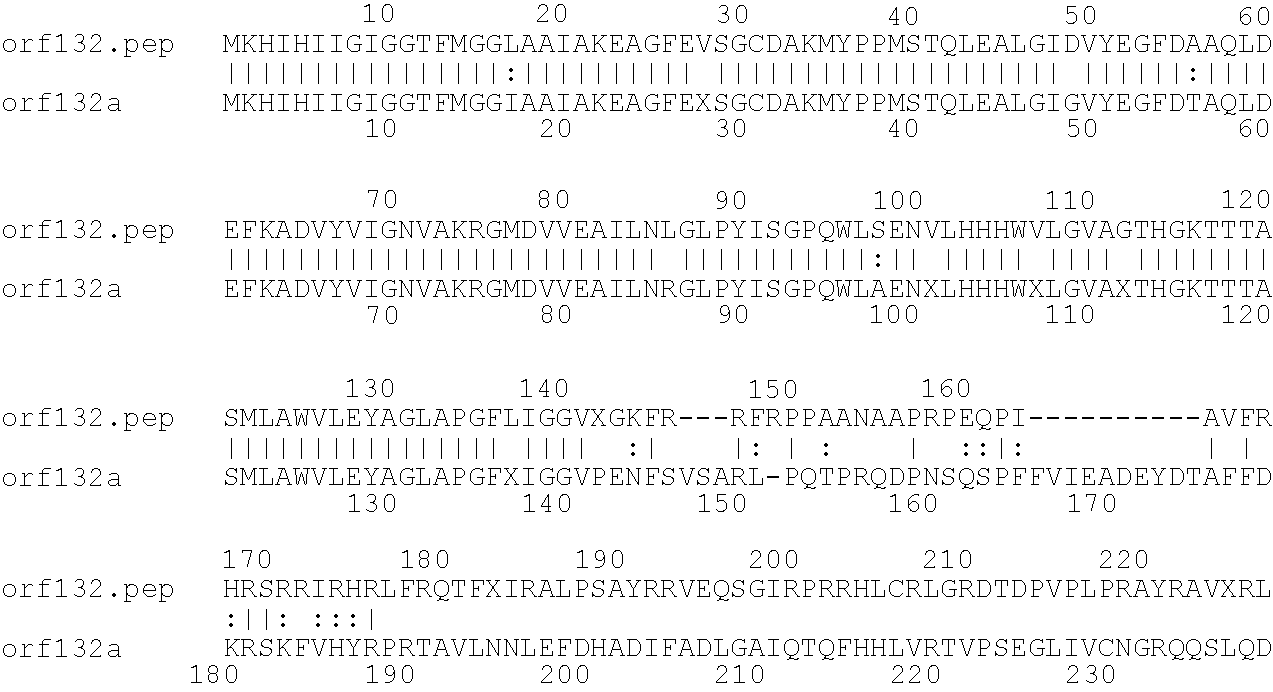
[1757]The complete length ORF132a nucleotide sequence <SEQ ID 869> is:
[0000]| 1 | ATGAAACACA TCCACATTAT CGGTATCGGC GGCACGTTTA TGGGTGGGAT | |
|
| 51 | TGCCGCCATT GCCAAAGAAG CAGGGTTTGA ANTCAGCGGT TGCGATGCGA |
|
| 101 | AGATGTATCC GCCGATGAGC ACCCAGCTCG AAGCCTTGGG CATAGGCGTG |
|
| 151 | TATGAAGGCT TCGACACCGC GCAGTTGGAC GAATTTAAAG CCGACGTTTA |
|
| 201 | CGTTATCGGC AATGTCGCCA AGCGCGGGAT GGATGTGGTT GAAGCGATTT |
|
| 251 | TGAACCGTGG GCTGCCTTAT ATTTCCGGCC CGCAATGGCT GGCTGAAAAC |
|
| 301 | NTGCTGCACC ATCATTGGNN ACTCGGCGTG GCGGNGACGC ACGGCAAAAC |
|
| 351 | GACCACCGCG TCTATGCTCG CGTGGGTTTT GGAATATGCC GGACTCGCAC |
|
| 401 | CGGGCTTCNT TATCGGCGGC GTACCGGAAA ACTTCAGCGT TTCCGCCCGC |
|
| 451 | CTGCCGCAAA CGCCGCGCCA AGACCCGAAC AGCCAATCGC CGTTTTTCGT |
|
| 501 | CATTGAAGCC GACGAATACG ACACCGCGTT TTTCGACAAA CGCTCCAAAT |
|
| 551 | TCGTGCATTA CCGTCCGCGT ACCGCCGTGT TGAACAATCT GGAATTCGAC |
|
| 601 | CACGCCGACA TCTTCGCCGA TTTGGGCGCG ATACAGACCC AGTTCCACCA |
|
| 651 | CCTCGTGCGT ACCGTGCCGT CTGAAGGCCT CATCGTCTGC AACGGACGGC |
|
| 701 | AGCAAAGCCT GCAAGACACT TTGGACAAAG GCTGCTGGAC GCCGGTGGAA |
|
| 751 | AAATTCGGCA CGGAACACGG CTGGCAGGCC GGCGAAGCCA ATGCCGATGG |
|
| 801 | CTCGTTCGAC GTGTTGCTTG ACGGCAAAAA AGCCGGACAC GTCGCTTGGA |
|
| 851 | GTTTGATGGG CGGACACAAC CGCATGAACG CGCTCGCNGT CATCGCCGCC |
|
| 901 | GCGCGTCATG CCGGAGTNGA CATTCAGACG GCCTGCGAAG CCTTGAGCAC |
|
| 951 | GTTTAAAAAC GTCAAACGCC GCATGGAAAT CAAAGGCACG GCAAACGGTA |
|
| 1001 | TCACCGTTTA CGACGACTTC GCCCACCATC CGACCGCTAT CGAAACCACG |
|
| 1051 | ATTCAAGGTT TGCGCCAGCG CGTCGGCGGC GCGCGCATCC TCGCCGTCCT |
|
| 1101 | CGAACCGCGT TCCAATACGA TGAAGCTGGG TACGATGAAA GCCGCCCTGC |
|
| 1151 | CCGCAAGCCT CAAAGAAGCC GACCAAGTGT TCTGNTACGC CGGCGGCGCG |
|
| 1201 | GACTGGGACG TTGCCGAAGC CCTCGCGCCT TTGGGCGGCA GGCTGCACGT |
|
| 1251 | CGGCAAAGAC TTCGATGCCT TCGTTGCCGA AATCGTGAAA AACGCCGAAG |
|
| 1301 | CAGGCGACCA TATTTTGGTG ATGAGCAACG GCGGTTTCGG CGGAATACAC |
|
| 1351 | ACCAAACTGC TGGACGCTTT GAGATAG |
[1758]This encodes a protein having amino acid sequence <SEQ ID 870>:
[0000]| 1 | MKHIHIIGIG GTFMGGIAAI AKEAGFEXSG CDAKMYPPMS TQLEALGIGV | |
|
| 51 | YEGFDTAQLD EFKADVYVIG NVAKRGMDVV EAILNRGLPY ISGPQWLAEN |
|
| 101 | XLHHHWXLGV AXTHGKTTTA SMLAWVLEYA GLAPGFXIGG VPENFSVSAR |
|
| 151 | LPQTPRQDPN SQSPFFVIEA DEYDTAFFDK RSKFVHYRPR TAVLNNLEFD |
|
| 201 | HADIFADLGA IQTQFHHLVR TVPSEGLIVC NGRQQSLQDT LDKGCWTPVE |
|
| 251 | KFGTEHGWQA GEANADGSFD VLLDGKKAGH VAWSLMGGHN RMNALAVIAA |
|
| 301 | ARHAGVDIQT ACEALSTFKN VKRRMEIKGT ANGITVYDDF AHHPTAIETT |
|
| 351 | IQGLRQRVGG ARILAVLEPR SNTMKLGTMK AALPASLKEA DQVFXYAGGA |
|
| 401 | DWDVAEALAP LGGRLHVGKD FDAFVAEIVK NAEAGDHILV MSNGGFGGIH |
|
| 451 | TKLLDALR* |
[1759]ORF132a and ORF132-1 show 93.9% identity in 458 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1760]ORF132 shows 89.6% identity over 259 aa overlap with a predicted ORF (ORF132ng) from N. gonorrhoeae:
[0000]
[1761]An ORF132ng nucleotide sequence <SEQ ID 871> was predicted to encode a protein having amino acid sequence <SEQ ID 872>:
[0000]| 1 | MKHIHIIGIG GTFMGGIAAI AKEAGFKVSG CDAKMYPPMS TQLEALGIGV | |
|
| 51 | HEGFDAAQLE EFQADIYVIG NVARRGMDVV EAILNRGLPY ISGPQWLAEN |
|
| 101 | VLHHHWVLGV AGTHGKTTTA SMLAWVLEYA GLAPGFLIGG VPGKFRRFRP |
|
| 151 | PTANAASRPE QQIAVFRHRS RRIRHRLFRQ TLQIRALSPA YRRVEQSGIR |
|
| 201 | PRRHLRRLGR DTDPVPPPRA HRTIRRPHRL QRTAAKPARY FGQRLLDAGG |
|
| 251 | KIRHRTRLAD W* |
[1762]Further work revealed the following gonococcal DNA sequence <SEQ ID 873>:
[0000]| 1 | ATGAAACACA TCCACATTAT CGGTATCGGC GGCACGTTTA TGGGCGGGAT | |
|
| 51 | TGCCGCCATT GCCAAAGAAG CCGGGTTCAA AGTCAGCGGT TGCGACGCGA |
|
| 101 | AGATGTATCC GCCGATGAGC ACCCAGCTCG AAGCCTTGGG CATAGGCGTA |
|
| 151 | CACGAAGGCT TCGATGCCGC GCAGTTGGAA GAATTTCAAG CCGATATTTA |
|
| 201 | CGTCATCGGC AATGTCGCCA GGCGCGGGAT GGATGTGGTC GAGGCGATTT |
|
| 251 | TGAACCGTGG GCTGCCTTAT ATTTCCGGCC CGCAATGGCT GGCTGAAAac |
|
| 301 | GTGCtgcacc atcaTTGGgt ACTCGGCGTG GcagggaCGC ACGGcaaAac |
|
| 351 | gaccaCcGcg tCCATGCTCG CCTGGGTCTT GGAATATGCC GGACTCGCGC |
|
| 401 | CGGGCTTCCT CATCGGCGGt gtaccggaAA ATTTCGGCGT TTCCGCCCGC |
|
| 451 | CTACCGCAAA CGCCGCGTCA AGACCCGAAC AGCAAATCGC CGTTTTTCGT |
|
| 501 | CATCGAAGCC GACGAATACG ACACCGCCTT TTTCGACAAA CGCTCCAAAT |
|
| 551 | TCGTGCATTA TCGCCCGCGT ACCGCCGTGT TGAACAATCT GGAATTCGAC |
|
| 601 | CACGCCGACA TCTTCGCCGA CTTGGGCGCG ATACAGACCC AGTTCCACCA |
|
| 651 | CCTCGTGCGC ACCGTACCAT CCGAAGGCCT CATCGTCTGC AACGGACAGC |
|
| 701 | AGCAAAGCCT GCAAGATACT TTGGACAAAG GCTGCTGGAC GCCGGTGGAA |
|
| 751 | AAATTCGGCA CCGGACACGG CTGGCAGATT GGTGAAGTCA ATGCCGACGG |
|
| 801 | CTCGTTCGAC GTATTGCTTG ACGGCAAAAA AGCCGGACAC GTCGCATGGG |
|
| 851 | ATTTGATGGG CGGACACAAC CGCATGAACG CGCTCGCCGT CATCGCTGCC |
|
| 901 | GCACGCCATG CCGGAGTCGA TGTTCAGACG GCCTGCGAAG CCTTGGGTGC |
|
| 951 | GTTTAAAAAC GTCAAACGCC GCATGGAAAT CAAAGGCACG GCAAACGGCA |
|
| 1001 | TCACCGTTTA CGACGATTTC GCCCACCACC CGACCGCCAT CGAAACCACG |
|
| 1051 | ATTCAAGGTT TGCGCCAACG TGTCGGCGGC GCGCGCATCC TCGCCGTCCT |
|
| 1101 | CGAGCCGCGT TCCAACACCA TGAAACTCGG CACGATGAAG TCCGCCCTGC |
|
| 1151 | CCGCAAGCCT CAAAGAAGCC GACCAAGTGT TCTGCTACGC CGGCGGCGCG |
|
| 1201 | GACTGGGACG TTGCCGAAGC CCTCGCGCCT TTGGGCTGCA GGCTGCGCGT |
|
| 1251 | CGGTAAAGAT TTCGATACCT TCGTTGCCGA AATTGTGAAA AACGCCCGAA |
|
| 1301 | CCGGCGACCA TATTTTGGTG ATGAGCAACG GCGGTTTCGG CGGAATACAC |
|
| 1351 | ACCAAACTGC TGGACGCTTT GAGATAG |
[1763]This corresponds to the amino acid sequence <SEQ ID 874; ORF132ng-1>:
[0000]| 1 | MKHIHIIGIG GTFMGGIAAI AKEAGFKVSG CDAKMYPPMS TQLEALGIGV | |
|
| 51 | HEGFDAAQLE EFQADIYVIG NVARRGMDVV EAILNRGLPY ISGPQWLAEN |
|
| 101 | VLHHHWVLGV AGTHGKTTTA SMLAWVLEYA GLAPGFLIGG VPENFGVSAR |
|
| 151 | LPQTPRQDPN SKSPFFVIEA DEYDTAFFDK RSKFVHYRPR TAVLNNLEFD |
|
| 201 | HADIFADLGA IQTQFHHLVR TVPSEGLIVC NGQQQSLQDT LDKGCWTPVE |
|
| 251 | KFGTGHGWQI GEVNADGSFD VLLDGKKAGH VAWDLMGGHN RMNALAVIAA |
|
| 301 | ARHAGVDVQT ACEALGAFKN VKRRMEIKGT ANGITVYDDF AHHPTAIETT |
|
| 351 | IQGLRQRVGG ARILAVLEPR SNTMKLGTMK SALPASLKEA DQVFCYAGGA |
|
| 401 | DWDVAEALAP LGCRLRVGKD FDTFVAEIVK NARTGDHILV MSNGGFGGIH |
|
| 451 | TKLLDALR* |
[1764]ORF132ng-1 and ORF132-1 show 93.2% identity in 458 aa overlap:
[0000]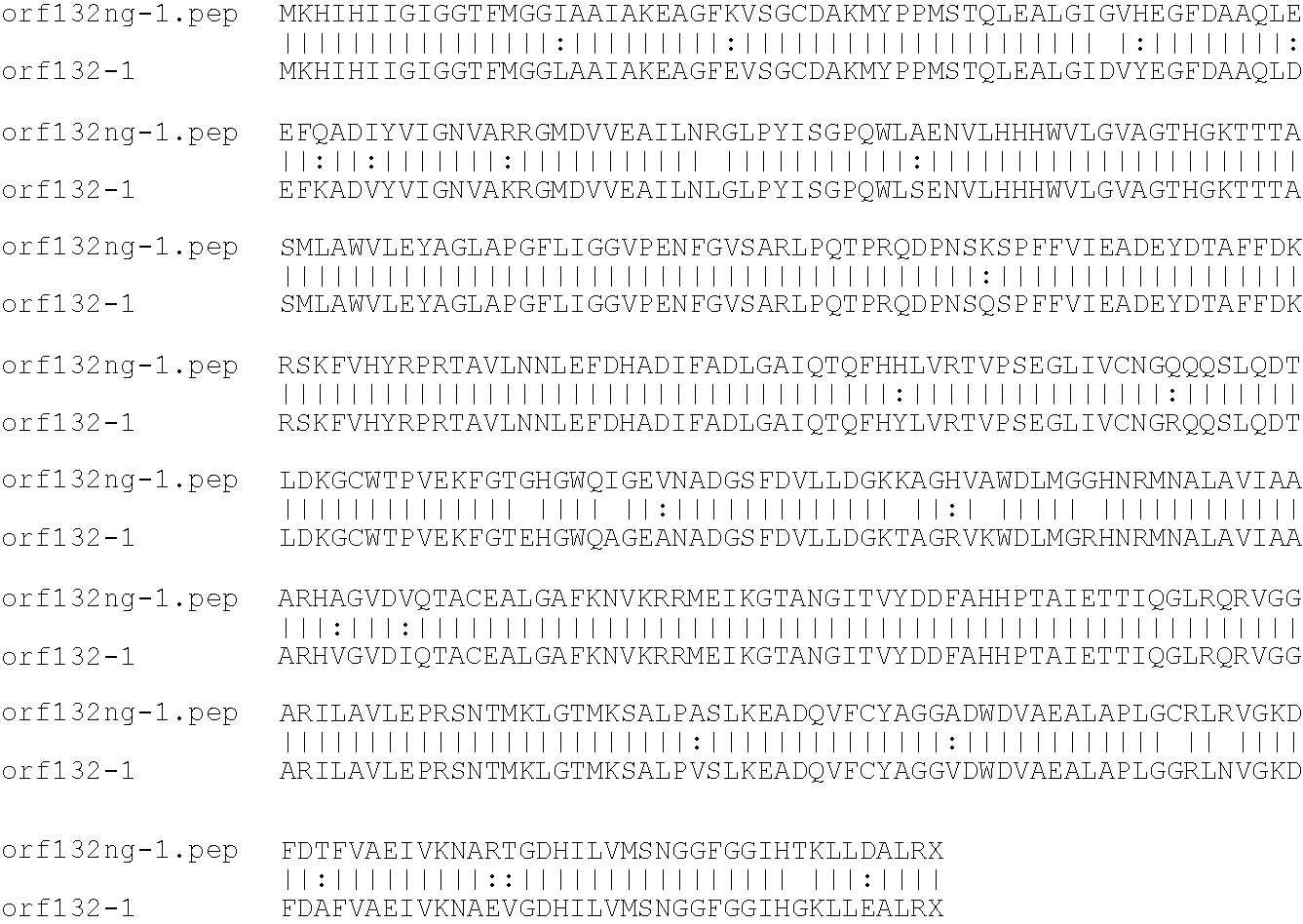
[1765]In addition, ORF132ng-1 is homologous to a hypothetical E. coli protein:
[0000]| pir||S56459 hypothetical protein o457 - Escherichia coli >gi|537075 | |
| (U14003) ORF_o457 [Escherichia coli] >gi|1790680 (AE000494), |
| hypothetical 48.5 kD protein in fbp-pmba intergenic region |
| [Escherichia coli] Length = 457 Score = 474 bits (1207), Expect = e−133 |
| Identities = 249/439 (56%), Positives = 294/439 (66%), Gaps = 13/439 (2%) |
|
| Query: | 22 | KEAGFKVSGCDAKMYPPMSTQLEALGIGVHEGFDAAQLEEFQADIYVIGNVARRGMDVVE | 81 | |
| | ++ G +V+G DA +YPPMST LE GI + +G+DA+QLE Q D+ +IGN RG VE |
| Sbjct: | 21 | RQLGHEVTGSDANVYPPMSTLLEKQGIELIQGYDASQLEP-QPDLVIIGNAMTRGNPCVE | 79 |
|
| Query: | 82 | AILNRGLPYISGPQWLAENVLHHHWVLGVAGTHGKTTTASMLAWVLEYAGLAPGFLIGGV | 141 |
| | A+L + +PY+SGPQWL + VL WVL VAGTHGKTTTA M W+LE G PGF+IGGV |
| Sbjct: | 80 | AVLEKNIPYMSGPQWLHDFVLADRWVLAVAGTHGKTTTAGMATWILEQCGYKPGFVIGGV | 139 |
|
| Query: | 142 | PENFGVSARLPQTPRQDPNSKSPFFVIEADEYDTAFFDKRSKFVHYRPRTAVLNNLEFDH | 201 |
| | P NF VSA L +S FFVIEADEYD AFFDKRSKFVHY PRT +LNNLEFDH |
| Sbjct: | 140 | PGNFEVSAHL---------GESDFFVIEADEYDCAFFDKRSKFVHYCPRTLILNNLEFDH | 190 |
|
| Query: | 202 | ADIFADLGAIQTQFHHLVRTVPSEGLIVCNGQQQSLQDTLDKGCWTPVEKFGTGHGWQIG | 261 |
| | ADIF DL AIQ QFHHLVR VP +G I+ +L+ T+ GCW+ E G WQ |
| Sbjct: | 191 | ADIFDDLKAIQKQFHHLVRIVPGOGRIIWPENDINLKQTMANGCWSEQELVGEQGHWQAK | 250 |
|
| Query: | 262 | EVNADGS-FDVLLDGKKAGHVAWDLMGGHNRMNALAVIAAARHAGVDVQTACEALGAFKN | 320 |
| | ++ D S ++VLLDG+K G V W L+G HN N L IAAARH GV A ALG+F N |
| Sbjct: | 251 | KLTTDASEWEVLLDGEKVGEVKWSLVGEHNMHNGLMAIAAARHVGVAPADAANALGSFIN | 310 |
|
| Query: | 321 | VKRRMEIKGTANGITVYDDFAHHPTAIETTIQGLRQRVGG-ARILAVLEPRSNTMKLGTM | 379 |
| | +RR+E++G ANG+TVYDDFAHHPTAI T+ LR +VGG ARI+AVLEPRSNTMK+G |
| Sbjct: | 311 | ARRRLELRGEANGVTVYDDFAHHPTAILATLAALRGKVGGTARIIAVLEPRSNTMKMGIC | 370 |
|
| Query: | 380 | KSALPASLKEADQVF-CYAGGADWDVAEALAPLGCRLRVGKDFDTFVAEIVKNARTGDHI | 438 |
| | K L SL AD+VF W VAE D DT +VK A+ GDHI |
| Sbjct: | 371 | KDDLAPSLGRADEVFLLQPAHIPWQVAEVAEACVQPAHWSGDVDTLADMVVKTAQPGDHI | 430 |
|
| Query: | 439 | LVMSNGGFGGIHTKLLDAL | 457 |
| | LVMSNGGFGGIH KLLD L |
| Sbjct: | 431 | LVMSNGGFGGIHQKLLDGL | 449 |
[1766]Based on this analysis, it was predicted that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1767]ORF132-1 (26.4 kDa) was cloned in pET and pGex vectors and expressed in E. coli, as described above. The products of protein expression and purification were analyzed by SDS-PAGE. FIG. 20A shows the results of affinity purification of the His-fusion protein, and FIG. 20B shows the results of expression of the GST-fusion in E. coli. Purified His-fusion protein was used to immunise mice, whose sera were used for FACS analysis (FIG. 20C) and ELISA (positive result). These experiments confirm that ORF132 is a surface-exposed protein, and that it is a useful immunogen.
Example 103
[1768]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 875>
[0000]| 1 | ..CCGGGCTATT ACGGCTCGGA TGACGAATTT AAGCGGGCAT TCGGAGAAAA | |
|
| 51 | CTCGCCGACA TmCAAGAAAC ATTGCAACCG GAGCTGCGGG ATTTATGAAC |
|
| 101 | CCGTATTGAA AAAATACGGC AAAAAGCGCG CCAACAACCA TTCGGTCAGC |
|
| 151 | ATTAGTGCGG ACTTCGGCGA TTATTTCATG CCGTTCGCCA GCTATTCGCG |
|
| 201 | CACACACCGT ATGCCCAACA TCCAAGAAAT GTATTTTTCC CAAATCGGCG |
|
| 251 | ACTCCGGCGT TCACACCGCC TTAAAACCAG AGCGCGCAAA CACTTGGCAA |
|
| 301 | TTTGGCTTCr ATACCTATAA AAAAGGATTG TTAAAACAAG ATGATACATT |
|
| 351 | AGGATTAAAA CTGGTCGGCT ACCGCAGCCG CATCGACAAC TACATCCACA |
|
| 401 | ACGTTTACGG GAAATGGTGG GATTTGAACG GGGATATTCC GAGCTGGGTC |
|
| 451 | AGCAGCACCG GGCTTGCCTA CACCATCCAA CATCGCrATT TCAwAGACAA |
|
| 501 | AGTGCATCAA nnnnnnnnnn nnnnnnnnnn nnnnTACGAT TATGGGCGTT |
|
| 551 | TTTTCACCAA CCTTTCTTAC GCCTATCAAA AAAGCACGCA ACCGACCAAC |
|
| 601 | TTCAGCGATG CGAGCGAATC GCCCAACAAT GCGTCCAAAG AAGACCAACT |
|
| 651 | CAAACAAGGT TATGGGTTGA GCAGGGTTTC CGCCCTGCCG CGAGATTACG |
|
| 701 | GACGTTTGGA AGTCGGTACG CGCTGGTTGG GCAACAAACT GACTTTGGGC |
|
| 751 | GGCGCGATGC GCTATTTCGG CAAGAGCATC CGCGCGACGG CTGAAGAACG |
|
| 801 | CTATATCGAC GGCACCAACG GGGGAAATAC CAGCAATTTC CGGCAACTGG |
|
| 851 | GCAAGCGTTC CATCAAACAA ACCGAAACTC TTGCCCGCCA GCCTTTGATT |
|
| 901 | TTwGATTTTa ACGCCGCTTA CGAGCCGAAG AAAAACCTTA TTTTCCGCGC |
|
| 951 | CGAAGTCAAA AATCTGTTCG ACAGGCGTTA TATCGATCCG CTCGATGCGG |
|
| 1001 | GCAATGATGC GGCAAC.GAG CGTTATTACA GCTCGTTCGA CCCGAAAGAC |
|
| 1051 | AAGGACrrAG ACGTAACGTG TAATGCTGAT AAAACGTTGT GCaACGGCAA |
|
| 1101 | ATACGGCGGC ACAAGCAAAA GCGTATTGAC CAATTTTGCA CGCGGACGCA |
|
| 1151 | CCTTTTTgAT GACGATGAGC TACAAGTTTT AA |
[1769]This corresponds to the amino acid sequence <SEQ ID 876; ORF133>:
[0000]| 1 | ..PGYYGSDDEF KRAFGENSPT XKKHCNRSCG IYEPVLKKYG KKRANNHSVS | |
|
| 51 | ISADFGDYFM PFASYSRTHR MPNIQEMYFS QIGDSGVHTA LKPERANTWQ |
|
| 101 | FGFXTYKKGL LKQDDTLGLK LVGYRSRIDN YIHNVYGKWW DLNGDIPSWV |
|
| 151 | SSTGLAYTIQ HRXFXDKVHQ XXXXXXXXYD YGRFFTNLSY AYQKSTQPTN |
|
| 201 | FSDASESPNN ASKEDQLKQG YGLSRVSALP RDYGRLEVGT RWLGNKLTLG |
|
| 251 | GAMRYFGKSI RATAEERYID GTNGGNTSNF RQLGKRSIKQ TETLARQPLI |
|
| 301 | XDFNAAYEPK KNLIFRAEVK NLFDRRYIDP LDAGNDAAXE RYYSSFDPKD |
|
| 351 | KDXDVTCNAD KTLCNGKYGG TSKSVLTNFA RGRTFLMTMS YKF* |
[1770]Further work revealed the further partial DNA sequence <SEQ ID 877>:
[0000]| 1 | GAGGCGCAGA TACAGGTTTT GGAAGATGTG CACGTCAAGG CGAAGCGCGT | |
|
| 51 | ACCGAAAGAC AAAAAAGTGT TTACCGATGC GCGTGCCGTA TCGACCCGTC |
|
| 101 | AGGATATATT CAAATCCAGC GAAAACCTCG ACAACATCGT ACGCAGCATC |
|
| 151 | CCCGGTGCGT TTACACAGCA AGATAAAAGC TCGGGCATTG TGTCTTTGAA |
|
| 201 | TATTCGCGGC GACAGCGGGT TCGGGCGGGT CAATACGATG GTGGACGGCA |
|
| 251 | TCACGCAGAC CTTTTATTCG ACTTCTACCG ATGCGGGCAG GGCAGGCGGT |
|
| 301 | TCATCTCAAT TCGGTGCATC TGTCGACAGC AATTTTATTG CCGGACTGGA |
|
| 351 | TGTCGTCAAA GGCAGCTTCA GCGGCTCGGC AGGCATCAAC AGCCTTGCCG |
|
| 401 | GTTCGGCGAA TCTGCGGACT TTAGGCGTGG ATGACGTCGT TCAGGGCAAT |
|
| 451 | AATACCTACG GCCTGCTGCT AAAAGGTCTG ACCGGCACCA ATTCAACCAA |
|
| 501 | AGGTAATGCG ATGGCGGCGA TAGGTGCGCG CAAATGGCTG GAAAGCGGAG |
|
| 551 | CATCTGTCGG TGTGCTTTAC GGGCACAGCA GGCGCAGCGT GGCGCAAAAT |
|
| 601 | TACCGCGTGG GCGGCGGCGG GCAGCACATC GGAAATTTTG GCGCGGAATA |
|
| 651 | TTTGGAACGG CGCAAGCAGC GATATTTTGT ACAAGAGGGT GCTTTGAAAT |
|
| 701 | TCAATTCCGA CAGCGGAAAA TGGGAGCGGG ATTTACAAAG GCAACAGTGG |
|
| 751 | AAATACAAGC CGTATAAAAA TTACAACAAC CAAGAACTAC AaAAATACAT |
|
| 801 | CGAAGAGCAT GACAAAAGCT GGCGGGAAAA CCTg.CaCCG CAATACGACA |
|
| 851 | TTACCCCCAT CGATCCGTCC AGCCTGAAGC AGCAGTCGGC AGGCAATCTG |
|
| 901 | TTTAAATTGG AATACGACGG CGTATTCAAT AAATACACGG CGCAATTTCG |
|
| 951 | CGATTTAAAC ACCAAAATCG GCAGCCGCAA AATCATCAAC CGCAATTATC |
|
| 1001 | AGTTCAATTA CGGTTTGTCT TTGAACCCGT ATACCAACCT CAATCTGACC |
|
| 1051 | GCAGCCTACA ATTCGGGCAG GCAGAAATAT CCGAAAGGGT CGAAGTTTAC |
|
| 1101 | AGGCTGGGGG CTTTTAAAGG ATTTTGAAAC CTACAACAAC GCGAAAATCC |
|
| 1151 | TCGACCTCAA CAACACCGCC ACCTTCCGGC TGCCCCGCGA AACCGAGTTG |
|
| 1201 | CAAACCACTT TGGGCTTCAA TTATTTCCAC AACGAATACG GCAAAAACCG |
|
| 1251 | CTTTCCTGAA GAATTGGGGC TGTTTTTCGA CGGTCCTGAT CAGGACAACG |
|
| 1301 | GGCTTTATTC CTATTTGGGG CGGTTTAAGG GCGATAAAGG GCTGCTGCCC |
|
| 1351 | CAAAAATCAA CCATTGTCCA ACCGGCCGGC AGCCAATATT TCAACACGTT |
|
| 1401 | CTACTTCGAT GCCGCGCTCA AAAAAGACAT TTACCGCTTA AACTACAGCA |
|
| 1451 | CCAATACCGT CGGCTACCGT TTCGGCGGCG AATATACGGG CTATTACGGC |
|
| 1501 | TCGGATGACG AATTTAAGCG GGCATTCGGA GAAAACTCGC CGACATACAA |
|
| 1551 | GAAACATTGC AACCGGAGCT GCGGGATTTA TGAACCCGTA TTGAAAAAAT |
|
| 1601 | ACGGCAAAAA GCGCGCCAAC AACCATTCGG TCAGCATTAG TGCGGACTTC |
|
| 1651 | GGCGATTATT TCATGCCGTT CGCCAGCTAT TCGCGCACAC ACCGTATGCC |
|
| 1701 | CAACATCCAA GAAATGTATT TTTCCCAAAT CGGCGACTCC GGCGTTCACA |
|
| 1751 | CCGCCTTAAA ACCAGAGCGC GCAAACACTT GGCAATTTGG CTTCAATACC |
|
| 1801 | TATAAAAAAG GATTGTTAAA ACAAGATGAT ACATTAGGAT TAAAACTGGT |
|
| 1851 | CGGCTACCGC AGCCGCATCG ACAACTACAT CCACAACGTT TACGGGAAAT |
|
| 1901 | GGTGGGATTT GAACGGGGAT ATTCCGAGCT GGGTCAGCAG CACCGGGCTT |
|
| 1951 | GCCTACACCA TCCAACATCG CAATTTCAAA GACAAAGTGC ACAAACACGG |
|
| 2001 | TTTTGAGTTG GAGCTGAATT ACGATTATGG GCGTTTTTTC ACCAACCTTT |
|
| 2051 | CTTACGCCTA TCAAAAAAGC ACGCAACCGA CCAACTTCAG CGATGCGAGC |
|
| 2101 | GAATCGCCCA ACAATGCGTC CAAAGAAGAC CAACTCAAAC AAGGTTATGG |
|
| 2151 | GTTGAGCAGG GTTTCCGCCC TGCCGCGAGA TTACGGACGT TTGGAAGTCG |
|
| 2201 | GTACGCGCTG GTTGGGCAAC AAACTGACTT TGGGCGGCGC GATGCGCTAT |
|
| 2251 | TTCGGCAAGA GCATCCGCGC GACGGCTGAA GAACGCTATA TCGACGGCAC |
|
| 2301 | CAACGGGGGA AATACCAGCA ATTTCCGGCA ACTGGGCAAG CGTTCCATCA |
|
| 2351 | AACAAACCGA AACTCTTGCC CGCCAGCCTT TGATTTTTGA TTTTTACGCC |
|
| 2401 | GCTTACGAGC CGAAGAAAAA CCTTATTTTC CGCGCCGAAG TCAAAAATCT |
|
| 2451 | GTTCGACAGG CGTTATATCG ATCCGCTCGA TGCGGGCAAT GATGCGGCAA |
|
| 2501 | CGCAGCGTTA TTACAGCTCG TTCGACCCGA AAGACAAGGA CGAAGACGTA |
|
| 2551 | ACGTGTAATG CTGATAAAAC GTTGTGCAAC GGCAAATACG GCGGCACAAG |
|
| 2601 | CAAAAGCGTA TTGACCAATT TTGCACGCGG ACGCACCTTT TTGATGACGA |
|
| 2651 | TGAGCTACAA GTTTTAA |
[1771]This corresponds to the amino acid sequence <SEQ ID 878; ORF133-1>:
[0000]| 1 | EAQIQVLEDV HVKAKRVPKD KKVFTDARAV STRQDIFKSS ENLDNIVRSI | |
|
| 51 | PGAFTQQDKS SGIVSLNIRG DSGFGRVNTM VDGITQTFYS TSTDAGRAGG |
|
| 101 | SSQFGASVDS NFIAGLDVVK GSFSGSAGIN SLAGSANLRT LGVDDVVQGN |
|
| 151 | NTYGLLLKGL TGTNSTKGNA MAAIGARKWL ESGASVGVLY GHSRRSVAQN |
|
| 201 | YRVGGGGQHI GNFGAEYLER RKQRYFVQEG ALKFNSDSGK WERDLQRQQW |
|
| 251 | KYKPYKNYNN QELQKYIEEH DKSWRENLXP QYDITPIDPS SLKQQSAGNL |
|
| 301 | FKLEYDGVFN KYTAQFRDLN TKIGSRKIIN RNYQFNYGLS LNPYTNLNLT |
|
| 351 | AAYNSGRQKY PKGSKFTGWG LLKDFETYNN AKILDLNNTA TFRLPRETEL |
|
| 401 | QTTLGFNYFH NEYGKNRFPE ELGLFFDGPD QDNGLYSYLG RFKGDKGLLP |
|
| 451 | QKSTIVQPAG SQYFNTFYFD AALKKDIYRL NYSTNTVGYR FGGEYTGYYG |
|
| 501 | SDDEFKRAFG ENSPTYKKHC NRSCGIYEPV LKKYGKKRAN NHSVSISADF |
|
| 551 | GDYFMPFASY SRTHRMPNIQ EMYFSQIGDS GVHTALKPER ANTWQFGFNT |
|
| 601 | YKKGLLKQDD TLGLKLVGYR SRIDNYIHNV YGKWWDLNGD IPSWVSSTGL |
|
| 651 | AYTIQHRNFK DKVHKHGFEL ELNYDYGRFF TNLSYAYQKS TQPTNFSDAS |
|
| 701 | ESPNNASKED QLKQGYGLSR VSALPRDYGR LEVGTRWLGN KLTLGGAMRY |
|
| 751 | FGKSIRATAE ERYIDGTNGG NTSNFRQLGK RSIKQTETLA RQPLIFDFYA |
|
| 801 | AYEPKKNLIF RAEVKNLFDR RYIDPLDAGN DAATQRYYSS FDPKDKDEDV |
|
| 851 | TCNADKTLCN GKYGGTSKSV LTNFARGRTF LMTMSYKF* |
[1772]Computer analysis of this amino acid sequence gave the following results:
[1773]Homology with the Probable TonB-Dependent Receptor HI121 of H. influenzae (Accession Number U32801)
[1774]ORF133 and HI121 show 57% aa identity in 363aa overlap:
[0000]| Orf133: | 31 | IYEPVLKKYGKKRANNHSVSISADFGDYFMPFASYSRTHRMPNIQEMYFSQIGDSGVHTA | 90 | |
| | I EP+L K G K+A NHS ++SA+ DYFMPF +YSRTHRMPNIQEM+FSQ+ ++GV+TA |
| HI121: | 563 | INEPILHKSGHKKAFNHSATLSAELSDYFMPFFTYSRTHRMPNIQEMFFSQVSNAGVNTA | 622 |
|
| Orf133: | 91 | LKPERANTWQFGFXTYKKGLLKQDDTLGLKLVGYRSRIDNYIHNVYGKWWDLNGDIPSWV | 150 |
| | LKPE+++T+Q GF TYKKGL QDD LG+KLVGYRS I NYIHNVYG WW +P+W |
| HI121: | 623 | LKPEQSDTYQLGFNTYKKGLFTQDDVLGVKLVGYRSFIKNYIHNVYGVWW--RDGMPTWA | 680 |
|
| Orf133: | 151 | SSTGLAYTIQHRXFXDKVHXXXXXXXXXYDYGRFFTNLSYAYQKSTQPTNFSDASESPNN | 210 |
| | S G YTI H+ + V YD GRFF N+SYAYQ++ QPTN++DAS PNN |
| HI121: | 681 | ESNGFKYTIAHQNYKPIVKKSGVELEINYDMGRFFANVSYAYQRTNQPTNYADASPRPNN | 740 |
|
| Orf133: | 211 | ASKEDQLKQGYGLSRVSALPRDYGRLEVGTRWLGNKLTLGGAMRYFGKSIRATAEERYID | 270 |
| | AS+ED LKQGYGLSRVS LP+DYGRLE+GTRW KLTLG A RY+GKS RAT EE YI+ |
| HI121: | 741 | ASQEDILKQGYGLSRVSMLPKDYGRLELGTRWFDQKLTLGLAARYYGKSKRATIEEEYIN | 800 |
|
| Orf133: | 271 | GTNGGNTSNFRQLGKRSIKQTETLARQPLIXDFNAAYEPKKNLIFRAEVKNLFDRRYIDP | 330 |
| | G+ + R+ ++K+TE + +QP+I D + +YEP K+LI +AEV+NL D+RY+DP |
| HI121: | 801 | GSR-FKKNTLRRENYYAVKKTEDIKKQPIILDLHVSYEPIKDLIIKAEVQNLLDKRYVDP | 859 |
|
| Orf133: | 331 | LDAGNDAAXERYYSSFDPKDKDXDVTCNADKTLCNGKYGGTSKSVLTNFARGRTFLMTMS | 390 |
| | LDAGNDAA +RYYSS + + C D + C GG+ K+VL NFARGRT++++++ |
| HI121: | 860 | LDAGNDAASQRYYSSL-----NNSIECAQDSSAC----GGSDKTVLYNFARGRTYILSLN | 910 |
|
| Orf133: | 391 | YKF | 393 |
| | YKF |
| HI121: | 911 | YKF | 913 |
Homology with a Predicted ORF from
N. meningitidis (Strain A)
[1775]ORF133 shows 90.8% identity over a 392aa overlap with an ORF (ORF133a) from strain A of N. meningitidis:
[0000]
[1776]A partial ORF133a nucleotide sequence <SEQ ID 879> is:
[0000]| 1 | AAAGACAAAA AAGTGTTTAC CGATGCGCGT GCCGTATCGA CCCGTCAGGA | |
|
| 51 | TATATTCAAA TCCANCGAAA ACCTCGACAA CATCGTACGC ANCATCCCCG |
|
| 101 | GTGCGTTTAC ACANCAANAT AAAAGCTCGG GCNTTGTGTC TTTGAATATT |
|
| 151 | CGCNGCGACA GCGGGTTCGG GCGGGTCAAT ACNATGGTNG ACGGCATCAC |
|
| 201 | NCANACCTTT TATTCGACTT CTACCGATGC GGGCAGGGCA GGCGGTTCAT |
|
| 251 | CTCAATTCGG TGCATCTGTC GACAGCAATT TTATNGCCGG ACTGGATGTC |
|
| 301 | GTCAAAGGCA GCTTCAGCGG CTCGGCAGGC ATCAACAGCC TTGCCGGTTC |
|
| 351 | GGCGAATCTG CGGACTTTAN GCGTGGATGA TGTCGTTCAG GGCAATANTA |
|
| 401 | CNTACGGCCT GCTGCTAAAA GGTCTGACCG GCACCAATTC AACCAAAGGT |
|
| 451 | AATGCGATGG CGGCGATAGG TGCGCGCAAA TGGCTGGAAA GCGGAGCATC |
|
| 501 | TGTCGGTGTG CTTTACGGGC ACAGCAGGCG CAGCGTGGCG CAAAATTACC |
|
| 551 | GCGTGGGCGG CGGCGGGCAG CACATCGGAA ATTTTGGCGC GGAATATCTG |
|
| 601 | GAACGACGCA AGCAACGATA TTTTGAGCAA GAAGGCGGGT TGAAATTCAA |
|
| 651 | TTCCAACAGC GGAAAATGGG AGCGGGATTT CCAAAAGTCG TACTGGAAAA |
|
| 701 | CCAAGTGGTA TCAAAAATAC GATGCCCCCC AAGAACTGCA AAAATACATC |
|
| 751 | GAAGGTCATG ATAAAAGCTG GCGGGAAAAC CTGGCGCCGC AATACGACAT |
|
| 801 | CACCCCCATC GATCCGTCCA GCCTGAAGCN GCAGTCGGCA GGCAACCTGT |
|
| 851 | TTAAATTGGA ATACGACGGC GTATTCAATA AATACACGGC GCAATTTCGC |
|
| 901 | GATTTAAACA CCAAAATCGG CAGCCGCAAA ATCATCAACC GCAATTATCA |
|
| 951 | ATTCAATTAC GGTTTGTCTT TGAACCCGTA TACCAACCTC AATCTGACCG |
|
| 1001 | CAGCCTACAA TTCGGGCAGG CAGAAATATC CGAAAGGGTC GAAGTTTACA |
|
| 1051 | GGCTGGGGGC TTTTNAAAGA TTTTGAAACC TACAACAACG CAAAAATCCT |
|
| 1101 | CGACCTCANC AACACCTCCA CCTTCCGGCT GCCCCGTGAA ACCGAGTTGC |
|
| 1151 | AAACCACTTT GGGCTTCAAT TATTTCCACA ACGAATACGG CAAAAACCGC |
|
| 1201 | TTTCCTGAAG AATTGGGGCT GTTTTTCGAC GGTCCGGATC ANGACAACGG |
|
| 1251 | GCTTTATTCC TATTTGGGGC GGTTTAAGGG CGATAAAGGG CTGCTGCCCC |
|
| 1301 | AAAAATCAAC CATTGTCCAA CCGGCCGGCA GCCAATATTT CAACACGTTC |
|
| 1351 | TACTTCGATG CCGCGCTCAA AAAAGACATT TACCGCTTAA ACTACAGCAC |
|
| 1401 | CAATACCGTC GGCTACCGTT TCGGCGGCNA ATATACGGGC TATTACNGCT |
|
| 1451 | CGGATGACGA ATTTAAGCGG GCATTCGGAG AAAACTCGCC GACATACANG |
|
| 1501 | AAACATTGCA ACCAGAGCTG CGGAATTTAT GAACCCGTAT TGAAAAAATA |
|
| 1551 | CGGCAAAAAG CGCGCCAACA ACCATTCGGT CAGCATTAGT GCGGACTTCG |
|
| 1601 | GCGATTATTT CATGCCGTTC GCCAGCTATT CGCGCACACA CCGTATGCCC |
|
| 1651 | AACATCCAAG AAATGTATTT TTCCCAAATC GGCGACTCCG GCGTTCACAC |
|
| 1701 | CGCCTTAAAA CCAGAGCGCG CAAACACTTG GCAATTTGGC TTCAATACCT |
|
| 1751 | ATAAAAAAGG ATTGTTAAAA CAAGATGATA TATTAGGATT AAAACTGGTC |
|
| 1801 | GGCTACCGCA GCCGCATCGA CNACTACATC CACAACGTTT ACGGGAAATG |
|
| 1851 | GTGGGATTTG AACGGGAATA TTCCGAGCTG GGTCAGCAGC ACCGGGCTTG |
|
| 1901 | CCTACACCAT CCAACACCGC AATTTCAAAG ACAAAGTGCA CAAACACGGT |
|
| 1951 | TTTGAGTTGG AGCTGAATTA CGATTATNGG CGTTTTTTCA CCAACCTTTC |
|
| 2001 | TTACGCCTAT CAAAAAAGCA CGCAACCGAC CAACTTCAGC GATGCGAGCG |
|
| 2051 | AATCGCCCAA CAATGCGTCC AAAGAAGACC AACTCAAACA AGGTTATGGG |
|
| 2101 | TTGAGCAGGG TTTCCGCCCT GCCGCGAGAT TACGGACGTT TGGAAGTCGG |
|
| 2151 | TACGCGCTGG TTGGGCAACA AACTGACTTT GGGCGGCGCG ATGCGCTATT |
|
| 2201 | TCGGCAAGAG CATCCGCGCG ACGGCTGAAG AACGCTATAT CGACGNCACC |
|
| 2251 | AATGGGGNAN NTACCAGCAA TTTCCGGCAA CTGGGCAAGC GTTCCATCAN |
|
| 2301 | ACAAACCGAA ACCCTTGCCC GCCAGCCTTT GATTTTTGAT TTNTACGCCG |
|
| 2351 | CTTACGAGCC GAAGAAAAAN CTTATTTTCC GCGCCGAAGT CAAAAATCTG |
|
| 2401 | TTCGACAGGC GTTATATCGA TCCGCTCGAT GCGGGCAATG ATGCGGCAAC |
|
| 2451 | GCAGCGTTAT TACAGTTCGT TCGACCCGAA AGACAAGGAC GAAGAAGTAA |
|
| 2501 | CGTGTAATGA TGATAACACG TTATGCAACG GCAAATACGG CGGCACAAGC |
|
| 2551 | AAAAGCGTAT TGACCAATTT TGCACGCGGA CNCACCTTTT TGATAACGAT |
|
| 2601 | GAGCTACAAG TTTTAA |
[1777]This encodes a protein having (partial) amino acid sequence <SEQ ID 880>:
[0000]| 1 | KDKKVFTDAR AVSTRQDIFK SXENLDNIVR XIPGAFTXQX KSSGXVSLNI | |
|
| 51 | RXDSGFGRVN TMVDGITXTF YSTSTDAGRA GGSSQFGASV DSNFXAGLDV |
|
| 101 | VKGSFSGSAG INSLAGSANL RTLXVDDVVQ GNXTYGLLLK GLTGTNSTKG |
|
| 151 | NAMAAIGARK WLESGASVGV LYGHSRRSVA QNYRVGGGGQ HIGNFGAEYL |
|
| 201 | ERRKQRYFEQ EGGLKFNSNS GKWERDFQKS YWKTKWYQKY DAPQELQKYI |
|
| 251 | EGHDKSWREN LAPQYDITPI DPSSLKXQSA GNLFKLEYDG VFNKYTAQFR |
|
| 301 | DLNTKIGSRK IINRNYQFNY GLSLNPYTNL NLTAAYNSGR QKYPKGSKFT |
|
| 351 | GWGLXKDFET YNNAKILDLX NTSTFRLPRE TELQTTLGFN YFHNEYGKNR |
|
| 401 | FPEELGLFFD GPDXDNGLYS YLGRFKGDKG LLPQKSTIVQ PAGSQYFNTF |
|
| 451 | YFDAALKKDI YRLNYSTNTV GYRFGGXYTG YYXSDDEFKR AFGENSPTYX |
|
| 501 | KHCNQSCGIY EPVLKKYGKK RANNHSVSIS ADFGDYFMPF ASYSRTHRMP |
|
| 551 | NIQEMYFSQI GDSGVHTALK PERANTWQFG FNTYKKGLLK QDDILGLKLV |
|
| 601 | GYRSRIDXYI HNVYGKWWDL NGNIPSWVSS TGLAYTIQHR NFKDKVHKHG |
|
| 651 | FELELNYDYX RFFTNLSYAY QKSTQPTNFS DASESPNNAS KEDQLKQGYG |
|
| 701 | LSRVSALPRD YGRLEVGTRW LGNKLTLGGA MRYFGKSIRA TAEERYIDXT |
|
| 751 | NGXXTSNFRQ LGKRSIXQTE TLARQPLIFD XYAAYEPKKX LIFRAEVKNL |
|
| 801 | FDRRYIDPLD AGNDAATQRY YSSFDPKDKD EEVTCNDDNT LCNGKYGGTS |
|
| 851 | KSVLTNFARG XTFLITMSYK F* |
[1778]ORF133a and ORF133-1 show 94.3% identity in 871 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1779]ORF133 shows 92.3% identity over 392 aa overlap with a predicted ORF (ORF133ng) from N. gonorrhoeae:
[0000]
[1780]The complete length ORF133ng nucleotide sequence <SEQ ID 881> is predicted to encode a protein having amino acid sequence <SEQ ID 882>:
[0000]| 1 | MRSSFRLKPI CFYLMGVMLY HHSYAEDAGR AGSEAQIQVL EDVHVKAKRV | |
|
| 51 | PKDKKVFTDA RAVSTRQDVF KSGENLDNIV RSIPGAFTQQ DKSSGIVSLN |
|
| 101 | IRGDSGFGRV NTMVDGITQT FYSTSTDAGR AGGSSQFGAS VDSNFIAGLD |
|
| 151 | VVKGSFSGSA GINSLAGSAN LRTLGVDDVV QGNNTYGLLL KGLTGTNSTK |
|
| 201 | GNAMAAIGAR KWLESGASVG VLYGHSRRGV AQNYRVGGGG QHIGNFGEEY |
|
| 251 | LERRKQQYFV QEGGLKFNAG SGKWERDLQR QYWKTKWYKK YEDPQELQKY |
|
| 301 | IEEHDKSWRE NLAPQYDITP IDPSGLKQQS AGNLLNLEYD GVFNKYTAQF |
|
| 351 | RDLNTRIGSR KIINRNYQFN YGLSLNPYTN LNLTAAYNSG RQKYPKGAKF |
|
| 401 | TGWGLLKDFE TYNNAKILDL NNTATFRLPR ETELQTTLGF NYFHNEYGKN |
|
| 451 | RFPEELGLFF DGPDQDNGLY SYLGRFKGDK GLLPQKSTIV QPAGSQYFNT |
|
| 501 | FYFDAALKKD IYRLNYSTNA INYRFGGEYT GYYGSENEFK RAFGENSPAY |
|
| 551 | KEHCDPSCGL YEPVLKKYGK KRANNHSVSI SADFGDYFMP FAGYSRTHRM |
|
| 601 | PNIQEMYFSQ IGDSGVHTAL KPERANTWQF GFNTYKKGLL KQDDILGLKL |
|
| 651 | VGYRSRIDNY IHNVYGKWWD LNGDIPSWVG STGLAYTIRH RNFKDKVHKH |
|
| 701 | GFELELNYDY GRFFTNLSYA YQKSTQPTNF SDASESPNNA SKEDQLKQGY |
|
| 751 | GLSRVSALPR DYGRLEVGTR WLGNKLTLGG AMRYFGKSIR ATAEERYIDG |
|
| 801 | TNGGNTSNVR QLGKRSIKQT ETLARQPLIF DFYAAYEPKK NLIFRAEVKN |
|
| 851 | LFDRRYIDPL DAGNDAATQR YYSSFDPKDK DEDVTCNADK TLCNGKYGGT |
|
| 901 | SKSVLTNFAR GRTFLMTMSY KF* |
[1781]A variant was also identified, being encoded by the gonococcal DNA sequence <SEQ ID 883>:
[0000]| 1 | ATGAGATCTT CTTTCCGGTT GAAGCCGATT TGTTTTTATC TTATGGGTGT | |
|
| 51 | TATGCTATAT CATCATAGTT ATGCCGAAGA TGCAGGGCGC GCGGGCAGCG |
|
| 101 | AGGCGCAGAT ACAGGTTTTG GAAGATGTGC ACGTCAAGGC GAAGCGCGTA |
|
| 151 | CCGAAAGACA AAAAAGTGTT TACCGATGCG CGTGCCGTAT CGACCCGTca |
|
| 201 | gGATGTGTTC AAATCCGGCG AAAACCTCGA CAACATCGTA CGCAGCATAC |
|
| 251 | CCGGTGCGTT TACACAGCAA GATAAAAGCT CGGGCATTGT GTCTTTGAAT |
|
| 301 | ATTCGCGGCG ACAGCGGGTT CGGGCGGGTC AATACGATGG TGGACGGCAT |
|
| 351 | CACGCAGACC TTTTATTCGA CTTCTACCGA TGCGGGCAGG GCAGGCGGTT |
|
| 401 | CATCTCAATT CGGTGCATCT GTCGACAGCA ATTTTATTGC CGGACTGGAT |
|
| 451 | GTCGTCAAAG GCAGCTTCAG CGGCTCGGCA GGCATCAACA GCCTTGCCGG |
|
| 501 | TTCGGCGAAT CTGCGGACTT TAGGCGTGGA TGACGTCGTT CAGGGCAATA |
|
| 551 | ATACCTACGG CCTGCTGCTA AAAGGTCTGA CCGGCACCAA TTCAACCAAA |
|
| 601 | GGTAATGCGA TGGCGGCGAT AGGTGCGCGC AAATGGCTGG AAAGCGGAGC |
|
| 651 | GTCTGTCGGT GTGCTTTACG GGCACAGCAG GCGCGGCGTG GCGCAAAATT |
|
| 701 | ACCGCGTGGG CGGCGGCGGG CAGCACATCG GAAATTTTGG TGAAGAATAT |
|
| 751 | CTGGAACGGC GCAAACAGCA ATATTTTGTA CAAGAGGGTG GTTTGAAATT |
|
| 801 | CAATGCCGGC AGCGGAAAAT GGGAACGGGA TTTGCAAAGG CAATACTGGA |
|
| 851 | AAACAAAGTG GTATAAAAAA TACGAAGACC CCCAAGAACT GCAAAAATAC |
|
| 901 | ATCGAAGAGC ATGATAAAAG CTGGCGGGAA AACCTGGCGC CGCAATACGA |
|
| 951 | CATCACCCCC ATCGATCCGT CCGGCCTGAA GCAGCAGTCG GCAGGCAATC |
|
| 1001 | TGTTTAAATT GGAATACGAC GGCGTATTCA ATAAATACAC GGCGCAATTT |
|
| 1051 | CGCGATTTAA ACACCAGAAT CGGCAGCCGC AAAATCATCA ACCGCAATTA |
|
| 1101 | TCAATTCAAT TACGGTTTGT CTTTGAACCC GTATACCAAC CTCAATCTGA |
|
| 1151 | CCGCAGCCTA CAATTCGGGC AGGCAGAAAT ATCCGAAAGG GGCGAAGTTT |
|
| 1201 | ACAGGCTGGG GGCTTTTAAA AGATTTTGAA ACCTACAACA ACGCGAAAAT |
|
| 1251 | CCTCGACCTC AACAACACCG CCACCTTCCG GCTGCCCCGC GAAACCGAGT |
|
| 1301 | TGCAAACCAC TTTGGGCTTC AATTATTTCC ACAACGAATA CGGCAAAAAC |
|
| 1351 | CGCTTTCCTG AAGAATTGGG GCTGTTTTTC GACGGTCCTG ATCAGGACAA |
|
| 1401 | CGGGCTTTAT TCCTATTTGG GGCGGTTTAA GGGCGATAAA GGGCTGTTGC |
|
| 1451 | CTCAAAAATC AACCATTGTC CAACCGGCCG GCAGCCAATA TTTCAACACG |
|
| 1501 | TTCTACTTCG ATGCCGCGCT CAAAAAAGAC ATTTACCGCT TAAACTACAG |
|
| 1551 | CACCAATGCA ATCAACTACC GTTTCGGCGG CGAATATACG GGCTATTACG |
|
| 1601 | GCTCGGAAAA CGAATTTAAG CGGGCATTCG GAGAAAACTC GCCGGCATAC |
|
| 1651 | AAGGAACATT GCGACCCGAG CTGCGGGCTT TATGAACCCG TATTGAAAAA |
|
| 1701 | ATACGGCAAA AAGCGCGCCA ACAACCATTC GGTCAGCATT AGTGCGGACT |
|
| 1751 | TCGGCGATTA TTTCATGCCG TTCGCCGGCT ATTCGCGCAC ACACCGTATG |
|
| 1801 | CCCAACATCC AAGAAATGTA TTTTTCCCAA ATCGGCGACT CCGGCGTTCA |
|
| 1851 | CACCGCCTTA AAACCAGAGC GCGCAAACAC TTGGCAATTT GGCTTCAATA |
|
| 1901 | CCTATAAAAA AGGATTGTTA AAACAAGATG ATATATTAGG ATTGAAACTG |
|
| 1951 | GTCGGCTACC GCAGCCGCAT TGACAACTAC ATCCACAACG TTTACGGGAA |
|
| 2001 | ATGGTGGGAT TTGAACGGGG ATATTCCGAG CTGGGTCGGC AGCACCGGGC |
|
| 2051 | TTGCCTACAC CATCCGACAC CGCAATTTCA AAGACAAAGT GCACAAACAC |
|
| 2101 | GGTTTTGAGC TGGAGCTGAA TTACGATTAT GGGCGTTTTT TCACCAACCT |
|
| 2151 | TTCTTACGCC TATCAAAAAA GCACGCAACC GACCAATTTC AGCGATGCGA |
|
| 2201 | GCGAATCGCC CAACAATGCC tccaaAGAAG ACCAACTCAA ACAAGGTTAT |
|
| 2251 | GGGCTGAGCA GGGTTTCCGC CCTGCCGCGA GATTACGGAC GTTTGGAAGT |
|
| 2301 | CGGTACGCGC TGGTTGGGCA ACAAACTGAC TTTGGGCGGC GCGAtgcGCT |
|
| 2351 | ATTTCGGCAA GAGCATCCGC GCGACGGCTG AAGAACGCTA TATCGACGGC |
|
| 2401 | ACCAACGGGG GAAATACCAG CAATGTCCGG CAACTGGGCA AGCGTTCCAT |
|
| 2451 | CAAACAAACC GAAACCCTTG CCCGACAGCC TTTGATTTTT GATTTTTACG |
|
| 2501 | CCGCTTACGA GCCGAAGAAA AACCTTATTT TCCGCGCCGA AGTCAAAAAC |
|
| 2551 | CTGTTCGACA GGCGTTATAT CGATCCGCTC GATGCGGGCA ATGATGCGGC |
|
| 2601 | AACGCAGCGT TATTACAGCT CGTTCGACCC GAAAGACAAG GACGAAGACG |
|
| 2651 | TAACGTGTAA TGCTGATAAA ACGTTGTGCA ACGGCAAATA CGGCGGCACA |
|
| 2701 | AGCAAAAGCG TATTGACCAA TTTCGCACGC GGACGCACCT TCTTGATGAC |
|
| 2751 | GATGAGCTAC AAGTTTTAA |
[1782]This corresponds to the amino acid sequence <SEQ ID 884; ORF133ng-1>:
[0000]| 1 | MRSSFRLKPI CFYLMGVMLY HHSYAEDAGR AGSEAQIQVL EDVHVKAKRV | |
|
| 51 | PKDKKVFTDA RAVSTRQDVF KSGENLDNIV RSIPGAFTQQ DKSSGIVSLN |
|
| 101 | IRGDSGFGRV NTMVDGITQT FYSTSTDAGR AGGSSQFGAS VDSNFIAGLD |
|
| 151 | VVKGSFSGSA GINSLAGSAN LRTLGVDDVV QGNNTYGLLL KGLTGTNSTK |
|
| 201 | GNAMAAIGAR KWLESGASVG VLYGHSRRGV AQNYRVGGGG QHIGNFGEEY |
|
| 251 | LERRKQQYFV QEGGLKFNAG SGKWERDLQR QYWKTKWYKK YEDPQELQKY |
|
| 301 | IEEHDKSWRE NLAPQYDITP IDPSGLKQQS AGNLFKLEYD GVFNKYTAQF |
|
| 351 | RDLNTRIGSR KIINRNYQFN YGLSLNPYTN LNLTAAYNSG RQKYPKGAKF |
|
| 401 | TGWGLLKDFE TYNNAKILDL NNTATFRLPR ETELQTTLGF NYFHNEYGKN |
|
| 451 | RFPEELGLFF DGPDQDNGLY SYLGRFKGDK GLLPQKSTIV QPAGSQYFNT |
|
| 501 | FYFDAALKKD IYRLNYSTNA INYRFGGEYT GYYGSENEFK RAFGENSPAY |
|
| 551 | KEHCDPSCGL YEPVLKKYGK KRANNHSVSI SADFGDYFMP FAGYSRTHRM |
|
| 601 | PNIQEMYFSQ IGDSGVHTAL KPERANTWQF GFNTYKKGLL KQDDILGLKL |
|
| 651 | VGYRSRIDNY IHNVYGKWWD LNGDIPSWVG STGLAYTIRH RNFKDKVHKH |
|
| 701 | GFELELNYDY GRFFTNLSYA YQKSTQPTNF SDASESPNNA SKEDQLKQGY |
|
| 751 | GLSRVSALPR DYGRLEVGTR WLGNKLTLGG AMRYFGKSIR ATAEERYIDG |
|
| 801 | TNGGNTSNVR QLGKRSIKQT ETLARQPLIF DFYAAYEPKK NLIFRAEVKN |
|
| 851 | LFDRRYIDPL DAGNDAATQR YYSSFDPKDK DEDVTCNADK TLCNGKYGGT |
|
| 901 | SKSVLTNFAR GRTFLMTMSY KF* |
[1783]ORF133ng-1 and ORF133-1 show 96.2% identity in 889 aa overlap:
[0000]

[1784]In addition, ORF133ng-1 is homologous to a TonB-dependent receptor in H. influenzae:
[0000]
[1785]The underlined motif in the gonococcal protein (also present in the meningococcal protein) is predicted to be an ATP/GTP-binding site motif A (P-loop), and the analysis suggests that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
Example 104
[1786]The following partial DNA sequence was identified in N. meningitidis <SEQ ID 885>
[0000]| 1 | ATGAACCTGA TTTCACGTTA CATCATCCGT CAAATGGCGG TTATGGCGGT | |
|
| 51 | TTACGCGCTC CTTGCCTTCC TCGCTTTGTA CAGCTTTTTT GAAATCCTGT |
|
| 101 | ACGAAACCGG CAACCTCGGC AAAGGCAGTT ACGGCATATG GGAAATGCTG |
|
| 151 | GGCTACACCG CCCTCAAAAT GCCCGCCCGC GCCTACGAAC TGATTCCCCT |
|
| 201 | CGCCGTCCTT ATCGGCGGAC TGGTCTCCCT CAGCCAGCTT GCCGCCGGCA |
|
| 251 | GCGAACTGAC CGTCATCAAA GCCAGCGGCA TGAGCACCAA AAAGCTGCTG |
|
| 301 | TTGATTCTGT CGCAGTTCGG TTTTATTTTT GCTATTGCCA CCGTCGCGCT |
|
| 351 | CGGCGAATGG GTTGCGCCCA CACTGAGCCA AAAAGCCGAA AACATCAAAG |
|
| 401 | CCGCCGCCAT CAACGGCAAA ATCAGCACCG GCAATACCGG CCTTTGGCTG |
|
| 451 | AAAGAAAAAA ACAGCGTGAT CAATGTGCGC GAAATGTTGC CCGACCAT.. |
[1787]This corresponds to the amino acid sequence <SEQ ID 886; ORF112>:
[0000]| 1 | MNLISRYIIR QMAVMAVYAL LAFLALYSFF EILYETGNLG KGSYGIWEML | |
|
| 51 | GYTALKMPAR AYELIPLAVL IGGLVSLSQL AAGSELTVIK ASGMSTKKLL |
|
| 101 | LILSQFGFIF AIATVALGEW VAPTLSQKAE NIKAAAINGK ISTGNTGLWL |
|
| 151 | KEKNSVINVR EMLPDH... |
[1788]Further work revealed further partial nucleotide sequence <SEQ ID 887>:
[0000]| 1 | ATGAACCTGA TTTCACGTTA CATCATCCGT CAAATGGCGG TTATGGCGGT | |
|
| 51 | TTACGCGCTC CTTGCCTTCC TCGCTTTGTA CAGCTTTTTT GAAATCCTGT |
|
| 101 | ACGAAACCGG CAACCTCGGC AAAGGCAGTT ACGGCATATG GGAAATGCTG |
|
| 151 | gGCTACACCG CCCTCAAAAT GCCCGCCCGC GCCTACGAAC TGATTCCCCT |
|
| 201 | CGCCGTCCTT ATCGGCGGAC TGGTCTCCCT CAGCCAGCTT GCCGCCGGCA |
|
| 251 | GCGAACTGAC CGTCATCAAA GCCAGCGGCA TGAGCACCAA AAAGCTGCTG |
|
| 301 | TTGATTCTGT CGCAGTTCGG TTTTATTTTT GCTATTGCCA CCGTCGCGCT |
|
| 351 | CGGCGAATGG GTTGCGCCCA CACTGAGCCA AAAAGCCGAA AACATCAAAG |
|
| 401 | CCGCCGCCAT CAACGGCAAA ATCAGCACCG GCAATACCGG CCTTTGGCTG |
|
| 451 | AAAGAAAAAA ACAGCrTkAT CAATGTGCGC GAAATGTTGC CCGACCATAC |
|
| 501 | GCTTTTGGGC ATCAAAATTT GGGCGCGCAA CGATAAAAAC GAATTGGCAG |
|
| 551 | AGGCAGTGGA AGCCGATTCC GCCGTTTTGA ACAGCGACGG CAGTTGGCAG |
|
| 601 | TTGAAAAACA TCCGCCGCAG CACGCTTGGC GAAGACAAAG TCGAGGTCTC |
|
| 651 | TATTGCGGCT GAAGAAAACT GGCCGATTTC CGTCAAACGC AACCTGATGG |
|
| 701 | ACGTATTGCT CGTCAAACCC GACCAAATGT CCGTCGGCGA ACTGACCACC |
|
| 751 | TACATCCGCC ACCTCCAAAA CAACAGCCAA AACACCCGAA TCTACGCCAT |
|
| 801 | CGCATGGTGG CGCAAATTGG TTTACCCCGC CGCAGCCTGG GTGATGGCGC |
|
| 851 | TCGTCGCCTT TGCCTTTACC CCGCAAACCA CCCGCCACGG CAATATGGGC |
|
| 901 | TTAAAACTCT TCGGCGGCAT CTGTsTCGGA TTGCTGTTCC ACCTTGCCGG |
|
| 951 | ACGGCTCTTT GGGTTTACCA GCCAACTCGG... |
[1789]This corresponds to the amino acid sequence <SEQ ID 888; ORF112-1>:
[0000]| 1 | MNLISRYIIR QMAVMAVYAL LAFLALYSFF EILYETGNLG KGSYGIWEML | |
|
| 51 | GYTALKMPAR AYELIPLAVL IGGLVSLSQL AAGSELTVIK ASGMSTKKLL |
|
| 101 | LILSQFGFIF AIATVALGEW VAPTLSQKAE NIKAAAINGK ISTGNTGLWL |
|
| 151 | KEKNSXINVR EMLPDHTLLG IKIWARNDKN ELAEAVEADS AVLNSDGSWQ |
|
| 201 | LKNIRRSTLG EDKVEVSIAA EENWPISVKR NLMDVLLVKP DQMSVGELTT |
|
| 251 | YIRHLQNNSQ NTRIYAIAWW RKLVYPAAAW VMALVAFAFT PQTTRHGNMG |
|
| 301 | LKLFGGICXG LLFHLAGRLF GFTSQL... |
[1790]Computer analysis of this amino acid sequence predicts two transmembrane domains and gave the following results:
[0000]Homology with a Predicted ORF from N. meningitidis (Strain A)
[1791]ORF112 shows 96.4% identity over a 166aa overlap with an ORF (ORF112a) from strain A of N. meningitidis:
[0000]
[1792]The ORF112a nucleotide sequence <SEQ ID 889> is:
[0000]| 1 | ATGAACCTGA TTTCACGTTA CATCATCCGT CAAATGGCGG TTATGGCGGT | |
|
| 51 | TTACGCGCTC CTTGCCTTCC TCGCTTTGTA CAGCTTTTTT GAAATCCTGT |
|
| 101 | ACGAAACCGG CAACCTCGGC AAAGGCAGTT ACGGCATATG GGAAATGNTG |
|
| 151 | GGNTACACCG CCCTCAAAAT GNCCGCCCGC GCCTACGAAC TGATGCCCCT |
|
| 201 | CGCCGTCCTT ATCGGCGGAC TGGTCTCTNT CAGCCAGCTT GCCGCCGGCA |
|
| 251 | GCGAACTGAN CGTCATCAAA GCCAGCGGCA TGAGCACCAA AAAGCTGCTG |
|
| 301 | TTGATTCTGT CGCAGTTCGG TTTTATTTTT GCTATTGCCA CCGTCGCGCT |
|
| 351 | CGGCGAATGG GTTGCGCCCA CACTGAGCCA AAAAGCCGAA AACATCAAAG |
|
| 401 | CCGCGGCCAT CAACGGCAAA ATCAGTACCG GCAATACCGG CCTTTGGCTG |
|
| 451 | AAAGAAAAAA ACAGCATTAT CAATGTGCGC GAAATGTTGC CCGACCATAC |
|
| 501 | CCTGCTGGGC ATTAAAATCT GGGCCCGCAA CGATAAAAAC GAACTGGCAG |
|
| 551 | AGGCAGTGGA AGCCGATTCC GCCGTTTTGA ACAGCGACGG CAGTTGGCAG |
|
| 601 | TTGAAAAACA TCCGCCGCAG CACGCTTGGC GAAGACAAAG TCGAGGTCTC |
|
| 651 | TATTGCGGCT GAAGAAAANT GGCCGATTTC CGTCAAACGC AACCTGATGG |
|
| 701 | ACGTATTGCT CGTCAAACCC GACCAAATGT CCGTCGGCGA ACTGACCACC |
|
| 751 | TACATCCGCC ACCTCCAAAN NNACAGCCAA AACACCCGAA TCTACGCCAT |
|
| 801 | CGCATGGTGG CGCAAATTGG TTTACCCCGC CGCAGCCTGG GTGATGGCGC |
|
| 851 | TCGTCGCCTT TGCCTTTACC CCGCAAACCA CCCGCCACGG CAATATGGGC |
|
| 901 | TTAAAANTCT TCGGCGGCAT CTGTCTCGGA TTGCTGTTCC ACCTTGCCGG |
|
| 951 | NCGGCTCTTC NGGTTTACCA GCCAACTCTA CGGCATCCCG CCCTTCCTCG |
|
| 1001 | NCGGCGCACT ACCTACCATA GCCTTCGCCT TGCTCGCCGT TTGGCTGATA |
|
| 1051 | CGCAAACAGG AAAAACGCTA A |
[1793]This encodes a protein having the amino acid sequence <SEQ ID 890>:
[0000]| 1 | MNLISRYIIR QMAVMAVYAL LAFLALYSFF EILYETGNLG KGSYGIWEMX | |
|
| 51 | GYTALKMXAR AYELMPLAVL IGGLVSXSQL AAGSELXVIK ASGMSTKKLL |
|
| 101 | LILSQFGFIF AIATVALGEW VAPTLSQKAE NIKAAAINGK ISTGNTGLWL |
|
| 151 | KEKNSIINVR EMLPDHTLLG IKIWARNDKN ELAEAVEADS AVLNSDGSWQ |
|
| 201 | LKNIRRSTLG EDKVEVSIAA EEXWPISVKR NLMDVLLVKP DQMSVGELTT |
|
| 251 | YIRHLQXXSQ NTRIYAIAWW RKLVYPAAAW VMALVAFAFT PQTTRHGNMG |
|
| 301 | LKXFGGICLG LLFHLAGRLF XFTSQLYGIP PFLXGALPTI AFALLAVWLI |
|
| 351 | RKQEKR* |
[1794]ORF112a and ORF112-1 show 96.3% identity in 326 aa overlap:
[0000]
[0000]Homology with a Predicted ORF from N. gonorrhoeae
[1795]ORF112 shows 95.8% identity over 166aa overlap with a predicted ORF (ORF112ng) from N. gonorrhoeae:
[0000]
[1796]The complete length ORF112ng nucleotide sequence <SEQ ID 891> is:
[0000]| 1 | ATGAACCTGA TTTCACGTTA CATCATCCGC CAAATGGCGG TTATGGCGGT | |
|
| 51 | TTACGCGCTC CTTGCCTTCC TCGCTTTGTA CAGCTTTTTT GAAATCCTGT |
|
| 101 | ACGAAACCGG CAACCTCGGC AAAGGCAGTT ACGGCATATG GGAAATGCTG |
|
| 151 | GGCTACACCG CCCTCAAAAT GCCCGCCCGC GCCTACGAAC TCATGCCCCT |
|
| 201 | CGCCGTCCTC ATCGGCGGAC TGGCCTCTCT CAGCCAGCTT GCCGCCGGCA |
|
| 251 | GCGAACTGGC CGTCATCAAA GCCAGCGGCA TGAGCACCAA AAAGCTGCTG |
|
| 301 | TTGATTCTGT CTCAGTTCGG TTTTATTTTT GCTATTGCCG CCGTCGCGCT |
|
| 351 | CGGCGAATGG GTTGCGCCCA CGCTGAGCCA AAAAGCCGAA AACATCAAag |
|
| 401 | cCGCCGCCAt taacggCAAA ATCAGCAccg gcAATACCGG CCTTTggcTG |
|
| 451 | AAAGAAAAAa ccAGCATTAT CAATGTGcGc GGAATGTTGC CCGACCATAC |
|
| 501 | GCTTTTGGGC ATCAAAATTT GGGCGCGCAA CGATAAAAAC GAATTGGCAG |
|
| 551 | AGGCAGTGGA AGCCGATTCC GCCGTTTTGA ACAGCGACGG CAGCTGGCAG |
|
| 601 | TTGAAAAACA TCCGCCGCAG CATCATGGGT ACAGACAAAA TCGAAACATC |
|
| 651 | cgCCGCCGCC GAAGAAACTT gGCCGATTGC CGTCAGACGC AACCTGATGG |
|
| 701 | ACGTATTGCT CGTCAAGCCC GACCAAATGT CCGTCGGCGA GCTGACCACC |
|
| 751 | TACATCCGCC ACCTCCAAAA CAACAGCCAA AACACCCAAA TCTACGCCAT |
|
| 801 | CGCATGGTGG CGTAAACTCG TTTACCCCGT CGCCGCATGG GTCATGGCGC |
|
| 851 | TCGTTGCCTT CGCCTTTACG CCGCAAACCA CGCGCCACGG CAATATGGGC |
|
| 901 | TTAAAACTCT TCGGCGGCAT CTGTCTCGGA TTGCTGTTCC ACCTTGCCGG |
|
| 951 | CAGGCTCTTC GGGTTTACCA GCCAACTCTA CGGCACCCCA CCCTTCCTCG |
|
| 1001 | CCGGCGCACT GCCTACCATA GCCTTCGCCT TGCTCGCTGT TTGGCTGATA |
|
| 1051 | CGCAAACAGG AAAAACGTTG A |
[1797]This encodes a protein having amino acid sequence <SEQ ID 892>:
[0000]| 1 | MNLISRYIIR QMAVMAVYAL LAFLALYSFF EILYETGNLG KGSYGIWEML | |
|
| 51 | GYTALKMPAR AYELMPLAVL IGGLASLSQL AAGSELAVIK ASGMSTKKLL |
|
| 101 | LILSQFGFIF AIAAVALGEW VAPTLSQKAE NIKAAAINGK ISTGNTGLWL |
|
| 151 | KEKTSIINVR GMLPDHTLLG IKIWARNDKN ELAEAVEADS AVLNSDGSWQ |
|
| 201 | LKNIRRSIMG TDKIETSAAA EETWPIAVRR NLMDVLLVKP DQMSVGELTT |
|
| 251 | YIRHLQNNSQ NTQIYAIAWW RKLVYPVAAW VMALVAFAFT PQTTRHGNMG |
|
| 301 | LKLFGGICLG LLFHLAGRLF GFTSQLYGTP PFLAGALPTI AFALLAVWLI |
|
| 351 | RKQEKR* |
[1798]ORF112ng and ORF112-1 show 94.2% identity in 326 aa overlap:
[0000]
[1799]This analysis suggests that these proteins from N. meningitidis and N. gonorrhoeae, and their epitopes, could be useful antigens for vaccines or diagnostics, or for raising antibodies.
[1800]It will be appreciated that the invention has been described by means of example only, and that modifications may be made whilst remaining within the spirit and scope of the invention.
[0000] |
| PCR primers |
| ORF | Primer | Sequence | Restriction sites |
|
| ORF 1 | Forward | CGCGGATCCGCTAGC-GGACACACTTATTTCGG | BamHI-NheI | |
| Reverse | CCCGCTCGAG-CCAGCGGTAGCCTAATT | XhoI |
|
| ORF 2 | Forward | GCGGATCCCATATG-TTTGATTTCGGTTTGGG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GACGGCATAACGGCG | XhoI |
|
| ORF 2-1 | Forward | GCGGATCCCATATG-TTTGATTTCGGTTTGGG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TGATTTACGGACGCGCA | XhoI |
|
| ORF 4 | Forward | GCGGATCCCATATG-TGCGGAGGTCAAAAAGAC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTGGCTGCGCCTTC | XhoI |
|
| ORF 5 | Forward | GGAATTCCATATGGCCATGG-TGGAAGGCGCACAACC | NdeI-NcoI |
| Forward | CGGGATCC-ATGGAAGGCGCACAAC | BamHI |
| Reverse | CCCGCTCGAG-GACTGTGCAAAAACGG | XhoI |
|
| ORF 6 | Forward | CGCGGATCCCATATG-ACCCGTCAATCTCTGCA | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TGCGCCGAACACTTTC | XhoI |
|
| ORF 7 | Forward | CGCGGATCCGCTAGC-GCGCTGCTTTTTGTTCC | BamHI-NheI |
| Reverse | CCCGCTCGAG-TTTCAAAATATATTTGCGGA | XhoI |
|
| ORF 8 | Forward | GCGGATCCCATATG-GCTCAACTGCTTCGTAC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AGCAGGCTTTGGCGC | XhoI |
|
| ORF 9 | Forward | CGCGGATCCCATATG-CCGAAGGAAGTCGGAAA | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTCCGAGGTTTTCGGG | XhoI |
|
| ORF 10 | Forward | GCGGATCCCATATG-GACACAAAAGAAATCCTC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TAATGGGAAACCTTGTTTT | XhoI |
|
| ORF 11 | Forward | GCGGATCCCATATG-GCGGTCAACCTCTACG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GGAAACGACTTCGCC | XhoI |
|
| ORF 13 | Forward | CGCGGATCCCATATG-GCTCTGCTTTCCGCGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AGGGTGTGTGATAATAAG | XhoI |
|
| ORF 15 | Forward | GGAATTCCATATGGCCATGG-GCGGGACACTGACAG | NdeI-NcoI |
| Forward | CGGGATCC-TGCGGGACACTGACAGG | BamHI |
| Reverse | CCCGCTCGAG-AGGTTGGCCTTGTCTATG | XhoI |
|
| ORF 17 | Forward | GGAATTCCATATGGCCATGG-TTGCCGGCCTGTTCG | NdeI-NcoI |
| Forward | CGGGATCC-ATTGCCGGCCTGTTCG | BamHI |
| Reverse | CCCGCTCGAG-AAGCAGGTTGTACAGC | XhoI |
|
| ORF 18 | Forward | GCGGATCCCATATG-ATTTTGCTGCATTTGGAT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TCTTCCAATTTCTGAAAGC | XhoI |
|
| ORF 19 | Forward | GGAATTCCATATGGCCATGG-TCGCCAGTGTTTTTACC | NdeI-NcoI |
| Forward | CGGGATCC-TTCGCCAGTGTTTTTACCG | BamHI |
| Reverse | CCCGCTCGAG-GGTGTTTTTGAAGCTGCC | XhoI |
|
| ORF 20 | Forward | GGAATTCCATATGGCCATGG-TCGGCGCGGGTATG | NdeI-NcoI |
| Forward | CGGGATCC-TTCGGCGCGGGTATG | BamHI |
| Reverse | CCCGCTCGAG-CGGCGAGCGAGAGCA | XhoI |
|
| ORF 22 | Forward | GGAATTCCATATGGCCATGG-TGATTAAAATCAAAAAAGGTCT | NdeI-NcoI |
| Forward | CGGGATCC-ATGATTAAAATCAAAAAAGGTCTAAACC | BamHI |
| Reverse | CCCGCTCGAG-ATTATGATAGCGGCCC | XhoI |
|
| ORF 23 | Forward | CGCGGATCCCATATG-GATGTTTCTGTTTCAGAC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTAAACCGATAGGTAAACG | XhoI |
|
| ORF 24 | Forward | GGAATTCCATATGGCCATGG-TGATGCCGGAAATGGTG | NdeI-NcoI |
| Forward | CGGGATCC-ATGATGCCGGAAATGGTG | BamHI |
| Reverse | CCCGCTCGAG-TGTCAGCGTGGCGCA | XhoI |
|
| ORF 25 | Forward | GCGGATCCCATATG-TATCGCAAACTGATTGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATCGATGGAATAGCCG | XhoI |
|
| ORF 26 | Forward | GCGGATCCCATATG-CAGCTGATCGACTATTC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GACATCGGCGCGTTTT | XhoI |
|
| ORF 27 | Forward | GGAATTCCATATGGCCATGG-AGACCTATTCTGTTTA | NdeI-NcoI |
| Forward | CGGGATCC-CAGACCTATTCTGTTTATTTTAATC | BamHI |
| Reverse | CCCGCTCGAG-GGGTTCGATTAAATAACCAT | XhoI |
|
| ORF 28 | Forward | GGAATTCCATATGGCCATGG-ACGGCTGTACGTTGATGT | NdeI-NcoI |
| Forward | CGGGATCC-AACGGCTGTACGTTGATG | BamHI |
| Reverse | CCCGCTCGAG-TTTGTCAGAGGAATTCGCG | XhoI |
|
| ORF 29 | Forward | GCGGATCCCATATG-AACGGTTTGGATGCCCG | BamHI-NdeI |
| Forward | CGCGGATCCGCTAGC-AACGGTTTGGATGCCCG | BamHI-NheI |
| Reverse | CCCGCTCGAG-TTTGTCTAAGTTCCTGATATG | XhoI |
|
| ORF 32 | Forward | CGCGGATCCCATATG-AATACTCCTCCTTTTG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GCGTATTTTTTGATGCTTTG | XhoI |
|
| ORF 33 | Forward | GCGGATCCCATATG-ATTGATAGGGATCGTATG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTGATCTTTCAAACGGCC | XhoI |
|
| ORF 35 | Forward | GCGGATCCCATATG-TTCAGAGCTCAGCTT | BamHI-NdeI |
| Forward | CGCGGATCCGCTAGC-TTCAGAGCTCAGCTT | BamHI-NheI |
| Reverse | CCCGCTCGAG-AAACAGCCATTTGAGCGA | XhoI |
|
| ORF 37 | Forward | GCGGATCCCATATG-GATGACGTATCGGATTTT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATAGCCCGCTTTCAGG | XhoI |
|
| ORF 58 | Forward | CGCGGATCCGCTAGC-TCCGAACGCGAGTGGAT | BamHI-NheI |
| Reverse | CCCGCTCGAG-AGCATTGTCCAAGGGGAC | XhoI |
|
| ORF 65 | Forward | GGAATTCCATATGGCCATGG-TGCTGTATCTGAATCAAG | NdeI-NcoI |
| Forward | CGGGATCC-TTGCTGTATCTGAATCAAGG | BamHI |
| Reverse | CCCGCTCGAG-CCGCATCGGCAGACA | XhoI |
|
| ORF 66 | Forward | GCGGATCCCATATG-TACGCATTTACCGCCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TGGATTTTGCAGAGATGG | XhoI |
|
| ORF 72 | Forward | CGCGGATCCCATATG-AATGCAGTAAAAATATCTGA | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GCCTGAGACCTTTGCAA | XhoI |
|
| ORF 73 | Forward | GCGGATCCCATATG-AGATTTTTCGGTATCGG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTCATCTTTTTCATGTTCG | XhoI |
|
| ORF 75 | Forward | GCGGATCCCATATG-TCTGTCTTTCAAACGGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTGTTTTTGCAAGACAG | XhoI |
|
| ORF 76 | Forward | GATCAGCTAGCCATATG-AAACAGAAAAAAACCGC | NheI-NdeI |
| Reverse | CGGGATCC-TTACGGTTTGACACCGTT | BamHI |
|
| ORF 79 | Forward | CGCGGATCCCATATG-GTTTCCGCCGCCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GTGCTGATGCGCTTCG | XhoI |
|
| ORF 83 | Forward | GCGGATCCCATATG-AAAACCCTGCTGCTGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GCCGCCTTTGCGGC | XhoI |
|
| ORF 84 | Forward | GCGGATCCCATATG-GCAGAGATCTGTTTG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GTTTGCCGATCCGACCA | XhoI |
|
| ORF 85 | Forward | CGCGGATCCCATATG-GCGGTTTGGGGCGGA | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TCGGCGCGGCGGGC | XhoI |
|
| ORF 89 | Forward | GGAATTCCATATGGCCATGG-CCATACCTTCTTATCA | NdeI-NcoI |
| Forward | CGGGATCC-GCCATACCTTCTTATCAGAG | BamHI |
| Reverse | CCCGCTCGAG-TTTTTTGCGATTAGAAAAAGC | XhoI |
|
| ORF 97 | Forward | GCGGATCCCATATG-CATCCTGCCAGCGAAC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTCGCCTACGGTTTTTTG | XhoI |
|
| ORF 98 | Forward | GCGGATCCCATATG-ACGGTAACTGCGG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTGTTGTTCGGGCAAATC | XhoI |
|
| ORF 100 | Forward | GCGGATCCCATATG-TCGGGCATTTACACCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ACGGGTTTCGGCGGAA | XhoI |
|
| ORF 101 | Forward | GCGGATCCCATATG-ATTTATCAAAGAAACCTC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTTCCGCCTTTCAATGT | XhoI |
|
| ORF 102 | Forward | GCGGATCCCATATG-GCAGGGCTGTTTTACC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AAACGGTTTGAACACGAC | XhoI |
|
| ORF 103 | Forward | GCGGATCCCATATG-AACCACGACATCAC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-CAGCCACAGGACGGC | XhoI |
|
| ORF 104 | Forward | GCGGATCCCATATG-ACGTGGGGAACGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GCGGCGTTTGAACGGC | XhoI |
|
| ORF 105 | Forward | GCGGATCCCATATG-ACCAAATTTCAAACCCCTC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TAAACGAATGCCGTCCAG | XhoI |
|
| ORF 106 | Forward | GCGGATCCCATATG-AGGATAACCGACGGCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTGTTCCCGATGATGTT | XhoI |
|
| ORF 109 | Forward | GCGGATCCCATATG-GAAGATTTATATATAATACTCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATCAGCTTCGAACCGAAG | XhoI |
|
| ORF110 | Forward | AAAGAATTC-ATGAGTAAATCCCGTAGATCTCCC | EcoRI |
| Reverse | AAACTGCAG-GGAAAACCACATCCGCACTCTGCC | PstI |
|
| ORF111 | Forward | AAAGAATTC-GCACCGCAAAAGGCAAAAACCGCA | EcoRI |
| Reverse | AAACTGCAG-TCTGCGCGTTTTCGGGCAGGGTGG | PstI |
|
| ORF113 | Forward | AAAGAATTC-ATGAACAAAACCCTCTATCGTGTGATTTTCAACCG | EcoRI |
| Reverse | AAACTGCAG-TTACGAATGCCTGCTTGCTCGACCGTACTG | PstI |
|
| ORF115 | Forward | AAAGAATTC-TTGCTTGTGCAAACAGAAAAAGACGG | EcoRI |
| Reverse | AAAAAAGTCGAC-CTATTTTTTAGGGGCTTTTGCTTGTTTGAAAAGCCTGCC | SalI |
|
| ORF119 | Forward | AAAGAATTC-TACAACATGTATCAGGAAAACCAATACCG | EcoRI |
| Reverse | AAACTGCAG-TTATGAAAACAGGCGCAGGGCGGTTTTGCC | PstI |
|
| ORF120 | Forward | AAAGAATTC-GCAAGGCTACCCCAATCCGCCGTG | EcoRI |
| Reverse | AAACTGCAG-CGGTTTGGCTGCCTGGCCGTTGAT | PstI |
|
| ORF121 | Forward | AAAGAATTC-GCCTTGGTCTGGCTGGTTTTCGC | EcoRI |
| Reverse | AAACTGCAG-TCATCCGCCACCCCACCTCGGCCATCCATC | PstI |
|
| ORF122 | Forward | AAAAAAGTCGAC-ATGTCTTACCGCGCAAGCAGTTCTCC | SalI |
| Reverse | AAACTGCAG-TCAGGAACACAAACGATGACGAATATCCGTATC | PstI |
|
| ORF125 | Forward | AAAGAATTC-GCGCTGTTTTTTGCGGCGGCGTAT | EcoRI |
| Reverse | AAACTGCAG-CGCCGTTTCAAGACGAAAAAGTCG | PstI |
|
| ORF126 | Forward | AAAGAATTC-GCGGAAACGGTCGAAG | EcoRI |
| Reverse | AAACTGCAG-TTAATCTTGTCTTCCGATATAC | PstI |
|
| ORF127 | Forward | AAAGAATTC-ATGACTGATAATCGGGGGTTTACG | EcoRI |
| Reverse | AAAAAAGTCGAC-CTTAAGTAACTTGCAGTCCTTATC | SalI |
|
| ORF128 | Forward | AAAGAATTC-ATGCAAGCTGTCCGCTACAGGCC | EcoRI |
| Reverse | AAACTGCAG-CTATTGCAATGCGCCGCCGCGGGAATGTTTGAGCAGGCG | PstI |
|
| ORF129 | Forward | AAAGAATTC-ATGGATTTTCGTTTTGACATTATTTACGAATACCG | EcoRI |
| Reverse | AAACTGCAG-TTATTTTTTGATGAAATTTTGGGGCGG | PstI |
|
| ORF130 | Forward | AAAGAATTC-GCAGTACTTGCCATTCTCGGTGCG | EcoRI |
| Reverse | AAACTGCAG-CTCCGGATCGTCTGTAAACGCATT | PstI |
|
| ORF 131 | Forward | GCGGATCCCATATG-GAAATTCGGGCAATAAAAT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-CCAGCGGACGCGTTC | XhoI |
|
| ORF 132 | Forward | GCGGATCCCATATG-AAAGAAGCGGGGTTTG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-CCAATCTGCCAGCCGT | XhoI |
|
| ORF 133 | Forward | CGCGGATCCCATATG-GAAGATGCAGGGCGCG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AAACTTGTAGCTCATCGT | XhoI |
|
| ORF 134 | Forward | GCGGATCCCATATG-TCTGTGCAAGCAGTATTG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATCCTGTGCCAATGCG | XhoI |
|
| ORF 135 | Forward | GCGGATCCCATATG-CCGTCTGAAAAAGCTTT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AAATACCGCTGAGGATG | XhoI |
|
| ORF 136 | Forward | CGCGGATCCGCTAGC-ATGAAGCGGCGTATAGCC | BamHI-NheI |
| Reverse | CCCGCTCGAG-TTCCGAATATTTGGAACTTTT | XhoI |
|
| ORF 137 | Forward | CGCGGATCCCATATG-GGCACGGCGGGAAATA | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATAACGGTATGCCGCC | XhoI |
|
| ORF 138 | Forward | GCGGATCCCATATG-TTTCGTTTACAATTCAGGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-CGGCGTTTTATAGCGG | XhoI |
|
| ORF 139 | Forward | GCGGATCCCATATG-GCTTTTTTGGCGGTAATG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TAACGTTTCCGTGCGTTT | XhoI |
|
| ORF 140 | Forward | GCGGATCCCATATG-TTGCCCACAGGCAGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-GACGATGGCAAACAGC | XhoI |
|
| ORF 141 | Forward | GCGGATCCCATATG-CCGTCTGAAGCAGTCT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-ATCTGTTGTTTTTAAAATATT | XhoI |
|
| ORF 142 | Forward | GCGGATCCCATATG-GATAATTCTGGTAGTGAAG | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AAACGTATAGCCTACCT | XhoI |
|
| ORF 143 | Forward | GCGGATCCCATATG-GATACCGCTTTGAACCT | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AATGGCTTCCGCAATATG | XhoI |
|
| ORF 144 | Forward | GCGGATCCCATATG-ACCTTTTTACAACGTTTGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-AGATTGTTGTTGTTTTTTCG | XhoI |
|
| ORF 147 | Forward | GCGGATCCCATATG-TCTGTCTTTCAAACGGC | BamHI-NdeI |
| Reverse | CCCGCTCGAG-TTTGTTTTTGCAAGACAG | XhoI |
|
| NB: |
| restriction sites are underlined |
| for ORFs 110-130, where the ORF itself carries an EcoRI site (eg. ORF122), a SalI site was used in the forward primer instead. Similarly, where the ORF carries a PstI site (eg. ORFs 115 and 127), a SalI site was used in the reverse primer. |
[0000] |
| Summary of cloning, expression and purification |
| | His-fusion | GST-fusion | |
| ORF | PCR/cloning | expression | expression | Purification |
|
| orf 1 | + | + | + | His-fusion |
| orf 2 | + | + | + | GST-fusion |
| orf 2.1 | + | n.d. | + | GST-fusion |
| orf 4 | + | + | + | His-fusion |
| orf 5 | + | n.d. | + | GST-fusion |
| orf 6 | + | + | + | GST-fusion |
| orf 7 | + | + | + | GST-fusion |
| orf 8 | + | n.d. | n.d. |
| orf 9 | + | + | + | GST-fusion |
| orf 10 | + | n.d. | n.d. |
| orf 11 | + | n.d. | n.d. |
| orf 13 | + | n.d. | + | GST-fusion |
| orf 15 | + | + | + | GST-fusion |
| orf 17 | + | n.d. | n.d. |
| orf 18 | + | n.d. | n.d. |
| orf 19 | + | n.d. | n.d. |
| orf 20 | + | n.d. | n.d. |
| orf 22 | + | + | + | GST-fusion |
| orf 23 | + | + | + | His-fusion |
| orf 24 | + | n.d. | n.d. |
| orf 25 | + | + | + | His-fusion |
| orf 26 | + | n.d. | n.d. |
| orf 27 | + | + | + | GST-fusion |
| orf 28 | + | + | + | GST-fusion |
| orf 29 | + | n.d. | n.d. |
| orf 32 | + | + | + | His-fusion |
| orf 33 | + | n.d. | n.d. |
| orf 35 | + | n.d. | n.d. |
| orf 37 | + | + | + | GST-fusion |
| orf 58 | + | n.d. | n.d. |
| orf 65 | + | n.d. | n.d. |
| orf 66 | + | n.d. | n.d. |
| orf 72 | + | + | n.d. | His-fusion |
| orf 73 | + | n.d. | + | n.d. |
| orf 75 | + | n.d. | n.d. |
| orf 76 | + | + | n.d. | His-fusion |
| orf 79 | + | + | n.d. | His-fusion |
| orf 83 | + | n.d. | + | n.d. |
| orf 84 | + | n.d. | n.d. |
| orf 85 | + | n.d. | + | GST-fusion |
| orf 89 | + | n.d. | + | GST-fusion |
| orf 97 | + | + | + | GST-fusion |
| orf 98 | + | n.d. | n.d. |
| orf 100 | + | n.d. | n.d. |
| orf 101 | + | n.d. | n.d. |
| orf 102 | + | n.d. | n.d. |
| orf 103 | + | n.d. | n.d. |
| orf 104 | + | n.d. | n.d. |
| orf 105 | + | n.d. | n.d. |
| orf 106 | + | + | + | His-fusion |
| orf 109 | + | n.d. | n.d. |
| orf 110 | + | n.d. | n.d. |
| orf 111 | + | + | n.d. | His-fusion |
| orf 113 | + | + | n.d. | His-fusion |
| orf 115 | n.d. | n.d. | n.d. |
| orf 119 | + | + | n.d. | His-fusion |
| orf 120 | + | + | n.d. | His-fusion |
| orf 121 | + | n.d. | n.d. |
| orf 122 | + | + | n.d. | His-fusion |
| orf 125 | + | + | n.d. | His-fusion |
| orf 126 | + | + | n.d. | His-fusion |
| orf 127 | + | + | n.d. | His-fusion |
| orf 128 | + | n.d. | n.d. |
| orf 129 | + | + | n.d. | His-fusion |
| orf 130 | + | n.d. | n.d. |
| orf 131 | + | + | + | n.d. |
| orf 132 | + | + | + | His-fusion |
| orf 133 | + | n.d. | + | GST-fusion |
| orf 134 | + | n.d. | n.d. |
| orf 135 | + | n.d. | n.d. |
| orf 136 | + | n.d. | n.d. |
| orf 137 | + | n.d. | + | GST-fusion |
| orf 138 | + | n.d. | + | GST-fusion |
| orf 139 | + | n.d. | n.d. |
| orf 140 | + | n.d. | n.d. |
| orf 141 | + | n.d. | n.d. |
| orf 142 | + | n.d. | n.d. |
| orf 143 | + | n.d. | n.d. |
| orf 144 | + | n.d. | + | n.d. |
| orf 147 | + | n.d. | n.d. |
|